Antibiotics Trigger Host Innate Immune Response via Microbiota–Brain Communication in C. elegans
Abstract
1. Introduction
2. Results
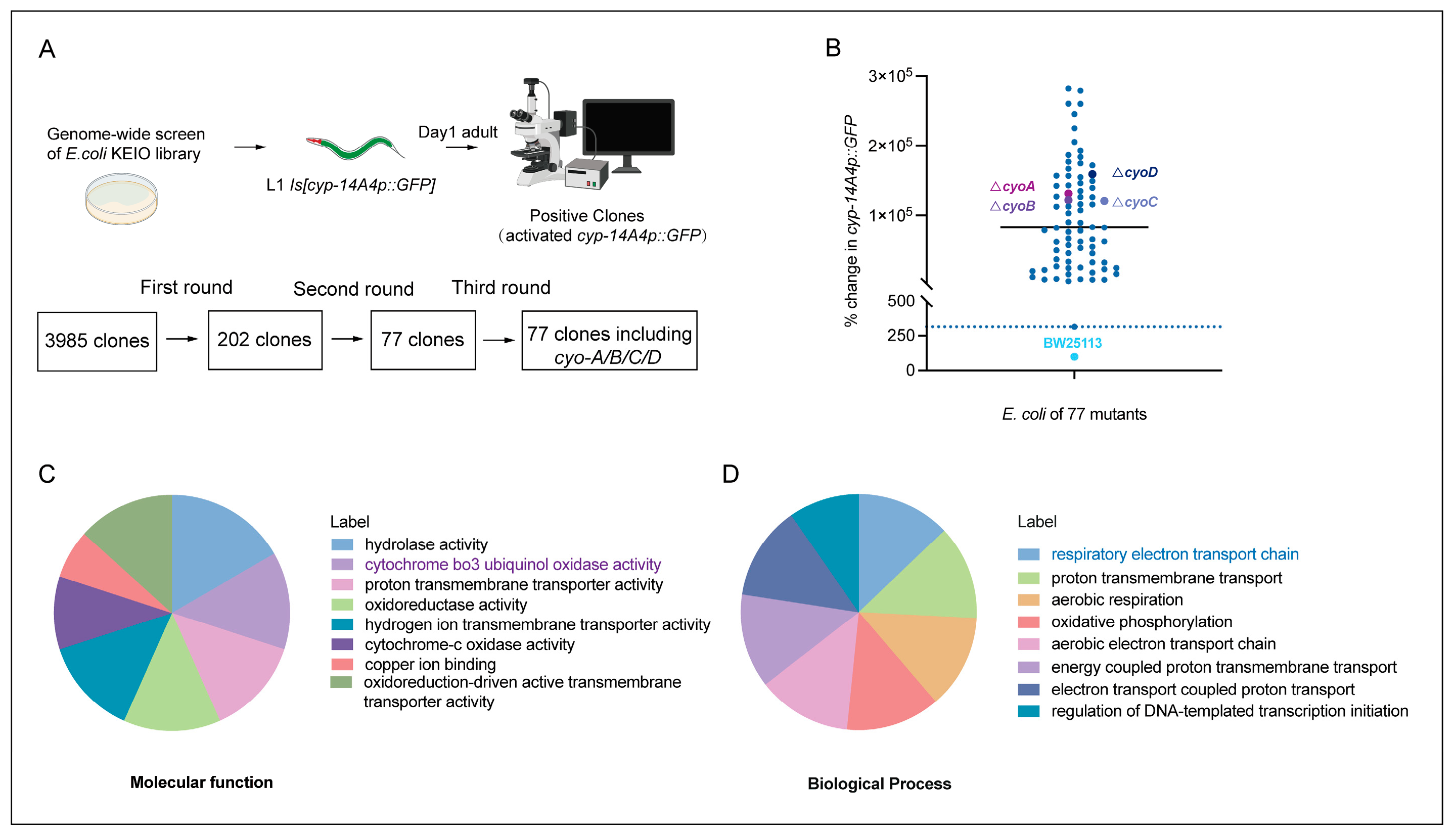
2.1. Genome-Wide Screening to Identify E. coli Genes Whose Inactivation Activates Detoxification and Immune Response in C. elegans
2.2. Q203 Antibiotic Potentially Activates the C. elegans Innate Immune Response and Detoxification Response through Targeting the Electron Transport Chain of Bacteria
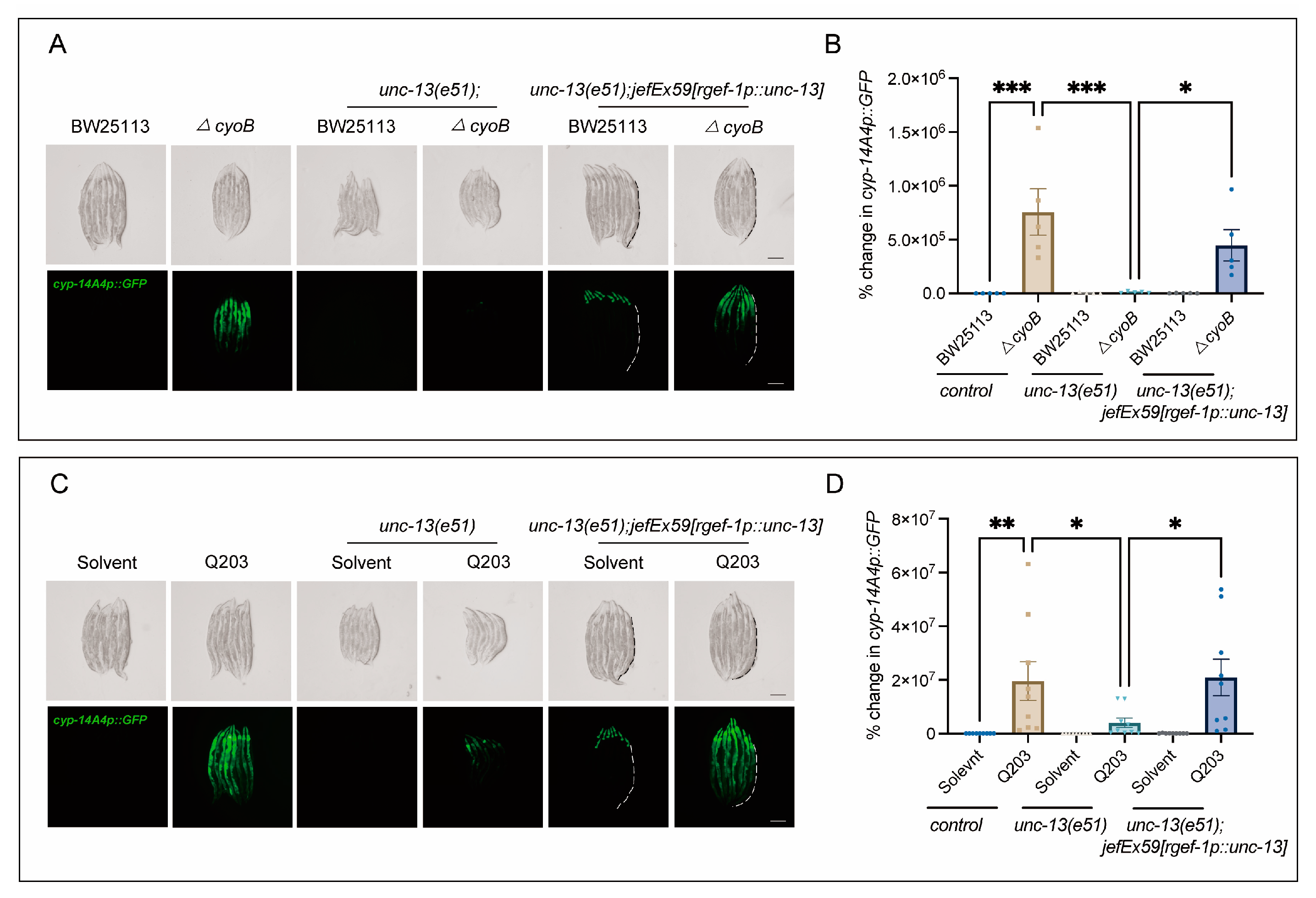
2.3. Host Brain Activity Is Required for ΔcyoB E. coli and Q203 to Activate C. elegans Detoxification and Immune Response
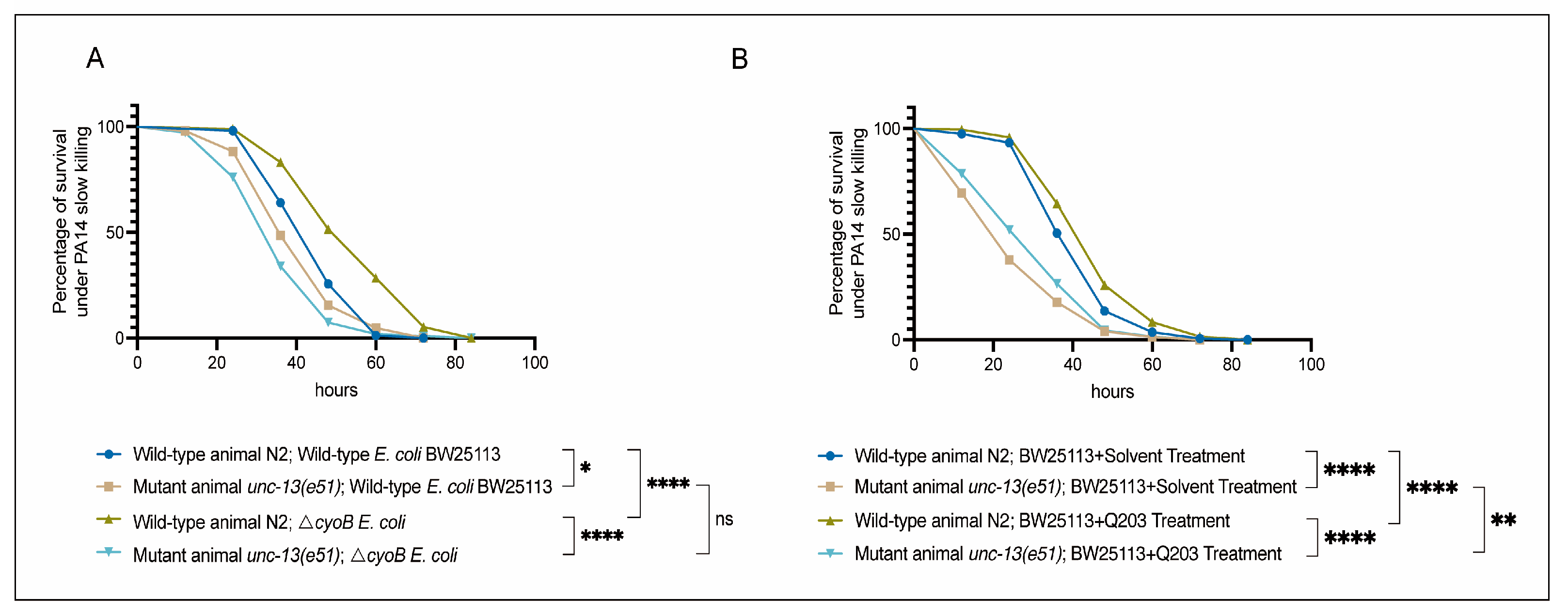
2.4. Brain-Mediated Activation of Immune Response by ΔcyoB E. coli and Q203 Enhances the Ability of C. elegans to Resist Pathogenic Invasion
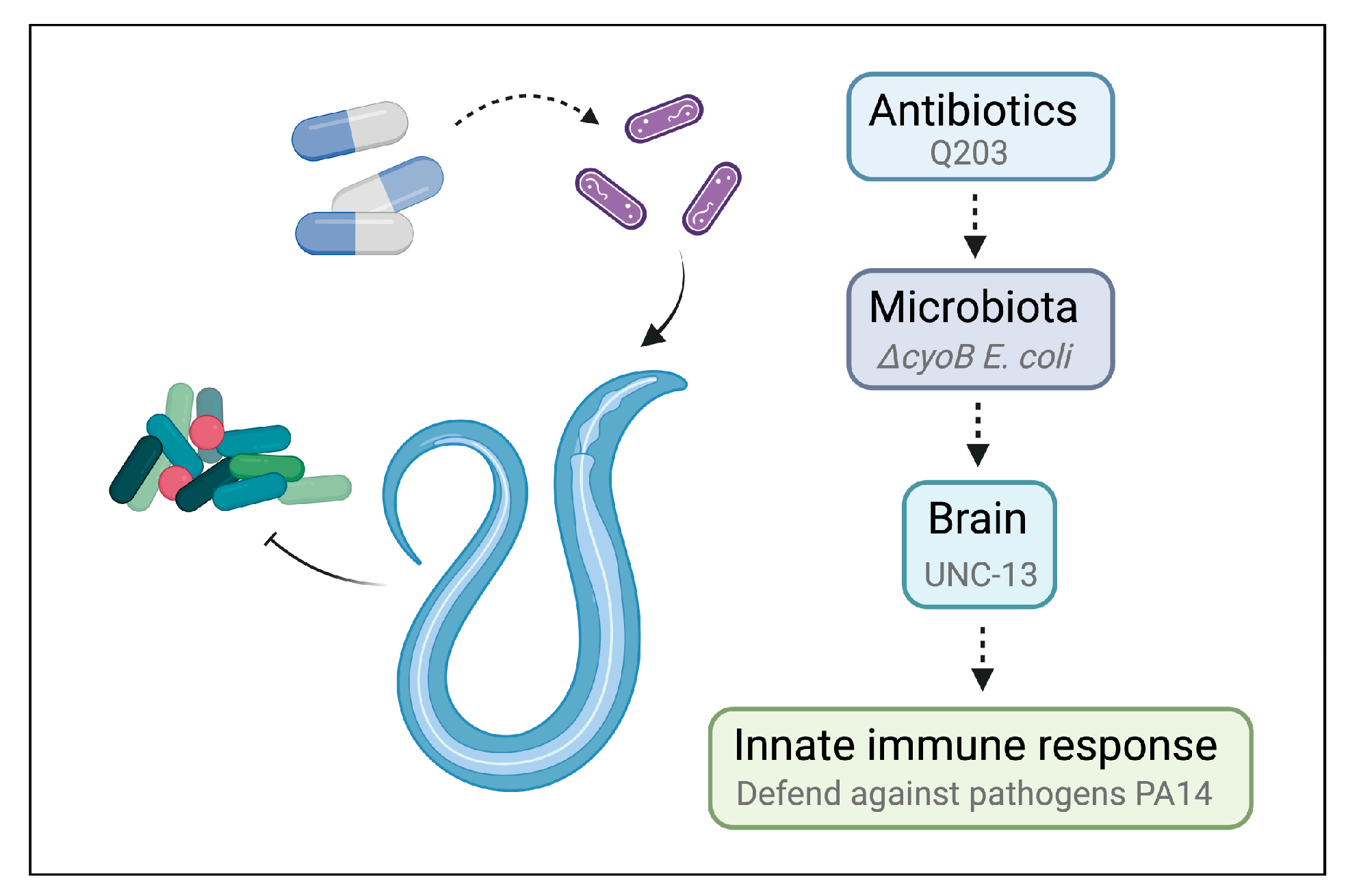
3. Discussion
4. Materials and Methods
4.1. Strains and Maintenance
4.2. Screening for E. coli K-12 Mutants That Activate cyp-14A4 Expression of C. elegans and GO Analysis
4.3. Q203 Supplementation
4.4. Imaging and Analysis of the Expression of Transgenic Reporter cyp-14A4p::GFP
4.5. qPCR Analyses
4.6. RNA Interference in C. elegans
4.7. Quantification of Bacterial Growth
4.8. Generation of Transgenic Worm Strains
4.9. PA14 Slow-Killing Assay
4.10. Macromolecular Docking
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- GBD 2019 Antimicrobial Resistance Collaborators. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Jo, E.K. Interplay between host and pathogen: Immune defense and beyond. Exp. Mol. Med. 2019, 51, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Fleming, D.; Moustafa, D.A.; Dolan, S.K.; Szymanik, K.H.; Redman, W.K.; Ramos, A.; Diggle, F.L.; Sullivan, C.S.; Goldberg, J.B.; et al. A Pseudomonas aeruginosa small RNA regulates chronic and acute infection. Nature 2023, 618, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Pamer, E.G. Antibiotics, microbiota, and immune defense. Trends Immunol. 2012, 33, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. The science of antibiotic discovery. Cell 2020, 181, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Berti, A.; Rose, W.; Nizet, V.; Sakoulas, G. Antibiotics and innate immunity: A cooperative effort toward the successful treatment of infections. Open Forum Infect. Dis. 2020, 7, ofaa302. [Google Scholar] [CrossRef] [PubMed]
- Hodille, E.; Rose, W.; Diep, B.A.; Goutelle, S.; Lina, G.; Dumitrescu, O. The role of antibiotics in modulating virulence in Staphylococcus aureus. Clin. Microbiol. Rev. 2017, 30, 887–917. [Google Scholar] [CrossRef] [PubMed]
- Craven, R.R.; Gao, X.; Allen, I.C.; Gris, D.; Bubeck Wardenburg, J.; McElvania-Tekippe, E.; Ting, J.P.; Duncan, J.A. Staphylococcus aureus alpha-hemolysin activates the NLRP3-inflammasome in human and mouse monocytic cells. PLoS ONE 2009, 4, e7446. [Google Scholar] [CrossRef]
- Li, R.S.; Liu, J.; Wen, C.; Shi, Y.; Ling, J.; Cao, Q.; Wang, L.; Shi, H.; Huang, C.Z.; Li, N. Transformable nano-antibiotics for mechanotherapy and immune activation against drug-resistant Gram-negative bacteria. Sci. Adv. 2023, 9, eadg9601. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, M.J.; Gao, B. Hepatocytes: A key cell type for innate immunity. Cell. Mol. Immunol. 2016, 13, 301–315. [Google Scholar] [CrossRef]
- Olson, J.; Nonejuie, P.; Dam, Q.; Dhand, A.; Pogliano, J.; Yeaman, M.R.; Hensler, M.E.; Bayer, A.S.; Nizet, V. Nafcillin enhances innate immune-mediated killing of methicillin-resistant. J. Mol. Med. 2014, 92, 139–149. [Google Scholar]
- Dhand, A.; Bayer, A.S.; Pogliano, J.; Yang, S.J.; Bolaris, M.; Nizet, V.; Wang, G.; Sakoulas, G. Use of antistaphylococcal β-Lactams to increase daptomycin activity in eradicating persistent bacteremia due to methicillin-resistant: Role of enhanced daptomycin binding. Clin. Infect. Dis. 2011, 53, 158–163. [Google Scholar] [CrossRef]
- Dhand, A.; Sakoulas, G. Daptomycin in combination with other antibiotics for the treatment of complicated methicillin-resistant bacteremia. Clin. Ther. 2014, 36, 1303–1316. [Google Scholar] [CrossRef]
- Yang, J.H.; Bhargava, P.; McCloskey, D.; Mao, N.; Palsson, B.O.; Collins, J.J. Antibiotic-induced changes to the host metabolic environment inhibit drug efficacy and alter immune function. Cell Host Microbe 2017, 22, 757–765. [Google Scholar] [CrossRef]
- Kelly, D.; Campbell, J.I.; King, T.P.; Grant, G.; Jansson, E.A.; Coutts, A.G.; Pettersson, S.; Conway, S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat. Immunol. 2004, 5, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Poojara, L.; Acharya, D.K.; Patel, J.; Rawal, R. Gut–Brain Axis: Role of the Gut Microbiome on Human Health in Microbiome-Gut-Brain Axis, 1st ed.; Springer: Singapore, 2022; pp. 187–211. [Google Scholar]
- Morais, L.H.; Schreiber, H.L.t.; Mazmanian, S.K. The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef]
- Mao, K.; Ji, F.; Breen, P.; Sewell, A.; Han, M.; Sadreyev, R.; Ruvkun, G. Mitochondrial dysfunction in activates mitochondrial relocalization and nuclear hormone receptor-dependent detoxification genes. Cell Metab. 2019, 29, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.Y.M.; Pan, Y.; Alshagga, M.; Lim, W.; Cin, K.; Alshehade, S.A.; Alshawsh, M. CYP14 family in Caenorhabditis elegans: Mitochondrial function, detoxification, and lifespan. J. Appl. Toxicol. 2024, 1–10. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Zhou, S.; Wang, W.; Zhou, X.; Zhang, Y.; Lai, Y.; Tang, Y.; Xu, J.; Li, D.; Lin, J.; Yang, X.; et al. Structure of cytochrome in complex with Q203 and TB47, two anti-TB drug candidates. elife 2021, 10, e69418. [Google Scholar] [CrossRef] [PubMed]
- Chepuri, V.; Lemieux, L.; Au, D.C.; Gennis, R.B. The sequence of the Cyo operon indicates substantial structural similarities between the cytochrome-o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome-c oxidases. J. Biol. Chem. 1990, 265, 11185–11192. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Yamato, I.; Anraku, Y.; Lemieux, L.; Gennis, R.B. Expression of cyoA and cyoB demonstrates that the co-binding heme component of the Escherichia coli cytochrome-o complex is in subunit I. J. Biol. Chem. 1990, 265, 11193–11197. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.Q.; Hanh, B.T.B.; Jeon, S.; Heo, B.E.; Park, Y.; Choudhary, A.; Moon, C.; Jang, J. Synergistic effect of Q203 combined with PBTZ169 against. Antimicrob. Agents Chemother. 2022, 66, e0044822. [Google Scholar] [CrossRef]
- Pethe, K.; Bifani, P.; Jang, J.; Kang, S.; Park, S.; Ahn, S.; Jiricek, J.; Jung, J.; Jeon, H.K.; Cechetto, J.; et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med. 2013, 19, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Liu, P.; Li, D.; Li, W.; Wang, D. Mitochondrial unfolded protein response to microgravity stress in nematode Caenorhabditis elegans. Sci. Rep. 2019, 9, 16474. [Google Scholar] [CrossRef] [PubMed]
- Menke, A.; Nitschke, F.; Hellmuth, A.; Helmel, J.; Wurst, C.; Stonawski, S.; Blickle, M.; Weiß, C.; Weber, H.; Hommers, L.; et al. Stress impairs response to antidepressants via HPA axis and immune system activation. Brain Behav. Immun. 2021, 93, 132–140. [Google Scholar] [CrossRef]
- Wei, P.L.; Keller, C.; Li, L.J. Neuropeptides in gut-brain axis and their influence on host immunity and stress. Comput. Struct. Biotechnol. J. 2020, 18, 843–851. [Google Scholar] [CrossRef]
- Pavlov, V.A.; Tracey, K.J. Neural regulation of immunity: Molecular mechanisms and clinical translation. Nat. Neurosci. 2017, 20, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Richmond, J.E.; Davis, W.S.; Jorgensen, E.M. UNC-13 is required for synaptic vesicle fusion in. Nat. Neurosci. 1999, 2, 959–964. [Google Scholar] [CrossRef]
- Stefanakis, N.; Carrera, I.; Hobert, O. Regulatory logic of pan-neuronal gene expression in C. elegans. Neuron 2015, 87, 733–750. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Cezairliyan, B.O.; Ausubel, F.M.; Powell, J.R. Pseudomonas aeruginosa PA14 pathogenesis in Caenorhabditis elegans. Methods Mol. Biol. 2014, 1149, 653–669. [Google Scholar]
- Tan, M.W.; Mahajan-Miklos, S.; Ausubel, F.M. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 1999, 96, 715–720. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, P.; Zheng, Z.; Afridi, M.I.; Zhang, S.; Wan, Z.; Zhang, X.; Stingelin, L.; Wang, Y.; Tu, H. GABAergic signaling between enteric neurons and intestinal smooth muscle promotes innate immunity and gut defense in Caenorhabditis elegans. Immunity 2023, 56, 1515–1532. [Google Scholar] [CrossRef]
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the balance: Antibiotic effects on host-microbiota mutualism. Nat. Rev. Microbiol. 2011, 9, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef]
- Ivanov, I.I.; de Llanos Frutos, R.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Anuforom, O.; Wallace, G.R.; Piddock, L.V. The immune response and antibacterial therapy. Med. Microbiol. Immunol. 2015, 204, 151–159. [Google Scholar] [CrossRef]
- Bhattarai, Y.; Muniz Pedrogo, D.A.; Kashyap, P.C. Irritable bowel syndrome: A gut microbiota-related disorder? Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G52–G62. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Tam, N.M.; Jogova, M.; Robertson, M.L.; Li, Y.; Lupp, C.; Finlay, B.B. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect. Immun. 2008, 76, 4726–4736. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Alford, M.; Akhoundsadegh, N.; Drayton, M.; Straus, S.K.; Hancock, R.E.W. Multifunctional antibiotic-host defense peptide conjugate kills bacteria, eradicates biofilms, and modulates the innate immune response. J. Med. Chem. 2021, 64, 16854–16863. [Google Scholar] [CrossRef] [PubMed]
- Von Wulffen, J.; RecogNice-Team; Sawodny, O.; Feuer, R. Transition of an anaerobic Escherichia coli culture to aerobiosis: Balancing mRNA and protein levels in a demand-directed dynamic flux balance analysis. PLoS ONE 2016, 11, e0158711. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, X.; Olmedo, M.; Holdorf, A.D.; Shang, Y.; Artal-Sanz, M.; Yilmaz, L.S.; Walhout, A.J.M. A delicate balance between bacterial iron and reactive oxygen species supports optimal C. elegans development. Cell Host Microbe 2019, 26, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Paganini, D.; Zimmermann, M.B. The effects of iron fortification and supplementation on the gut microbiome and diarrhea in infants and children: A review. Am. J. Clin. Nutr. 2017, 106, 1688–1693. [Google Scholar] [CrossRef]
- Su, C.C.; Lyu, M.; Morgan, C.E.; Bolla, J.R.; Robinson, C.V.; Yu, E.W. A ‘Build and Retrieve’ methodology to simultaneously solve cryo-EM structures of membrane proteins. Nat. Methods 2021, 18, 69–75. [Google Scholar] [CrossRef]

| Gene Name | Primers | Sequence |
|---|---|---|
| rpl-26 | primer F | CTTCGAAGGTCGCTATCACCA |
| primer R | TTGACGGTCTCGTCAGTGTG | |
| cyp-14A4 | primer F | CGTTTGTAACGCAAGGCGAA |
| primer R | CGTTTCCAACGCAAAGCTGA | |
| nlp-29 | primer F | TATGGAAGAGGATATGGAGGATATG |
| primer R | TCCATGTATTTACTTTCCCCATCC | |
| irg-1 | primer F | TGAAACTTGTGGAGGCCTCA |
| primer R | TGGCATCTAGTTTCCAGGCT |
| Docking Box | x | y | z | |
|---|---|---|---|---|
| 6wti | 101 Å × 101 Å × 101 Å | 161.597 | 168.208 | 157.094 |
| CyoB | 62 Å × 70 Å × 60 Å | 170.031 | 161.868 | 151.459 |
| CyoA | 106 Å × 64 Å × 56 Å | 160.696 | 182.206 | 141.716 |
| CyoC | 88 Å × 64 Å × 56 Å | 160.696 | 170.206 | 180.716 |
| CyoD | 81 Å × 56 Å × 65 Å | 160.696 | 180.206 | 180.716 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Li, G.; Tang, H. Antibiotics Trigger Host Innate Immune Response via Microbiota–Brain Communication in C. elegans. Int. J. Mol. Sci. 2024, 25, 8866. https://doi.org/10.3390/ijms25168866
Wu Y, Li G, Tang H. Antibiotics Trigger Host Innate Immune Response via Microbiota–Brain Communication in C. elegans. International Journal of Molecular Sciences. 2024; 25(16):8866. https://doi.org/10.3390/ijms25168866
Chicago/Turabian StyleWu, Yangyang, Guanqun Li, and Hongyun Tang. 2024. "Antibiotics Trigger Host Innate Immune Response via Microbiota–Brain Communication in C. elegans" International Journal of Molecular Sciences 25, no. 16: 8866. https://doi.org/10.3390/ijms25168866
APA StyleWu, Y., Li, G., & Tang, H. (2024). Antibiotics Trigger Host Innate Immune Response via Microbiota–Brain Communication in C. elegans. International Journal of Molecular Sciences, 25(16), 8866. https://doi.org/10.3390/ijms25168866





