Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo
Abstract
1. Introduction
2. Results
2.1. Narrow Down Chromosome 11 Candidate Region for WM-7-Derived ToLCNDV Resistance
2.1.1. Genotyping-by-Sequencing
2.1.2. Cosegregation Analysis in the F3 Population
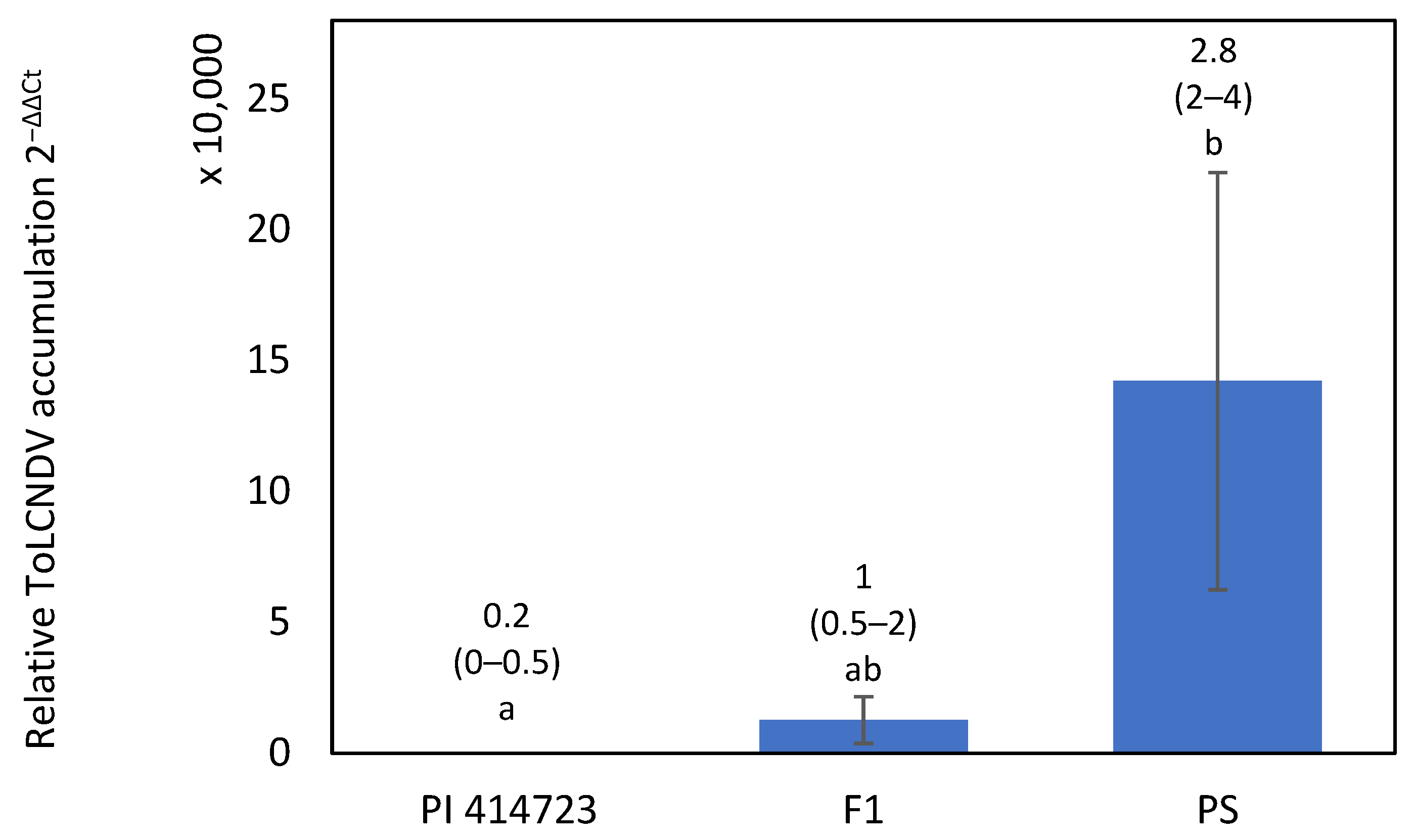
2.2. Fine Mapping of PI 414723-Derived Resistance
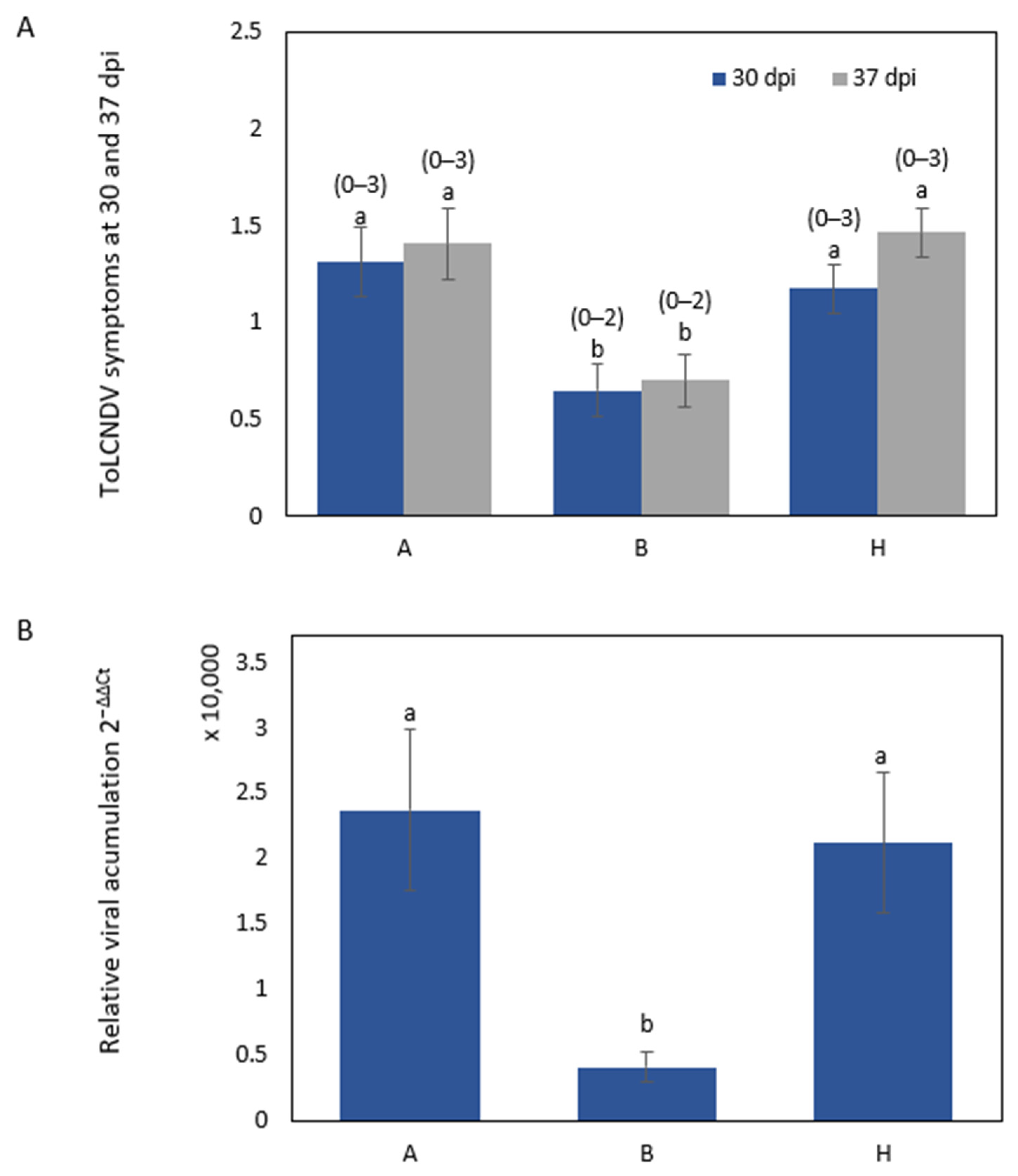
2.2.1. Phenotypic Variation in ToLCNDV Resistance in the F2 Population
2.2.2. QTL Analysis of F2 Population
2.2.3. Validation of the Markers Linked to the WM-7 Candidate Region in PI 414723 F2 Populations with a Different Ibericus Genetic Background
2.2.4. QTLs Mapping and Expression Analysis Based on BSR-seq
- Bulk construction and sequencing
- 2.
- Differential expression analysis
3. Discussion
3.1. WM-7
3.2. PI 414723, Source of ToLCNDV Resistance
3.3. QTL Analysis
3.4. Additional Regions of Interest
3.5. Differential Expression Analysis
4. Materials and Methods
4.1. Plant Material
4.1.1. WM-7
4.1.2. PI 414723
4.2. Inoculation Method and Disease Assessment
4.2.1. Inoculation Method
4.2.2. Disease Assessment
- Symptoms evaluation
- 2.
- Viral accumulation analysis
- 3.
- Tissue printing
- 4.
- DNA extraction
4.3. Molecular Markers Analysis
4.3.1. WM-7
4.3.2. PI 414723
4.4. Data Analysis
4.5. QTL Analysis
4.6. F2 PI 414723 × PS ‘Piñonet’ RNA-BSR-seq Analysis
4.6.1. BSR-seq Data Analysis
4.6.2. Transcript-Level Expression Analysis
4.6.3. Expression Differences Analysis
4.7. DNA Primase Large Subunit Sequencing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Srivastava, K.M.; Hallan, V.; Raizada, R.K.; Chandra, G.; Singh, B.P.; Sane, P.V. Molecular cloning of Indian tomato leaf curl virus genome following a simple method of concentrating the supercoiled replicative form of viral DNA. J. Virol. Methods 1995, 51, 297–304. [Google Scholar] [CrossRef]
- Padidam, R.; Beachy, R.N.; Fauquet, C.M. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J. Gen. Virol. 1995, 76 Pt 1, 25–35. [Google Scholar] [CrossRef] [PubMed]
- Jyothsna, P.; Haq, Q.M.I.I.; Singh, P.; Sumiya, K.V.; Praveen, S.; Rawat, R.; Briddon, R.W.; Malathi, V.G. Infection of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl. Microbiol. Biotechnol. 2013, 97, 5457–5471. [Google Scholar] [CrossRef]
- Juárez, M.; Tovar, R.; Fiallo-Olivé, E.; Aranda, M.A.; Gosálvez, B.; Castillo, P.; Moriones, E.; Navas-Castillo, J. First Detection of Tomato leaf curl New Delhi virus Infecting Zucchini in Spain. Plant Dis. 2014, 98, 857. [Google Scholar] [CrossRef]
- EPPO. EEPPO Global Database. Tomato Leaf Curl New Delhi Virus (ToLCNDV). 2021. Available online: https://gd.eppo.int/taxon/TOLCND (accessed on 20 January 2023).
- Simón, A.; Ruiz, L.; Velasco, L.; Janssen, D. Absolute Quantification of Tomato leaf curl New Delhi virus Spain strain, ToLCNDV-ES: Virus Accumulation in a Host-Specific Manner. Plant Dis. 2018, 102, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Font San Ambrosio, M.I.; Alfaro Fernández, A. “El virus de Nueva Delhi” (Tomato leaf curl New Delhi virus, ToLCNDV) amplía su gama de hospedantes en los cultivos españoles. Phytoma 2015, 272, 25–32. [Google Scholar]
- FAO. FAOSTAT Database. Food and Agriculture Organization of the United Nations. 2022. Available online: https://www.fao.org/faostat/es/#home (accessed on 30 January 2024).
- MAPA Superfícies y Producciones Anuales de Cultivos. Ministerio de Agricultura, Pesca y Alimentación (MAPA). 2021. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/agricultura/ (accessed on 3 December 2023).
- Rodríguez, E.; Téllez, M.M.; Janssen, D. Whitefly Control Strategies against Tomato Leaf Curl New Delhi Virus in Greenhouse Zucchini. Int. J. Environ. Res. Public Health 2019, 16, 2673. [Google Scholar] [CrossRef] [PubMed]
- Lapidot, M.; Legg, J.P.; Wintermantel, W.M.; Polston, J.E. Management of Whitefly-Transmitted Viruses in Open-Field Production Systems. In Advances in Virus Research, 1st ed.; Academic Press: Waltham, MA, USA, 2014; Volume 90, pp. 147–206. [Google Scholar]
- Islam, S.; Munshi, A.D.; Mandal, B.; Kumar, R.; Behera, T.K. Genetics of resistance in Luffa cylindrica Roem. against Tomato leaf curl New Delhi virus. Euphytica 2010, 174, 83–89. [Google Scholar] [CrossRef]
- Siskos, L.; Cui, L.; Wang, C.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A new challenge in melon resistance breeding: The ToLCNDV case. Euphytica 2022, 218, 129. [Google Scholar] [CrossRef]
- Martín-Hernández, A.M.; Picó, B. Natural Resistances to Viruses in Cucurbits. Agronomy 2020, 11, 23. [Google Scholar] [CrossRef]
- López, C.; Ferriol, M.; Picó, M.B. Mechanical transmission of Tomato leaf curl New Delhi virus to cucurbit germplasm: Selection of tolerance sources in Cucumis melo. Euphytica 2015, 204, 679–691. [Google Scholar] [CrossRef]
- Sáez, C.; Esteras, C.; Martínez, C.; Ferriol, M.; Dhillon, N.P.S.; López, C.; Picó, B. Resistance to tomato leaf curl New Delhi virus in melon is controlled by a major QTL located in chromosome 11. Plant Cell Rep. 2017, 36, 1571–1584. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, M.J.; Díaz, A.; Dhillon, N.P.S.; Reddy, U.K.; Picó, B.; Monforte, A.J. Re-evaluation of the role of Indian germplasm as center of melon diversification based on genotyping-by-sequencing analysis. BMC Genom. 2019, 20, 448. [Google Scholar] [CrossRef]
- Romay, G.; Pitrat, M.; Lecoq, H.; Wipf-Scheibel, C.; Millot, P.; Girardot, G.; Desbiez, C. Resistance Against Melon Chlorotic Mosaic Virus and Tomato Leaf Curl New Delhi Virus in Melon. Plant Dis. 2019, 103, 2913–2919. [Google Scholar] [CrossRef] [PubMed]
- Sáez, C.; Flores-León, A.; Montero-Pau, J.; Sifres, A.; Dhillon, N.P.S.; López, C.; Picó, B. RNA-Seq Transcriptome Analysis Provides Candidate Genes for Resistance to Tomato Leaf Curl New Delhi Virus in Melon. Front. Plant Sci. 2022, 12, 798858. [Google Scholar] [CrossRef] [PubMed]
- Sáez, C.; Martínez, C.; Montero-Pau, J.; Esteras, C.; Sifres, A.; Blanca, J.; Ferriol, M.; López, C.; Picó, B. A Major QTL Located in Chromosome 8 of Cucurbita moschata Is Responsible for Resistance to Tomato Leaf Curl New Delhi Virus. Front. Plant Sci. 2020, 11, 207. [Google Scholar] [CrossRef] [PubMed]
- Siskos, L.; Antoniou, M.; Riado, J.; Enciso, M.; Garcia, C.; Liberti, D.; Esselink, D.; Baranovskiy, A.G.; Tahirov, T.H.; Visser, R.G.F.; et al. DNA primase large subunit is an essential plant gene for geminiviruses, putatively priming viral ss-DNA replication. Front. Plant Sci. 2023, 14, 1130723. [Google Scholar] [CrossRef] [PubMed]
- Pitrat, M.; Lecoq, H. Inheritance of zucchini yellow mosaic virus resistance in Cucumis melo L. Euphytica 1984, 33, 57–61. [Google Scholar] [CrossRef]
- Danin-Poleg, Y.; Paris, H.S.; Cohen, S.; Rabinowitch, H.D.; Karchi, Z. Oligogenic inheritance of resistance to zucchini yellow mosaic virus in melons. Euphytica 1997, 93, 331–337. [Google Scholar] [CrossRef]
- Esteras, C.; Formisano, G.; Roig, C.; Díaz, A.; Blanca, J.; Garcia-Mas, J.; Gómez-Guillamón, M.L.; López-Sesé, A.I.; Lázaro, A.; Monforte, A.J.; et al. SNP genotyping in melons: Genetic variation, population structure, and linkage disequilibrium. Theor. Appl. Genet. 2013, 126, 1285–1303. [Google Scholar] [CrossRef]
- Leida, C.; Moser, C.; Esteras, C.; Sulpice, R.; Lunn, J.E.; de Langen, F.; Monforte, A.J.; Picó, B. Variability of candidate genes, genetic structure and association with sugar accumulation and climacteric behavior in a broad germplasm collection of melon (Cucumis melo L.). BMC Genet. 2015, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Perpiñá, G.; Esteras, C.; Gibon, Y.; Monforte, A.J.; Picó, B. A new genomic library of melon introgression lines in a cantaloupe genetic background for dissecting desirable agronomical traits. BMC Plant Biol. 2016, 16, 154. [Google Scholar] [CrossRef]
- Pérez-de-Castro, A.; Esteras, C.; Alfaro-Fernández, A.; Daròs, J.A.; Monforte, A.J.; Picó, B.; Gómez-Guillamón, M.L. Fine mapping of wmv1551, a resistance gene to Watermelon mosaic virus in melon. Mol. Breed. 2019, 39, 93. [Google Scholar] [CrossRef]
- López-Martín, M.; Pérez-de-Castro, A.; Picó, B.; Gómez-Guillamón, M.L. Advanced Genetic Studies on Powdery Mildew Resistance in TGR-1551. Int. J. Mol. Sci. 2022, 23, 12553. [Google Scholar] [CrossRef]
- Padmanabha, K.; Choudhary, H.; Mishra, G.P.; Mandal, B.; Solanke, A.U.; Mishra, D.C.; Yadav, R.K. Identifying new sources of resistance to tomato leaf curl New Delhi virus from Indian melon germplasm by designing an improved method of field screening. Genet. Resour. Crop Evol. 2023, 71, 1911–1933. [Google Scholar] [CrossRef]
- Chen, L.; Sun, D.; Zhang, X.; Shao, D.; Lu, Y.; An, Y. Transcriptome analysis of yellow passion fruit in response to cucumber mosaic virus infection. PLoS ONE 2021, 16, e0247127. [Google Scholar] [CrossRef]
- Lantican, D.V.; Nocum, J.D.L.; Manohar, A.N.C.; Mendoza, J.V.S.; Gardoce, R.R.; Lachica, G.C.; Gueco, L.S.; Dela Cueva, F.M. Comparative RNA-seq analysis of resistant and susceptible banana genotypes reveals molecular mechanisms in response to banana bunchy top virus (BBTV) infection. Sci. Rep. 2023, 13, 18719. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, N.; Basak, J. Comparative transcriptome profiling of a resistant vs. susceptible Vigna mungo cultivar in response to Mungbean yellow mosaic India virus infection reveals new insight into MYMIV resistance. Curr. Plant Biol. 2018, 15, 8–24. [Google Scholar] [CrossRef]
- Wang, R.; Du, Z.; Bai, Z.; Liang, Z. The interaction between endogenous 30S ribosomal subunit protein S11 and Cucumber mosaic virus LS2b protein affects viral replication, infection and gene silencing suppressor activity. PLoS ONE 2017, 12, e0182459. [Google Scholar] [CrossRef]
- Li, S. Regulation of ribosomal proteins on viral infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef]
- Gilbert, R.Z.; Kyle, M.M.; Munger, H.M.; Gray, S.M. Inheritance of Resistance to Watermelon Mosaic Virus in Cucumis melo L. HortScience 1994, 29, 107–110. [Google Scholar] [CrossRef]
- Anagnostou, K.; Jahn, M.; Perl-Treves, R. Inheritance and linkage analysis of resistance to zucchini yellow mosaic virus, watermelon mosaic virus, papaya ringspot virus and powdery mildew in melon. Euphytica 2000, 116, 265–270. [Google Scholar] [CrossRef]
- Prakash, V.; Singh, A.; Singh, A.K.; Dalmay, T.; Chakraborty, S. Tobacco RNA-dependent RNA polymerase 1 affects the expression of defence-related genes in Nicotiana benthamiana upon Tomato leaf curl Gujarat virus infection. Planta 2020, 252, 11. [Google Scholar] [CrossRef]
- Willmann, M.R.; Endres, M.W.; Cook, R.T.; Gregory, B.D. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arab. Book 2011, 9, e0146. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-J.; Carter, S.A.; Cole, A.B.; Cheng, N.-H.; Nelson, R.S. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 2004, 101, 6297–6302. [Google Scholar] [CrossRef] [PubMed]
- Leibman, D.; Kravchik, M.; Wolf, D.; Haviv, S.; Weissberg, M.; Ophir, R.; Paris, H.S.; Palukaitis, P.; Ding, S.; Gaba, V.; et al. Differential expression of cucumber RNA-dependent RNA polymerase 1 genes during antiviral defence and resistance. Mol. Plant Pathol. 2018, 19, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Verlaan, M.G.; Hutton, S.F.; Ibrahem, R.M.; Kormelink, R.; Visser, R.G.F.; Scott, J.W.; Edwards, J.D.; Bai, Y. The Tomato Yellow Leaf Curl Virus Resistance Genes Ty-1 and Ty-3 Are Allelic and Code for DFDGD-Class RNA–Dependent RNA Polymerases. PLoS Genet. 2013, 9, e1003399. [Google Scholar] [CrossRef]
- Shao, F.; Lu, S. Identification, Molecular Cloning and Expression Analysis of Five RNA-Dependent RNA Polymerase Genes in Salvia miltiorrhiza. PLoS ONE 2014, 9, e95117. [Google Scholar] [CrossRef]
- Shrestha, S.; Fu, Y.; Michael, V.N.; Meru, G. Whole Genome Re-sequencing and Bulk Segregant Analysis Reveals Chromosomal Location for Papaya Ringspot Virus W Resistance in Squash. Front. Plant Sci. 2022, 13, 848631. [Google Scholar] [CrossRef]
- Pan, L.-L.; Chen, Q.-F.; Zhao, J.-J.; Guo, T.; Wang, X.-W.; Hariton-Shalev, A.; Czosnek, H.; Liu, S.-S. Clathrin-mediated endocytosis is involved in Tomato yellow leaf curl virus transport across the midgut barrier of its whitefly vector. Virology 2017, 502, 152–159. [Google Scholar] [CrossRef]
- Mercer, J.; Schelhaas, M.; Helenius, A. Virus Entry by Endocytosis. Annu. Rev. Biochem. 2010, 79, 803–833. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Cui, X.; Chen, H.; Renaud, J.B.; Yu, K.; Chen, X.; Wang, A. Dynamin-Like Proteins of Endocytosis in Plants Are Coopted by Potyviruses To Enhance Virus Infection. J. Virol. 2018, 92, 23. [Google Scholar] [CrossRef]
- Xia, W.-Q.; Liang, Y.; Chi, Y.; Pan, L.-L.; Zhao, J.; Liu, S.-S.; Wang, X.-W. Intracellular trafficking of begomoviruses in the midgut cells of their insect vector. PLoS Pathog. 2018, 14, e1006866. [Google Scholar] [CrossRef]
- Silva, F.D.A.; Raimundo, G.S.; Fontes, E.P.B. Begomovirus–host protein-protein interactions in intracellular virus movement. In Geminivirus: Detection, Diagnosis and Management; Elsevier: Amsterdam, The Netherlands, 2022; pp. 347–356. [Google Scholar]
- Nersissian, A.M.; Valentine, J.S.; Immoos, C.; Hill, M.G.; Hart, P.J.; Williams, G.; Herrmann, R.G. Uclacyanins, stellacyanins, and plantacyanins are distinct subfamilies of phytocyanins: Plant-specific mononuclear blue copper proteins. Protein Sci. 1998, 7, 1915–1929. [Google Scholar] [CrossRef]
- Liu, J.; Fan, H.; Wang, Y.; Han, C.; Wang, X.; Yu, J.; Li, D.; Zhang, Y. Genome-Wide microRNA Profiling Using Oligonucleotide Microarray Reveals Regulatory Networks of microRNAs in Nicotiana benthamiana during Beet Necrotic Yellow Vein Virus Infection. Viruses 2020, 12, 310. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.-L.; Gou, W.; Han, X.-L.; Qin, C.; Zhang, L.-X.; Abomohra, A.; Ashraf, M. Cloning and Functional Analysis of Phosphoethanolamine Methyltransferase Promoter from Maize (Zea mays L.). Int. J. Mol. Sci. 2018, 19, 191. [Google Scholar] [CrossRef] [PubMed]
- Ismayil, A.; Haxim, Y.; Wang, Y.; Li, H.; Qian, L.; Han, T.; Chen, T.; Jia, Q.; Yihao Liu, A.; Zhu, S.; et al. Cotton Leaf Curl Multan virus C4 protein suppresses both transcriptional and post-transcriptional gene silencing by interacting with SAM synthetase. PLoS Pathog. 2018, 14, e1007282. [Google Scholar] [CrossRef]
- De, S.; Pollari, M.; Varjosalo, M.; Mäkinen, K. Association of host protein VARICOSE with HCPro within a multiprotein complex is crucial for RNA silencing suppression, translation, encapsidation and systemic spread of potato virus A infection. PLoS Pathog. 2020, 16, e1008956. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Huang, X.; Xia, R.; Zhao, Q.; Lai, J.; Teng, K.; Li, Y.; Liang, L.; Du, Q.; et al. BSCTV C2 Attenuates the Degradation of SAMDC1 to Suppress DNA Methylation-Mediated Gene Silencing in Arabidopsis. Plant Cell 2011, 23, 273–288. [Google Scholar] [CrossRef]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the actin cytoskeleton during viral infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [Google Scholar] [CrossRef]
- Román, B.; Gómez, P.; Janssen, D.; Ruiz, L. Insights into the Key Genes in Cucumis melo and Cucurbita moschata ToLCNDV Resistance. Horticulturae 2023, 9, 231. [Google Scholar] [CrossRef]
- Sáez, C.; Martínez, C.; Ferriol, M.; Manzano, S.; Velasco, L.; Jamilena, M.; López, C.; Picó, B. Resistance to Tomato leaf curl New Delhi virus in Cucurbita spp. Ann. Appl. Biol. 2016, 169, 91–105. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sáez, C.; Ambrosio, L.G.M.; Miguel, S.M.; Valcárcel, J.V.; Díez, M.J.; Picó, B.; López, C. Resistant Sources and Genetic Control of Resistance to ToLCNDV in Cucumber. Microorganisms 2021, 9, 913. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.; Doyle, J. Isolation of plant DNA from fresh tissue. Focus 1987, 12, 13–15. [Google Scholar]
- Flores-León, A.; Peréz Moro, C.; Martí, R.; Beltran, J.; Roselló, S.; Cebolla-Cornejo, J.; Picó, B. Spanish Melon Landraces: Revealing Useful Diversity by Genomic, Morphological, and Metabolomic Analysis. Int. J. Mol. Sci. 2022, 23, 7162. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing—Free bayes—Variant Calling—Longranger. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap® 4 Software for the Calculation of Genetic Linkage Maps in Experimental Populations; Kyazma, B.V., Ed.; ScienceOpen: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kendall, K.; George, M. Kruskal-Wallis Test. In The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008; pp. 288–290. [Google Scholar]
- Joehanes, R.; Nelson, J.C. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 2008, 24, 2788–2789. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 20 September 2022).
- Melonomics Database (v 4.0). Available online: www.melonomics.net (accessed on 15 October 2023).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R Package for Bulk Segregant Analysis with Next-Generation Sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef] [PubMed]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat. Biotechnol. 2015, 33, 243–246. [Google Scholar] [CrossRef]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ibeas, D.; Blanca, J.; Roig, C.; González-To, M.; Picó, B.; Truniger, V.; Gómez, P.; Deleu, W.; Caño-Delgado, A.; Arús, P.; et al. MELOGEN: An EST database for melon functional genomics. BMC Genom. 2007, 8, 306. [Google Scholar] [CrossRef]




| Markers | Position (bp) 1 | Chr 2 | F2-WM-7 × RC |
|---|---|---|---|
| SNPCmND1 | 23,984,243 | 2 | B |
| SNPCmND2 | 25,291,938 | 2 | B |
| SNPCmND3 | 25,448,714 | 2 | B |
| SNPCmND4 | 25,611,353 | 2 | B |
| SNPCmND5bis | 25,904,727 | 2 | B |
| SNPCmND6 | 26,504,936 | 2 | B |
| SNPCmND7 | 30,249,798 | 11 | H |
| SNPCmND9 | 30,276,354 | 11 | H |
| SNPCmND11 | 30,280,636 | 11 | H |
| SNPCmND13bis | 30,347,863 | 11 | H |
| SNPCmND15 | 30,377,414 | 11 | A |
| SNPCmND14 | 30,395,841 | 11 | A |
| SNPCmND16bis | 30,403,862 | 11 | A |
| SNPCmND17 | 30,410,536 | 11 | A |
| SNPCmND19 | 30,441,821 | 11 | A |
| SNPCmND20 | 30,458,337 | 11 | A |
| SNPCmND22 | 30,472,365 | 11 | A |
| SNPCmND23 | 30,482,001 | 11 | A |
| SNPCmND25 | 30,537,322 | 11 | A |
| SNPCmND26bis | 10,175,361 | 12 | A |
| SNPCmND27 | 11,965,753 | 12 | A |
| SNPCmND28bis | 13,551,907 | 12 | A |
| SNPCmND29 | 14,425,696 | 12 | A |
| SNPCmND30 | 15,368,098 | 12 | A |
| Marker | Position (bp) | 8 | 4 | 11 | 17 | 14 | 7 | 4b |
|---|---|---|---|---|---|---|---|---|
| S11_30159412 | 30,159,412 | B | B | H | A | B | H | H |
| S11_30191060 | 30,191,060 | B | B | H | A | H | H | H |
| S11_30191187 | 30,191,187 | B | B | H | A | H | H | H |
| S11_30191253 | 30,191,253 | B | B | H | A | H | H | H |
| S11_30191271 | 30,191,271 | B | B | H | A | H | H | H |
| S11_30191293 | 30,191,293 | B | B | H | A | H | H | H |
| S11_30197124 | 30,197,124 | B | B | H | A | H | H | H |
| S11_30197480 | 30,197,480 | B | B | H | A | A | H | H |
| S11_30197584 | 30,197,584 | B | B | H | A | H | H | H |
| S11_30197679 | 30,197,679 | B | B | H | A | H | H | H |
| S11_30197706 | 30,197,706 | B | B | H | A | H | H | H |
| S11_30197725 | 30,197,725 | B | B | H | A | H | H | H |
| S11_30197739 | 30,197,739 | B | B | H | A | H | H | H |
| S11_30202708 | 30,202,708 | B | B | H | A | H | H | H |
| S11_30202743 | 30,202,743 | B | B | H | A | H | H | H |
| S11_30202810 | 30,202,810 | B | B | H | A | H | H | H |
| S11_30202831 | 30,202,831 | B | B | H | A | H | H | H |
| S11_30216956 | 30,216,956 | B | B | H | A | H | H | H |
| S11_30217175 | 30,217,175 | B | B | H | A | H | H | H |
| S11_30221819 | 30,221,819 | B | B | H | A | H | H | H |
| S11_30221970 | 30,221,970 | B | B | H | A | H | H | H |
| S11_30227392 | 30,227,392 | B | B | H | A | H | H | H |
| S11_30261314 | 30,261,314 | B | B | H | A | H | H | H |
| S11_30287824 | 30,287,824 | B | B | H | A | H | A | H |
| S11_30287838 | 30,287,838 | B | B | H | A | H | A | H |
| S11_30287875 | 30,287,875 | B | B | H | A | H | A | H |
| S11_30307415 | 30,307,415 | B | B | H | A | H | H | H |
| S11_30307457 | 30,307,457 | B | B | H | A | H | H | H |
| S11_30339408 | 30,339,408 | B | B | H | A | H | H | H |
| S11_30358926 | 30,358,926 | B | B | H | A | H | H | H |
| S11_30359039 | 30,359,039 | B | B | H | A | H | H | H |
| S11_30401768 | 30,401,768 | A | A | A | A | A | A | A |
| Phenotype | R | R | S | S | S | S | S | |
| Composite Interval Mapping | Kruskal-Wallis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Trait 1 | Chr 2 | Interval 1.5 (cM) 3 | Nearest Marker 4 | LOD 5 | Add 6 | Dom 7 | d/a 8 | R 9 | Mean PI 414723 10 | Mean PS 11 |
| 9 DPI | 11 | 103.8–131.2 29,176,476–30,819,884 bp | CMPSNP315 | 8 | 0.799 | 0.651 | 0.815 | 20.2 | 0.545 | 1.917 |
| 30 DPI VT | 11 | 103.8–131.2 29,176,476–30,819,884 bp | CMPSNP315 | 4.98 | 0.612 | 0.409 | 1.252 | 13.7 | 1.256 | 2.392 |
| G-Prime Method Analysis | QTLseq Method Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Bulks | QTL Name | Chr 1 | Start 2 | End 3 | Max G-Prime 4 | Pos | Start 6 | End 7 | Peak | Pos Peak |
| Comparation | MaxG-Prime 5 | ∆SNP 8 | ∆SNP 9 | |||||||
| 1 vs. 3 | 1vs3.chr2 | chr02 | 3,877,068 | 6,945,099 | 33.58 | 5,228,546 | 4,405,764 | 5,962,440 | −0.42 | 5,355,437 |
| 1vs3.chr9 | chr09 | 633,982 | 13,389,235 | 51.76 | 6,725,288 | 2,480,077 | 9,259,367 | −0.51 | 6,752,980 | |
| 10,249,392 | 10,603,982 | −0.39 | 10,506,551 | |||||||
| 1 vs. 4 | 1vs4.chr11 | chr11 | 29,046,233 | 31,900,318 | 112.82 | 31,900,318 | 30,571,554 | 31,399,076 | 0.59 | 30,904,594 |
| 1 vs. 5 | 1vs5.chr11 | chr11 | 27,931,306 | 31,900,318 | 132.74 | 29,907,303 | 30,752,546 | 31,146,927 | 0.58 | 30,904,594 |
| 2 vs. 4 | 2vs4.chr2 | chr02 | 12,066,407 | 26,957,490 | 100.79 | 23,226,160 | 12,635,695 | 15,275,207 | 0.48 | 14,509,192 |
| 18,855,678 | 22,821,114 | 0.54 | 21,096,983 | |||||||
| 24,325,575 | 25,333,267 | 0.53 | 24,858,784 | |||||||
| 2vs4.chr5 | chr05 | 23,851,469 | 25,788,776 | 41.77 | 24,970,077 | 24,710,571 | 25,611,014 | 0.43 | 24,970,077 | |
| 2vs4.chr11 | chr11 | 25,706,075 | 31,900,318 | 158.85 | 30,403,683 | 30,571,554 | 31,417,092 | 0.58 | 30,904,594 | |
| 2 vs. 5 | 2vs5.chr2 | chr02 | 18,022,512 | 25,261,922 | 98.31 | 21,866,865 | 16,291,849 | 22,874,524 | 0.67 | 21,150,416 |
| 24,345,760 | 25,203,049 | 0.47 | 24,858,784 | |||||||
| 2vs5.chr8 | chr08 | 3,341,340 | 3,834,866 | 46.44 | 3,655,973 | 2,826,416 | 3,746,350 | 0.43 | 3,132,745 | |
| 2vs5.chr11 | chr11 | 26,303,416 | 31,900,318 | 144.67 | 30,404,623 | 30,442,913 | 31,415,732 | 0.60 | 30,904,594 | |
| 3 vs. 4 | 3vs4.chr2 | chr02 | 3,580,909 | 6,532,819 | 25.49 | 4,547,718 | 4,442,145 | 4,444,641 | 0.38 | 4,442,145 |
| 3vs4.chr8.1 | chr08 | 8,323,310 | 13,456,090 | 39.19 | 11,123,496 | 8,838,130 | 12,710,777 | −0.44 | 10,503,405 | |
| 3vs4.chr8.2 | chr08 | 14,028,052 | 23,893,540 | 41 | 18,282,774 | 18,282,774 | 19,070,463 | −0.43 | 18,734,123 | |
| 3vs4.chr11 | chr11 | 16,997,817 | 31,804,037 | 109.9 | 29,814,321 | 30,571,554 | 31,500,362 | 0.55 | 30,814,948 | |
| 3 vs. 5 | 3vs5.chr2.1 | chr02 | 9,669,827 | 10,218,866 | 33.58 | 9,936,717 | 9,936,717 | 10,062,220 | 0.37 | 9,936,717 |
| 3vs5.chr2.2 | chr02 | 21,065,243 | 21,339,968 | 32.26 | 21,122,957 | 20,979,784 | 21,339,968 | 0.39 | 21,277,282 | |
| 3vs5.chr11 | chr11 | 24,679,309 | 31,804,037 | 150.28 | 29,324,169 | 30,571,554 | 31,293,094 | 0.51 | 30,818,944 | |
| Gene Name | Position (bp) | Description | QTL |
|---|---|---|---|
| MELO3C015406.2 | 1,367,327–1,381,672 | RNA-dependent RNA polymerase | - |
| MELO3C010318.2 | 17,006,652–17,008,337 | Clathrin assembly protein. Putative | 2vs4.chr2 |
| MELO3C010326.2 | 17,122,601–17,123,944 | umecyanin-like | 2vs4.chr2 |
| MELO3C029682.2 | 18,969,012–18,975,891 | Transcriptional corepressor LEUNIG | overlapping region between QTLs 2vs4.chr2 and 2vs5.chr2 |
| MELO3C017424.2 | 23,325,650–23,329,684 | Transcription factor bHLH35 | overlapping region between QTLs 2vs4.chr2 and 2vs5.chr2 |
| MELO3C017356.2 | 23,965,259–23,968,861 | Phosphoethanolamine n-methyltransferase | overlapping region between QTLs 2vs4.chr2 and 2vs5.chr2 |
| MELO3C017295.2 | 24,499,946–24,508,159 | Actin-related protein 4 | overlapping region between QTLs 2vs3.chr2, 2vs4.chr2, 2vs5.chr2 and 3vs4.5 |
| MELO3C017283.2 | 24,603,206–24,603,619 | Transmembrane protein, putative | overlapping region between QTLs 2vs4.chr2 and 2vs5.chr2 |
| MELO3C017106.2 | 25,821,307–25,826,094 | RNA-dependent RNA polymerase | 2vs4.chr2 |
| MELO3C017185.2 | 25,323,802–25,325,467 | NAC domain protein | 2vs4.chr2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Moro, C.; Sáez, C.; Sifres, A.; López, C.; Dhillon, N.P.S.; Picó, B.; Pérez-de-Castro, A. Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo. Int. J. Mol. Sci. 2024, 25, 8880. https://doi.org/10.3390/ijms25168880
Pérez-Moro C, Sáez C, Sifres A, López C, Dhillon NPS, Picó B, Pérez-de-Castro A. Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo. International Journal of Molecular Sciences. 2024; 25(16):8880. https://doi.org/10.3390/ijms25168880
Chicago/Turabian StylePérez-Moro, Clara, Cristina Sáez, Alicia Sifres, Carmelo López, Narinder P. S. Dhillon, Belén Picó, and Ana Pérez-de-Castro. 2024. "Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo" International Journal of Molecular Sciences 25, no. 16: 8880. https://doi.org/10.3390/ijms25168880
APA StylePérez-Moro, C., Sáez, C., Sifres, A., López, C., Dhillon, N. P. S., Picó, B., & Pérez-de-Castro, A. (2024). Genetic Dissection of ToLCNDV Resistance in Resistant Sources of Cucumis melo. International Journal of Molecular Sciences, 25(16), 8880. https://doi.org/10.3390/ijms25168880







