Transient Adaptation of Toxoplasma gondii to Exposure by Thiosemicarbazone Drugs That Target Ribosomal Proteins Is Associated with the Upregulated Expression of Tachyzoite Transmembrane Proteins and Transporters
Abstract
:1. Introduction
2. Results
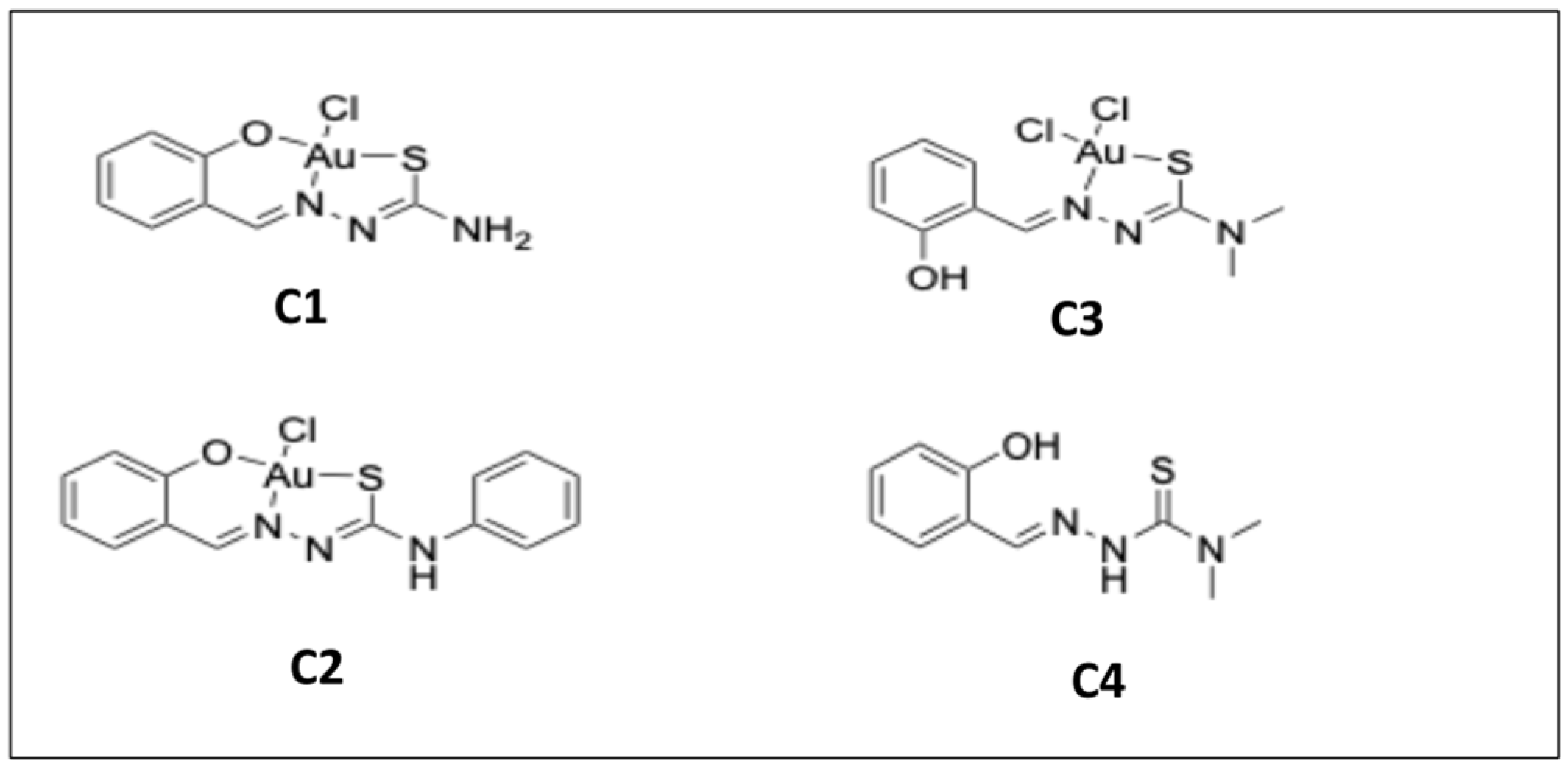
2.1. In Vitro Activities of Three Gold Complexes and Their Salicyl-TSC Ligand (C4) against T. gondii-tachyzoites
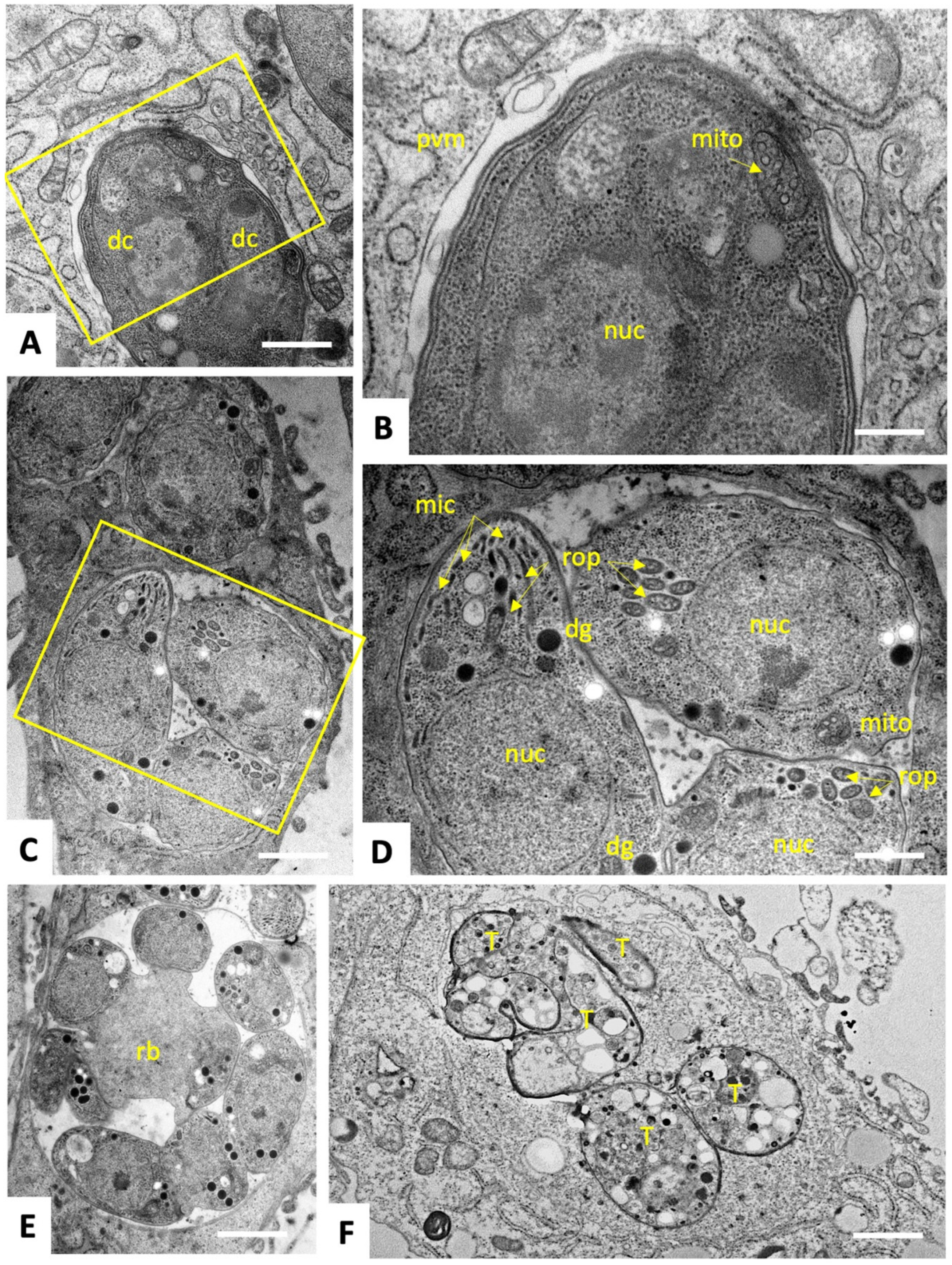
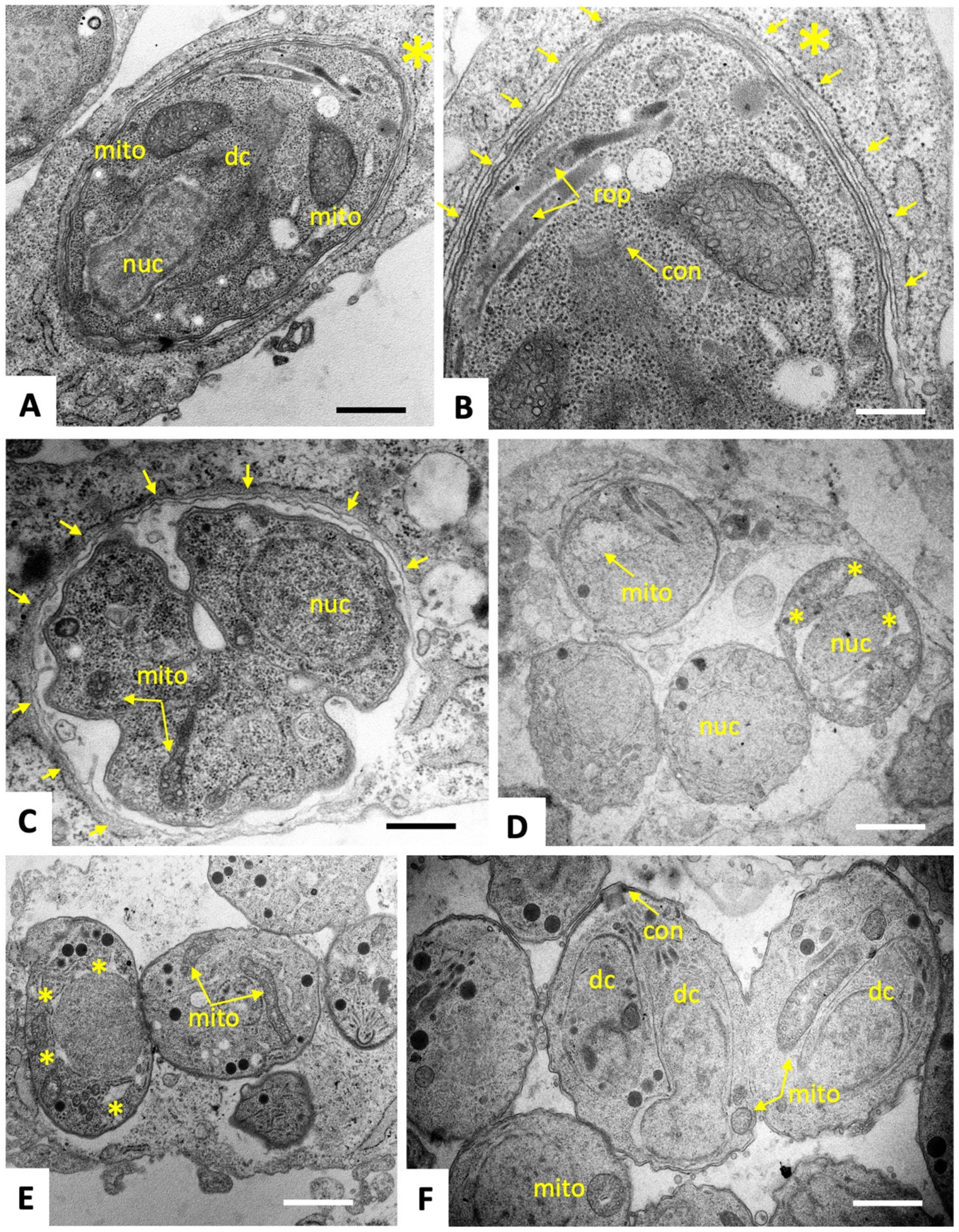
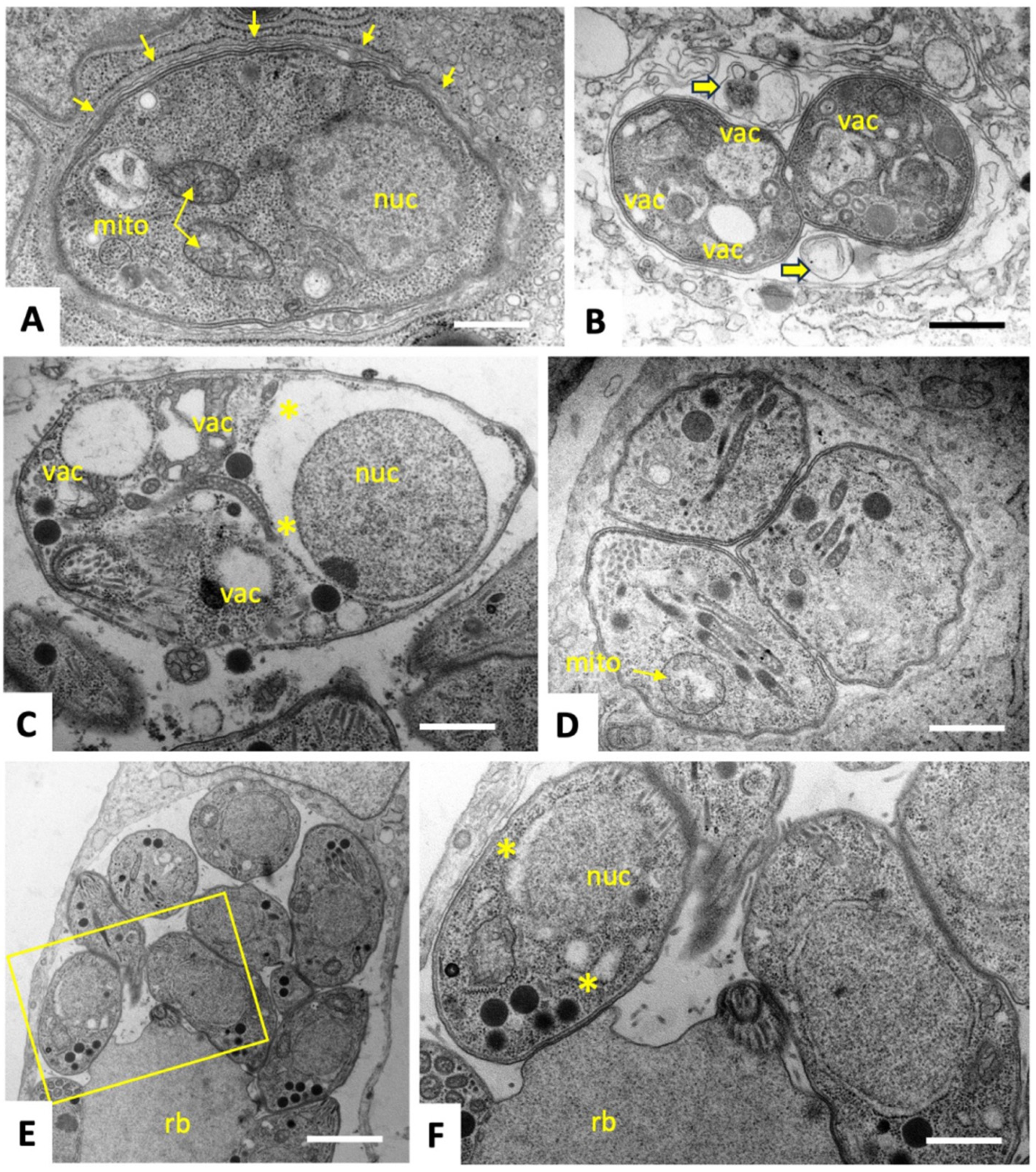
2.2. C3 and C4 Induce Only Minor and Transient Ultrastructural Alterations in and Do Not Affect the Mitochondrial Membrane Potential T. gondii ME49 Tachyzoites
2.3. Identification of C3- and C4-Binding Proteins in Extracts of T. gondii ME49 tachyzoites
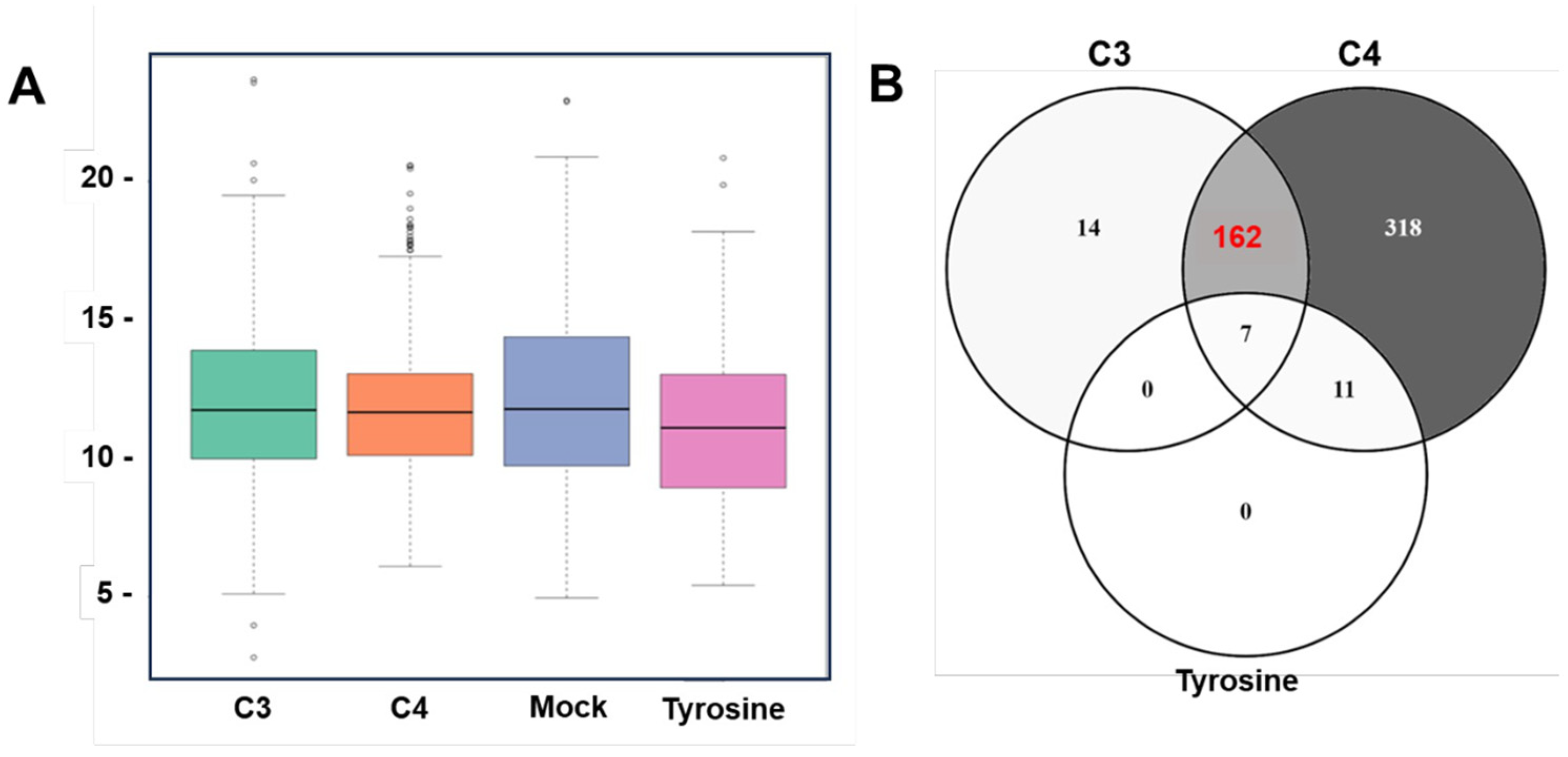
2.4. Long-Term Treatments and Generation of Clones of Drug-Adapted T. gondii Tachyzoites
2.5. Proteomic Analysis of T. gondii ME49 Clones Recovered after In Vitro Adaptation to C3 and C4
3. Discussion
4. Materials and Methods
4.1. Equipment and Reagents Used in Cell Culture
4.2. Compounds
4.3. Host Cells and Parasites
4.4. In Vitro Assessment of Toxicity and Efficacy of Compounds in HFF and T. gondii tachyzoites
4.5. Transmission Electron Microscopy (TEM)
4.6. Evaluation of the MMP by Tetramethyl Rhodamine, Ethyl Ester (TMRE) Uptake Assay
4.7. Drug Treatments for Extended Periods of Time and Generation of C3 and C4 Adapted T. gondii ME49 Clonal Strains
4.8. Differential Affinity Chromatography (DAC)
4.9. Proteomics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Almeria, S.; Dubey, J.P. Foodborne Transmission of Toxoplasma Gondii Infection in the Last Decade. An Overview. Res. Vet. Sci. 2021, 135, 371–385. [Google Scholar]
- Attias, M.; Teixeira, D.E.; Benchimol, M.; Vommaro, R.C.; Crepaldi, P.H.; De Souza, W. The Life-Cycle of Toxoplasma Gondii Reviewed Using Animations. Parasit. Vectors 2020, 13, 1–13. [Google Scholar]
- Lindsay, D.S.; Dubey, J.P. Neosporosis, Toxoplasmosis, and Sarcocystosis in Ruminants: An Update. Vet. Clin. N. Am.-Food Anim. Pract. 2020, 36, 205–222. [Google Scholar]
- Dubey, J.P.; Murata, F.H.A.; Cerqueira-Cézar, C.K.; Kwok, O.C.H.; Villena, I. Congenital Toxoplasmosis in Humans: An Update of Worldwide Rate of Congenital Infections. Parasitology 2021, 148, 1406–1416. [Google Scholar] [PubMed]
- Dubey, J.P. Toxoplasmosis of Animals and Humans, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2010; Volume 3. [Google Scholar]
- Di Cristina, M.; Marocco, D.; Galizi, R.; Proietti, C.; Spaccapelo, R.; Crisanti, A. Temporal and Spatial Distribution of Toxoplasma Gondii Differentiation into Bradyzoites and Tissue Cyst Formation in Vivo. Infect. Immun. 2008, 76, 3491–3501. [Google Scholar] [CrossRef] [PubMed]
- Konstantinovic, N.; Guegan, H.; Stäjner, T.; Belaz, S.; Robert-Gangneux, F. Treatment of Toxoplasmosis: Current Options and Future Perspectives. Food Waterborne Parasitol. 2019, 15, e00036. [Google Scholar] [PubMed]
- Montazeri, M.; Mehrzadi, S.; Sharif, M.; Sarvi, S.; Shahdin, S.; Daryani, A. Activities of Anti-Toxoplasma Drugs and Compounds against Tissue Cysts in the Last Three Decades (1987 to 2017), a Systematic Review. Parasitol. Res. 2018, 117, 3045–3057. [Google Scholar] [PubMed]
- Murata, Y.; Sugi, T.; Weiss, L.M.; Kato, K. Identification of Compounds That Suppress Toxoplasma Gondii Tachyzoites & Bradyzoites. PLoS ONE 2017, 12, e0178203. [Google Scholar] [CrossRef]
- Heloisa, B.; Dinorah, G. The Wide Pharmacological Versatility of Semicarbazones, Thiosemicarbazones and Their Metal Complexes. Mini-Rev. Med. Chem. 2004, 4, 31–39. [Google Scholar]
- Khan, T.; Raza, S.; Lawrence, A.J. Medicinal Utility of Thiosemicarbazones with Special Reference to Mixed Ligand and Mixed Metal Complexes: A Review. Russ. J. Coord. Chem./Koord. Khimiya 2022, 48, 877–895. [Google Scholar] [CrossRef]
- Pelosi, G. Thiosemicarbazone Metal Complexes: From Structure to Activity. Open Crystallogr. J. 2010, 3, 16–28. [Google Scholar]
- Scaccaglia, M.; Rega, M.; Vescovi, M.; Pinelli, S.; Tegoni, M.; Bacci, C.; Pelosi, G.; Bisceglie, F. Gallium(III)-Pyridoxal Thiosemicarbazone Derivatives as Nontoxic Agents against Gram-Negative Bacteria. Metallomics 2022, 14, mfac070. [Google Scholar] [CrossRef] [PubMed]
- Krchniakova, M.; Paukovcekova, S.; Chlapek, P.; Neradil, J.; Skoda, J.; Veselska, R. Thiosemicarbazones and Selected Tyrosine Kinase Inhibitors Synergize in Pediatric Solid Tumors: NDRG1 Upregulation and Impaired Prosurvival Signaling in Neuroblastoma Cells. Front. Pharmacol. 2022, 13, 976955. [Google Scholar] [CrossRef]
- Kolesar, J.M.; Schelman, W.R.; Geiger, P.G.; Holen, K.D.; Traynor, A.M.; Alberti, D.B.; Thomas, J.P.; Chitambar, C.R.; Wilding, G.; Antholine, W.E. Electron Paramagnetic Resonance Study of Peripheral Blood Mononuclear Cells from Patients with Refractory Solid Tumors Treated with Triapine®. J. Inorg. Biochem. 2008, 102, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.; Montazeri, M.; Daryani, A.; Farshadfar, K.; Emami, S. Synthesis and in Vitro Anti-Toxoplasma Gondii Activity of a New Series of Aryloxyacetophenone Thiosemicarbazones. Mol. Divers. 2020, 24, 1223–1234. [Google Scholar] [CrossRef] [PubMed]
- Bekier, A.; Węglińska, L.; Paneth, A.; Paneth, P.; Dzitko, K. 4-Arylthiosemicarbazide Derivatives as a New Class of Tyrosinase Inhibitors and Anti-Toxoplasma Gondii Agents. J. Enzym. Inhib. Med. Chem. 2021, 36, 1145–1164. [Google Scholar] [CrossRef]
- De Aquino, T.M.; Liesen, A.P.; da Silva, R.E.A.; Lima, V.T.; Carvalho, C.S.; de Faria, A.R.; de Araújo, J.M.; de Lima, J.G.; Alves, A.J.; de Melo, E.J.T.; et al. Synthesis, Anti-Toxoplasma Gondii and Antimicrobial Activities of Benzaldehyde 4-Phenyl-3-Thiosemicarbazones and 2-[(Phenylmethylene)Hydrazono]-4-Oxo-3-Phenyl-5-Thiazolidineacetic Acids. Bioorg. Med. Chem. 2008, 16, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Dzitko, K.; Paneth, A.; Plech, T.; Pawełczyk, J.; Stączek, P.; Stefańska, J.; Paneth, P. 1,4-Disubstituted Thiosemicarbazide Derivatives Are Potent Inhibitors of Toxoplasma Gondii Proliferation. Molecules 2014, 19, 9926–9943. [Google Scholar] [CrossRef]
- Gomes, M.A.G.B.; Carvalho, L.P.; Rocha, B.S.; Oliveira, R.R.; De Melo, E.J.T.; Maria, E.J. Evaluating Anti-Toxoplasma Gondii Activity of New Serie of Phenylsemicarbazone and Phenylthiosemicarbazones in Vitro. Med. Chem. Res. 2013, 22, 3574–3580. [Google Scholar] [CrossRef]
- Paneth, A.; Weglinska, L.; Bekier, A.; Stefaniszyn, E.; Wujec, M.; Trotsko, N.; Dzitko, K. Systematic Identification of Thiosemicarbazides for Inhibition of Toxoplasma Gondii Growth In Vitro. Molecules 2019, 24, 614. [Google Scholar] [CrossRef]
- Tenório, R.P.; Carvalho, C.S.; Pessanha, C.S.; De Lima, J.G.; De Faria, A.R.; Alves, A.J.; De Melo, E.J.T.; Góes, A.J.S. Synthesis of Thiosemicarbazone and 4-Thiazolidinone Derivatives and Their in Vitro Anti-Toxoplasma Gondii Activity. Bioorg. Med. Chem. Lett. 2005, 15, 2575–2578. [Google Scholar] [CrossRef]
- Faa, G.; Gerosa, C.; Fanni, D.; Lachowicz, J.I.; Nurchi, V.M. Gold–Old Drug with New Potentials. Curr. Med. Chem. 2017, 25, 75–84. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, Y.; Xu, Z.; Ma, X.; Chen, X.; Liu, W. Repurposing of the Gold Drug Auranofin and a Review of Its Derivatives as Antibacterial Therapeutics. Drug Discov. Today 2022, 27, 1961–1973. [Google Scholar]
- Andrade, R.M.; Chaparro, J.D.; Capparelli, E.; Reed, S.L. Auranofin Is Highly Efficacious against Toxoplasma Gondii In Vitro and in an In Vivo Experimental Model of Acute Toxoplasmosis. PLoS Negl. Trop. Dis. 2014, 8, e2973. [Google Scholar] [CrossRef]
- Almeida, C.M.; Pedro, P.H.; Nascimento, É.C.M.; Martins, J.B.L.; Chagas, M.A.S.; Fujimori, M.; De Marchi, P.G.F.; França, E.L.; Honorio-França, A.C.; Gatto, C.C. Organometallic Gold (III) and Platinum (II) Complexes with Thiosemicarbazone: Structural Behavior, Anticancer Activity, and Molecular Docking. Appl. Organomet. Chem. 2022, 36, e6761. [Google Scholar] [CrossRef]
- Dominelli, B.; Correia, J.D.G.; Kühn, F.E. Medicinal Applications of Gold(I/III)-Based Complexes Bearing N-Heterocyclic Carbene and Phosphine Ligands. J. Organomet. Chem. 2018, 866, 153–164. [Google Scholar] [CrossRef]
- Dou, Z.; Carruthers, V.B. Cathepsin Proteases in Toxoplasma Gondii. Adv. Exp. Med. Biol. 2011, 712, 49–61. [Google Scholar] [CrossRef]
- Xue, J.; Jiang, W.; Chen, Y.; Gong, F.; Wang, M.; Zeng, P.; Xia, C.; Wang, Q.; Huang, K. Thioredoxin Reductase from Toxoplasma Gondii: An Essential Virulence Effector with Antioxidant Function. FASEB J. 2017, 31, 4447–4457. [Google Scholar] [CrossRef] [PubMed]
- Radisavljević, S.; Petrović, B. Gold(III) Complexes: An Overview on Their Kinetics, Interactions With DNA/BSA, Cytotoxic Activity, and Computational Calculations. Front. Chem. 2020, 8, 379. [Google Scholar]
- Linciano, P.; Moraes, C.B.; Alcantara, L.M.; Franco, C.H.; Pascoalino, B.; Freitas-Junior, L.H.; Macedo, S.; Santarem, N.; Cordeiro-da-Silva, A.; Gul, S.; et al. Aryl Thiosemicarbazones for the Treatment of Trypanosomatidic Infections. Eur. J. Med. Chem. 2018, 146, 423–434. [Google Scholar] [CrossRef]
- Houngue, H.D.; Aguida, B.S.; Kassehin, U.C.; Poupaert, J.H.; Gbaguidi, F.A.; Codjo, H. Biological Evaluation of a Series of Thiosemicarbazones Targeting the Large Subunit Ribosomal Protein EL42 from Human 80S Ribosomes. MOJ Bioorganic Org. Chem. 2017, 1, 6–12. [Google Scholar] [CrossRef]
- Hofflin, J.M.; Remington, J.S. Clindamycin in a Murine Model of Toxoplasmic Encephalitis. Antimicrob. Agents Chemother. 1987, 31, 492–496. [Google Scholar] [PubMed]
- Chew, W.K.; Segarra, I.; Ambu, S.; Mak, J.W. Significant Reduction of Brain Cysts Caused by Toxoplasma Gondii after Treatment with Spiramycin Coadministered with Metronidazole in a Mouse Model of Chronic Toxoplasmosis. Antimicrob. Agents Chemother. 2012, 56, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Anghel, N.; Imhof, D.; Hänggeli, K.; Uldry, A.C.; Braga-Lagache, S.; Heller, M.; Ojo, K.K.; Ortega-Mora, L.M.; Van Voorhis, W.C.; et al. Common Molecular Targets of a Quinolone Based Bumped Kinase Inhibitor in Neospora Caninum and Danio Rerio. Int. J. Mol. Sci. 2022, 23, 2381. [Google Scholar] [CrossRef] [PubMed]
- Hänggeli, K.P.A.; Hemphill, A.; Müller, N.; Heller, M.; Uldry, A.C.; Braga-Lagache, S.; Müller, J.; Boubaker, G. Comparative Proteomic Analysis of Toxoplasma Gondii RH Wild-Type and Four SRS29B (SAG1) Knock-Out Clones Reveals Significant Differences between Individual Strains. Int. J. Mol. Sci. 2023, 24, 10454. [Google Scholar] [CrossRef]
- Gc, K.; To, D.; Jayalath, K.; Abeysirigunawardena, S. Discovery of a Novel Small Molecular Peptide That Disrupts Helix 34 of Bacterial Ribosomal RNA. RSC Adv. 2019, 9, 40268–40276. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Yang, J.; Fu, J.; Chen, H.; Jia, H. The Dissection of SNAREs Reveals Key Factors for Vesicular Trafficking to the Endosome-like Compartment and Apicoplast via the Secretory System in Toxoplasma Gondii. mBio 2021, 12, 01380-21. [Google Scholar] [CrossRef]
- Fu, J.; Zhao, L.; Yang, J.; Chen, H.; Cao, S.; Jia, H. An Unconventional SNARE Complex Mediates Exocytosis at the Plasma Membrane and Vesicular Fusion at the Apical Annuli in Toxoplasma Gondii. PLoS Pathog. 2023, 19, e1011288. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E.W. Adaptive and Mutational Resistance: Role of Porins and Efflux Pumps in Drug Resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Ayaz, M.; Subhan, F.; Sadiq, A.; Ullah, F.; Ahmed, J.; Sewell, R.D. Cellular Efflux Transporters and the Potential Role of Natural Products in Combating Efflux Mediated Drug Resistance. Front. Biosci. Landmark 2017, 22, 732–756. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, Y.; Yu, X.Y.; Pan, X.; Zhang, Y.; Tu, J.; Song, Y.H.; Li, Y. Ribosome Biogenesis in Disease: New Players and Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 15. [Google Scholar] [PubMed]
- Rudra, P.; Hurst-Hess, K.; Lappierre, P.; Ghosha, P. High Levels of Intrinsic Tetracycline Resistance in Mycobacterium Abscessus Are Conferred by a Tetracycline-Modifying Monooxygenase. Antimicrob. Agents Chemother. 2018, 62, 00119-18. [Google Scholar] [CrossRef]
- Müller, J.; Schlange, C.; Heller, M.; Uldry, A.C.; Braga-Lagache, S.; Haynes, R.K.; Hemphill, A. Proteomic Characterization of Toxoplasma Gondii ME49 Derived Strains Resistant to the Artemisinin Derivatives Artemiside and Artemisone Implies Potential Mode of Action Independent of ROS Formation. Int. J. Parasitol. Drugs Drug Resist. 2023, 21, 1–12. [Google Scholar] [CrossRef]
- Ramseier, J.; Imhof, D.; Anghel, N.; Hänggeli, K.; Beteck, R.M.; Balmer, V.; Ortega-mora, L.M.; Sanchez-sanchez, R.; Ferre, I.; Haynes, R.K.; et al. Assessment of the Activity of Decoquinate and Its Quinoline–O–carbamate Derivatives against Toxoplasma Gondii in Vitro and in Pregnant Mice Infected with t. Gondii Oocysts. Molecules 2021, 26, 6393. [Google Scholar] [CrossRef]
- Imhof, D.; Anghel, N.; Winzer, P.; Balmer, V.; Ramseier, J.; Hänggeli, K.; Choi, R.; Hulverson, M.A.; Whitman, G.R.; Arnold, S.L.M.; et al. In Vitro Activity, Safety and in Vivo Efficacy of the Novel Bumped Kinase Inhibitor BKI-1748 in Non-Pregnant and Pregnant Mice Experimentally Infected with Neospora Caninum Tachyzoites and Toxoplasma Gondii Oocysts. Int. J. Parasitol. Drugs Drug Resist. 2021, 16, 90–101. [Google Scholar] [CrossRef]
- Barna, F.; Debache, K.; Vock, C.A.; Küster, T.; Hemphill, A. In Vitro Effects of Novel Ruthenium Complexes in Neospora Caninum and Toxoplasma Gondii Tachyzoites. Antimicrob. Agents Chemother. 2013, 57, 5747–5754. [Google Scholar] [CrossRef]
- Kropf, C.; Debache, K.; Rampa, C.; Barna, F.; Schorer, M.; Stephens, C.E.; Ismail, M.A.; Boykin, D.W.; Hemphill, A. The Adaptive Potential of a Survival Artist: Characterization of the in Vitro Interactions of Toxoplasma Gondii Tachyzoites with Di-Cationic Compounds in Human Fibroblast Cell Cultures. Parasitology 2012, 139, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Anghel, N.; Balmer, V.; Müller, J.; Winzer, P.; Aguado-Martinez, A.; Roozbehani, M.; Pou, S.; Nilsen, A.; Riscoe, M.; Doggett, J.S.; et al. Endochin-like Quinolones Exhibit Promising Efficacy against Neospora Caninum in Vitro and in Experimentally Infected Pregnant Mice. Front. Vet. Sci. 2018, 5, 285. [Google Scholar] [CrossRef]
- Aspinall, T.V.; Joynson, D.H.M.; Guy, E.; Hyde, J.E.; Sims, P.F.G. The Molecular Basis of Sulfonamide Resistance in Toxoplasma Gondii and Implications for the Clinical Management of Toxoplasmosis. J. Infect. Dis. 2002, 185, 1637–1643. [Google Scholar]
- Doliwa, C.; Escotte-Binet, S.; Aubert, D.; Velard, F.; Schmid, A.; Geers, R.; Villena, I. Induction of Sulfadiazine Resistance in Vitro in Toxoplasma Gondii. Exp. Parasitol. 2013, 133, 131–136. [Google Scholar] [CrossRef]
- Leggett, H.C.; Benmayor, R.; Hodgson, D.J.; Buckling, A. Experimental Evolution of Adaptive Phenotypic Plasticity in a Parasite. Curr. Biol. 2013, 23, 139–142. [Google Scholar] [CrossRef]
- Dixon, S.E.; Stilger, K.L.; Elias, E.V.; Naguleswaran, A.; Sullivan, W.J. A Decade of Epigenetic Research in Toxoplasma Gondii. Mol. Biochem. Parasitol. 2010, 173, 1–9. [Google Scholar]
- Radke, J.B.; Worth, D.; Hong, D.; Huang, S.; Sullivan, W.J.; Wilson, E.H.; White, M.W. Transcriptional Repression by ApiAP2 Factors Is Central to Chronic Toxoplasmosis. PLoS Pathog. 2018, 14, e1007035. [Google Scholar] [CrossRef]
- Müller, J.; Braga, S.; Heller, M.; Müller, N. Resistance Formation to Nitro Drugs in Giardia Lamblia: No Common Markers Identified by Comparative Proteomics. Int. J. Parasitol. Drugs Drug Resist. 2019, 9, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Barbasz, A.; Oćwieja, M. Gold Nanoparticles and Ions—Friends or Foes? As They Are Seen by Human Cells U-937 and HL-60. J. Exp. Nanosci. 2016, 11, 564–580. [Google Scholar] [CrossRef]
- Scaccaglia, M.; Pinelli, S.; Manini, L.; Ghezzi, B.; Nicastro, M.; Heinrich, J.; Kulak, N.; Mozzoni, P.; Pelosi, G.; Bisceglie, F. Gold(III) Complexes with Thiosemicarbazone Ligands: Insights into Their Cytotoxic Effects on Lung Cancer Cells. J. Inorg. Biochem. 2024, 251, 112438. [Google Scholar] [CrossRef]
- Theisen, T.C.; Boothroyd, J.C. Transcriptional Signatures of Clonally Derived Toxoplasma Tachyzoites Reveal Novel Insights into the Expression of a Family of Surface Proteins. PLoS ONE 2022, 17, e0262374. [Google Scholar] [CrossRef]
- Winzer, P.; Müller, J.; Aguado-Martínez, A.; Rahman, M.; Balmer, V.; Manser, V.; Ortega-Mora, L.M.; Ojo, K.K.; Fan, E.; Maly, D.J.; et al. In Vitro and in Vivo Effects of the Bumped Kinase Inhibitor 1294 in the Related Cyst-Forming Apicomplexans Toxoplasma Gondii and Neospora Caninum. Antimicrob. Agents Chemother. 2015, 59, 6361–6374. [Google Scholar] [CrossRef] [PubMed]
- Păunescu, E.; Boubaker, G.; Desiatkina, O.; Anghel, N.; Amdouni, Y.; Hemphill, A.; Furrer, J. The Quest of the Best—A SAR Study of Trithiolato-Bridged Dinuclear Ruthenium(II)-Arene Compounds Presenting Antiparasitic Properties. Eur. J. Med. Chem. 2021, 222, 113610. [Google Scholar] [CrossRef]
- Anghel, N.; Müller, J.; Serricchio, M.; Jelk, J.; Bütikofer, P.; Boubaker, G.; Imhof, D.; Ramseier, J.; Desiatkina, O.; Păunescu, E.; et al. Cellular and Molecular Targets of Nucleotide-Tagged Trithiolato-Bridged Arene Ruthenium Complexes in the Protozoan Parasites Toxoplasma Gondii and Trypanosoma Brucei. Int. J. Mol. Sci. 2021, 22, 10787. [Google Scholar] [CrossRef]
- Müller, J.; Boubaker, G.; Imhof, D.; Hänggeli, K.; Haudenschild, N.; Uldry, A.C.; Braga-Lagache, S.; Heller, M.; Ortega-Mora, L.M.; Hemphill, A. Differential Affinity Chromatography Coupled to Mass Spectrometry: A Suitable Tool to Identify Common Binding Proteins of a Broad-Range Antimicrobial Peptide Derived from Leucinostatin. Biomedicines 2022, 10, 2675. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Haynes, S.E.; Teo, G.C.; Avtonomov, D.M.; Polasky, D.A.; Nesvizhskii, A.I. Fast Quantitative Analysis of TimsTOF PASEF Data with MSFragger and IonQuant. Mol. Cell. Proteom. 2020, 19, 1575–1585. [Google Scholar] [CrossRef]
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Silva, J.C.; Gorenstein, M.V.; Li, G.Z.; Vissers, J.P.C.; Geromanos, S.J. Absolute Quantification of Proteins by LCMSE: A Virtue of Parallel MS Acquisition. Mol. Cell. Proteom. 2006, 5, 144–156. [Google Scholar] [CrossRef]
- Huber, W.; Von Heydebreck, A.; Sültmann, H.; Poustka, A.; Vingron, M. Variance Stabilization Applied to Microarray Data Calibration and to the Quantification of Differential Expression. Bioinformatics 2002, 18, S96–S104. [Google Scholar] [PubMed]
- Schwanhüusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global Quantification of Mammalian Gene Expression Control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef]
- Braga-Lagache, S.; Buchs, N.; Iacovache, M.I.; Zuber, B.; Jackson, C.B.; Heller, M. Robust Label-Free, Quantitative Profiling of Circulating Plasma Microparticle (MP) Associated Proteins. Mol. Cell. Proteom. 2016, 15, 3640–3652. [Google Scholar] [CrossRef]
- Alvarez-Jarreta, J.; Amos, B.; Aurrecoechea, C.; Bah, S.; Barba, M.; Barreto, A.; Basenko, E.Y.; Belnap, R.; Blevins, A.; Böhme, U.; et al. VEuPathDB: The Eukaryotic Pathogen, Vectorãnd Host Bioinf Ormatics Resour Ce Cent Er in 2023. Nucleic Acids Res. 2024, 52, D808–D816. [Google Scholar] [CrossRef] [PubMed]
- Silver, J.D.; Ritchie, M.E.; Smyth, G.K. Microarray Background Correction: Maximum Likelihood Estimation for the Normal-Exponential Convolution. Biostatistics 2009, 10, 352–363. [Google Scholar] [CrossRef]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting Significant Changes in Protein Abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef]
- Strimmer, K. A Unified Approach to False Discovery Rate Estimation. BMC Bioinform. 2008, 9, 1–14. [Google Scholar] [CrossRef]
- Uldry, A.C.; Maciel-Dominguez, A.; Jornod, M.; Buchs, N.; Braga-Lagache, S.; Brodard, J.; Jankovic, J.; Bonadies, N.; Heller, M. Effect of Sample Transportation on the Proteome of Human Circulating Blood Extracellular Vesicles. Int. J. Mol. Sci. 2022, 23, 4515. [Google Scholar] [CrossRef] [PubMed]
| T. gondii β-Gal | HFF | ||
|---|---|---|---|
| EC50 When Added Concomitantly to Infection (nM) | EC50 When Added 3 h Post-Infection (nM) | No Viability Impairment at (µM) | |
| Compound | |||
| C3 | 28 (27–30) | 23 (20–26) | 25 |
| C4 | 30 (40–20) | 20 (15–25) | 25 |
| T. gondii ME49 | |||
| C3 | 31 (16–62) | ||
| C4 | 13 (6–27) | ||
| ToxoDB ORF | Annotation | rAbu C3 |
|---|---|---|
| TGME49_262690 | ribosomal protein RPL27 | 741 |
| TGME49_244110 | nucleosome assembly protein (nap) protein | 324 |
| TGME49_309740 | LSM domain-containing protein | 100 |
| TGME49_321520 | hypothetical protein | 91 |
| TGME49_279390 | proliferation-associated protein 2G4, putative | 80 |
| TGME49_319870 | ubiquitin-conjugating enzyme subfamily protein | 77 |
| TGME49_313560 | 60S ribosomal protein L7a, putative | 71 |
| TGME49_248810 | nuclear factor NF7 | 54 |
| TGME49_231850 | serine-threonine phosophatase 2C (PP2C) | 51 |
| TGME49_209170 | hypothetical protein | 43 |
| TGME49_272400 | casein kinase ii regulatory subunit protein | 43 |
| TGME49_227810 | rhoptry kinase family protein ROP11 (incomplete catalytic triad) | 35 |
| TGME49_256050 | signal recognition particle 14kd protein | 28 |
| TGME49_305340 | corepressor complex CRC230 | 9 |
| ToxoDB ORF | Annotation | rAbu C4 |
|---|---|---|
| TGME49_213280 | SAG-related sequence SRS25 | 22,200 |
| TGME49_253690 | hypothetical protein (putative transmembrane protein) | 15,804 |
| TGME49_316710 | hypothetical protein | 4878 |
| TGME49_266060 | ribosomal protein RPSA | 3977 |
| TGME49_288245 | hypothetical protein | 2980 |
| TGME49_231160 | hypothetical protein | 2724 |
| TGME49_226570 | hypothetical protein | 1943 |
| TGME49_285870 | SAG-related sequence SRS20A | 1652 |
| TGME49_319560 | microneme protein MIC3 | 1600 |
| TGME49_289680 | Ras-related protein Rab11 | 1461 |
| TGME49_263630 | hypothetical protein | 1426 |
| TGME49_214410 | hypothetical protein | 1316 |
| TGME49_214770 | small GTP binding protein rab1a, putative | 1305 |
| TGME49_225555 | hypothetical protein | 1284 |
| TGME49_308020 | SAG-related sequence SRS57 | 1218 |
| TGME49_249900 | adenine nucleotide translocator, putative | 1171 |
| TGME49_233450 | SAG-related sequence SRS29A | 1156 |
| TGME49_308840 | SAG-related sequence SRS51 | 1153 |
| TGME49_245490 | microneme protein MIC8 | 1126 |
| TGME49_227920 | hypothetical protein | 1043 |
| ToxoDB ORF | Annotation | rAbu C3 | rAbu C4 |
|---|---|---|---|
| TGME49_309810 | ribosomal protein RPP2 | 3431 | 3473 |
| TGME49_249250 | ribosomal protein RPL35A | 3406 | 214 |
| TGME49_262670 | ribosomal protein RPL18A | 2651 | 522 |
| TGME49_313390 | ribosomal protein RPL6 | 2390 | 482 |
| TGME49_261570 | ribosomal protein RPL7A | 2348 | 372 |
| TGME49_205340 | ribosomal protein RPS12 | 2026 | 493 |
| TGME49_292130 | ribosomal protein RPL13A | 1678 | 447 |
| TGME49_238070 | glutaredoxin domain-containing protein | 1666 | 332 |
| TGME49_267060 | ribosomal protein RPL14 | 1540 | 317 |
| TGME49_320050 | ribosomal protein RPL5 | 1492 | 223 |
| TGME49_204020 | ribosomal protein RPL8 | 1487 | 114 |
| TGME49_238250 | ribosomal protein RPL36 | 1422 | 517 |
| TGME49_245680 | ribosomal protein RPL21 | 1362 | 219 |
| TGME49_260260 | ribosomal protein RPP1 | 1354 | 2505 |
| TGME49_314810 | ribosomal protein RPL7 | 1325 | 666 |
| TGME49_270380 | ribosomal protein RPS13 | 1265 | 1846 |
| TGME49_210690 | ribosomal protein RPS6 | 1242 | 759 |
| TGME49_215470 | ribosomal protein RPL10A | 1090 | 400 |
| TGME49_218820 | alba 2 | 1046 | 753 |
| TGME49_232230 | ribosomal protein RPL30 | 986 | 50 |
| EC50 (nM) | MIC (µM) | |
|---|---|---|
| C3 | ||
| T. gondii Me49 wildtype (WT) | 31 (16–62) | 25 |
| P1F3 | 45 (28–70) | 25 |
| P1E5 | 90 (54–150) | 25 |
| P2F5 | 51 (31–84) | 25 |
| C4 | ||
| T. gondii Me49 wildtype (WT) | 13 (6–27) | 25 |
| P2F3 | 11 (6–18) | 25 |
| P1G10 | 10 (4–23) | 25 |
| P3E2 | 6 (4–9) | 25 |
| ToxoDB ORF | Product Description | C3_Clone | Fold Change (FC) |
|---|---|---|---|
| Upregulated ↑ | |||
| TGME49_258700 | transporter, major facilitator family protein | P1F3 | 6.5 |
| TGME49_225480 | hypothetical protein | P1E5 | 6.4 |
| TGME49_248140 | hypothetical protein | P1E5 | 5.6 |
| TGME49_262120 | IQ calmodulin-binding motif domain-containing protein | P1E5 | 3.3 |
| TGME49_253360 | SNARE protein STX20 | P1E5 | 3.2 |
| TGME49_306300 | hypothetical protein | P1E5 | 3.2 |
| TGME49_257160 | hypothetical protein | P1E5 | 3.1 |
| TGME49_261740 | rhoptry protein ROP47 | P1E5 | 2.8 |
| TGME49_293480 | MoeA N-terminal region (domain I and II) domain-containing protein | P1F3 | 2.6 |
| TGME49_250950 | KRUF family protein | P1E5 | 2.5 |
| TGME49_203630 | ribosomal protein RPL44 | P1E5 | 2.4 |
| TGME49_315290 | hypothetical protein | P1E5 | 2.4 |
| TGME49_214410 | hypothetical protein | P1E5 | 2.3 |
| TGME49_268730 | glutaredoxin 1 | P1E5 | 2.3 |
| TGME49_268740 | hypothetical protein | P1E5 | 2.2 |
| TGME49_234180 | hypothetical protein | P1E5 | 2.1 |
| TGME49_238200 | alpha/beta hydrolase fold domain-containing protein | P2F5/P1E5 | 2/1.6 |
| TGME49_219690 | hypothetical protein | P1E5 | 2.0 |
| TGME49_202940 | hypothetical protein | P1E5 | 2.0 |
| TGME49_251180 | KRUF family protein | P1E5/P1F3/P2F5 | 2/1.7/1.5 |
| Downregulated ↓ | |||
| TGME49_248200 | ribosomal RNA (adenine(1779)-N(6)/adenine(1780)-N(6))-dimethyltransferase, putative | P1E5 | 0.1 |
| TGME49_209755 | cyst matrix protein MAG2 | P1E5 | 0.1 |
| TGME49_253350 | hypothetical protein | P1E5 | 0.3 |
| TGME49_297110 | kinesin motor domain-containing protein | P1E5 | 0.3 |
| TGME49_273780 | SWI2/SNF2-containing protein | P1E5 | 0.4 |
| TGME49_280570 | SAG-related sequence SRS35A | P1E5 | 0.4 |
| TGME49_272900 | DNA repair protein RAD51 | P1E5 | 0.4 |
| TGME49_281900 | SET domain containing lysine methyltransferase KMTox | P1E5 | 0.4 |
| TGME49_247390 | ATPase, AAA family protein | P1E5 | 0.4 |
| TGME49_280390 | HEAT repeat-containing protein | P1E5 | 0.5 |
| TGME49_320190 | SAG-related sequence SRS16B | P1E5 | 0.5 |
| TGME49_318470 | AP2 domain transcription factor AP2IV-4 | P1E5 | 0.5 |
| TGME49_297870 | inner membrane complex protein IMC36 | P1E5 | 0.5 |
| TGME49_214970 | DNA replication licensing factor, putative | P1E5 | 0.5 |
| TGME49_301170 | SAG-related sequence SRS19D | P1E5 | 0.5 |
| TGME49_244500 | Tubulin-tyrosine ligase family protein | P1E5 | 0.5 |
| TGME49_290970 | serine palmitoyltransferase SPT2 | P1E5 | 0.5 |
| TGME49_219700 | DNA replication licensing factor MCM4, putative | P1E5 | 0.5 |
| TGME49_237220 | DNA replication licensing factor Mcm7, putative | P1E5 | 0.5 |
| TGME49_262990 | hypothetical protein | P1E5 | 0.5 |
| ToxoDB ORF | Product Description | C4_Clone | Fold Change (FC) |
|---|---|---|---|
| Upregulated ↑ | |||
| TGME49_306300 | hypothetical protein | P1G10 | 7.2 |
| TGME49_258700 | transporter, major facilitator family protein | P2F3 | 4.2 |
| TGME49_248140 | hypothetical protein (putative transmembrane protein) | P3E2 | 4.1 |
| TGME49_234980 | hypothetical protein (putative transmembrane protein) | P2F3 | 3.8 |
| TGME49_277740 | zinc finger, C3HC4 type (RING finger) domain-containing protein | P2F3 | 2.8 |
| TGME49_268860 | enolase 1 | P2F3 | 2.8 |
| TGME49_265190 | Ulp1 protease family, C-terminal catalytic domain-containing protein | P2F3 | 2.7 |
| TGME49_276120 | histone lysine methyltransferase, SET, putative | P2F3 | 2.3 |
| TGME49_220170 | hypothetical protein | P2F3 | 2.2 |
| TGME49_200440 | hypothetical protein | P2F3 | 2.2 |
| TGME49_273060 | ribosomal protein S17, putative | P2F3 | 2.1 |
| TGME49_273550 | hypothetical protein | P2F3 | 2.1 |
| TGME49_293480 | MoeA N-terminal region (domain I and II) domain-containing protein | P3E2 | 2 |
| TGME49_238200 | alpha/beta hydrolase fold domain-containing protein | P2F3/P1G10 | 2.01/1.9 |
| TGME49_284580 | ribose-phosphate diphosphokinase subfamily protein | P2F3 | 2 |
| TGME49_211470 | Fcf2 pre-rRNA processing protein | P2F3 | 2 |
| TGME49_229260 | basal complex component BCC4 | P2F3 | 2 |
| TGME49_226640 | zinc binding protein, putative | P2F3 | 2 |
| TGME49_224932 | hypothetical protein | P2F3 | 1.9 |
| TGME49_297220 | AMP-binding enzyme domain-containing protein | P2F3 | 1.9 |
| Downregulated ↓ | |||
| TGME49_309580 | transporter, major facilitator family protein | P2F3 | 0.1 |
| TGME49_268220 | hypothetical protein | P2F3 | 0.2 |
| TGME49_305870 | DAD family protein | P1G10 | 0.4 |
| TGME49_305610 | hypothetical protein | P2F3 | 0.5 |
| TGME49_229140 | MaoC family domain-containing protein | P1G10 | 0.6 |
| TGME49_320588 | glycosyl hydrolases family 35 protein | P2F3 | 0.6 |
| Annotation | C3_Clone | FC | C4_Clone | FC | |
|---|---|---|---|---|---|
| Upregulated C3↑ C4 ↑ | |||||
| TGME49_306300 | hypothetical protein | P1E5 | 3.2 | P1G10 | 7.2 |
| TGME49_258700 | transporter, major facilitator family protein | P1F3 | 6.5 | P2F3 | 4.2 |
| TGME49_248140 | hypothetical protein (putative transmembrane protein) | P1E5 | 5.6 | P3E2 | 4.1 |
| TGME49_293480 | MoeA N-terminal region (domain I and II) domain-containing protein | P1F3 | 2.6 | P3E2 | 2.0 |
| TGME49_238200 | alpha/beta hydrolase fold domain-containing protein | P1E5/P2F5 | 1.62/2.03 | P2F3/P1G10 | 2.0/1.9 |
| TGME49_211470 | Fcf2 pre-rRNA processing protein | P2F5 | 1.8 | P2F3 | 2.0 |
| TGME49_297220 | AMP-binding enzyme domain-containing protein | P1E5 | 2.0 | P2F3 | 1.9 |
| TGME49_277920 | hypothetical protein (putative transmembrane protein) | P1E5 | 2.0 | P2F3 | 1.5 |
| TGME49_224932 | hypothetical protein (putative jmjC domain protein) | P1E5 | 1.6 | P2F3 | 1.9 |
| TGME49_269660 | TFIIH basal transcription factor complex helicase XPB subunit | P1E5 | 1.6 | P2F3 | 1.5 |
| TGME49_274100 | hypothetical protein | P2F5 | 1.5 | P2F3 | 1.6 |
| Downregulated C3↓ C4 ↓ | |||||
| TGME49_229140 | MaoC family domain-containing protein | P1F3 | 0.5 | P1G10 | 0.6 |
| Divergent C3↓ C4 ↑ | |||||
| TGME49_312500 | hypothetical protein | P1E5 | 0.6 | P2F3 | 1.5 |
| TGME49_315590 | macro domain-containing protein | P1E5 | 0.6 | P2F3 | 1.9 |
| TGME49_242720 | aspartyl protease ASP5 | P1E5 | 0.7 | P2F3 | 1.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semeraro, M.; Boubaker, G.; Scaccaglia, M.; Müller, J.; Vigneswaran, A.; Hänggeli, K.P.A.; Amdouni, Y.; Kramer, L.H.; Vismarra, A.; Genchi, M.; et al. Transient Adaptation of Toxoplasma gondii to Exposure by Thiosemicarbazone Drugs That Target Ribosomal Proteins Is Associated with the Upregulated Expression of Tachyzoite Transmembrane Proteins and Transporters. Int. J. Mol. Sci. 2024, 25, 9067. https://doi.org/10.3390/ijms25169067
Semeraro M, Boubaker G, Scaccaglia M, Müller J, Vigneswaran A, Hänggeli KPA, Amdouni Y, Kramer LH, Vismarra A, Genchi M, et al. Transient Adaptation of Toxoplasma gondii to Exposure by Thiosemicarbazone Drugs That Target Ribosomal Proteins Is Associated with the Upregulated Expression of Tachyzoite Transmembrane Proteins and Transporters. International Journal of Molecular Sciences. 2024; 25(16):9067. https://doi.org/10.3390/ijms25169067
Chicago/Turabian StyleSemeraro, Manuela, Ghalia Boubaker, Mirco Scaccaglia, Joachim Müller, Anitha Vigneswaran, Kai Pascal Alexander Hänggeli, Yosra Amdouni, Laura Helen Kramer, Alice Vismarra, Marco Genchi, and et al. 2024. "Transient Adaptation of Toxoplasma gondii to Exposure by Thiosemicarbazone Drugs That Target Ribosomal Proteins Is Associated with the Upregulated Expression of Tachyzoite Transmembrane Proteins and Transporters" International Journal of Molecular Sciences 25, no. 16: 9067. https://doi.org/10.3390/ijms25169067







