The Impact of the Major Endoribonucleases RNase E and RNase III and of the sRNA StsR on Photosynthesis Gene Expression in Rhodobacter sphaeroides Is Growth-Phase-Dependent
Abstract
:1. Introduction
2. Results and Discussion
2.1. Effect of the Endoribonucleases RNase E and RNase III and of the sRNA StsR on Growth and Levels of Photosynthetic Complexes in Exponential and Stationary Phase
2.2. RNA-seq Reveals Growth-Phase-Dependent Effects of RNase E, RNase III, and StsR on the Transcriptome
2.3. Growth-Phase-Dependent Effects of RNase E, RNase III, and StsR on Expression of Photosynthesis-Related Genes
2.4. Effect of Growth Conditions and Growth Phases on Other Functional Groups of Genes
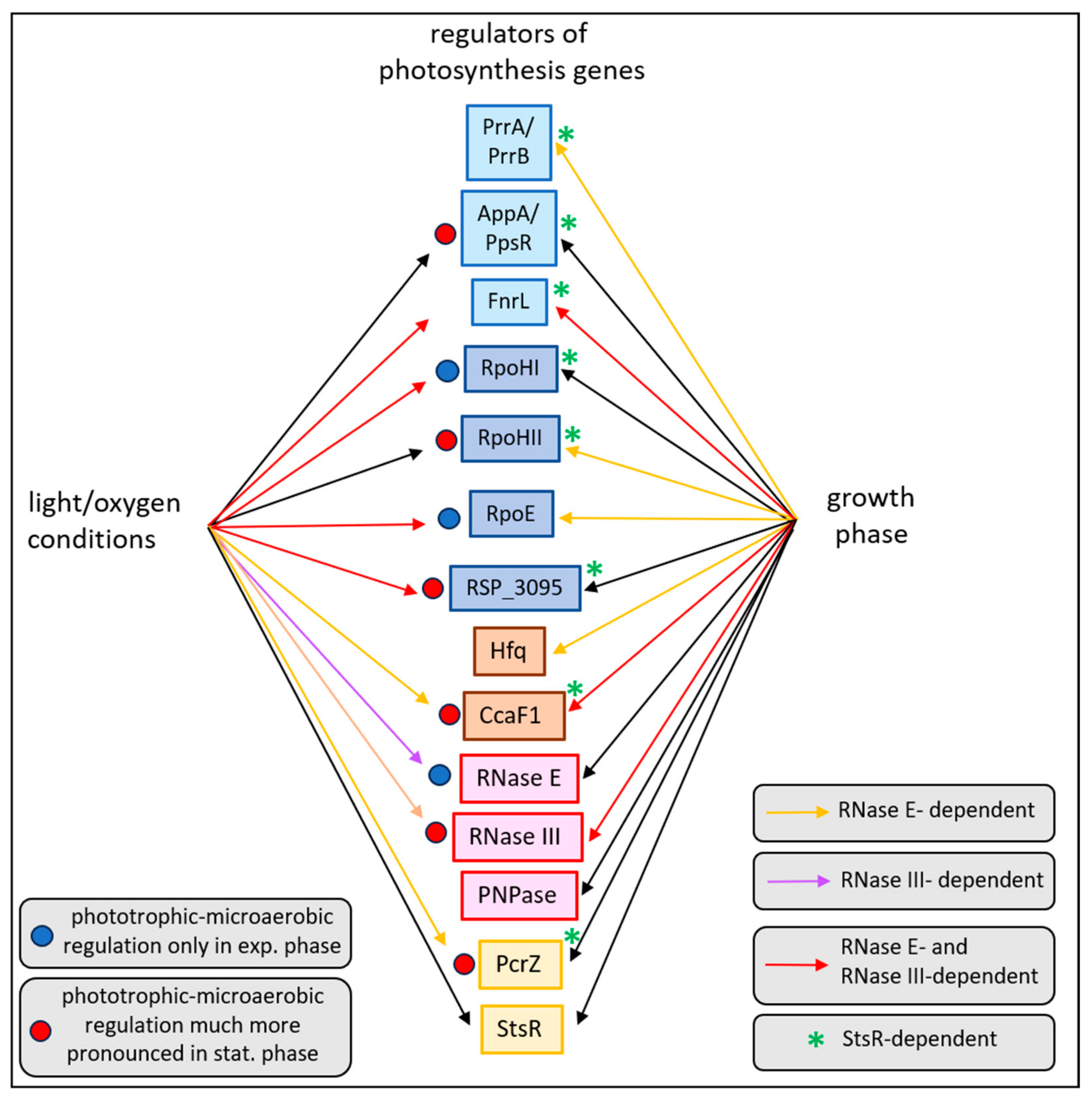
2.5. Influence of RNase E, RNase III, and StsR on the Growth-Phase-Dependent Expression of Selected Photosynthesis Genes and Their Regulators
3. Materials and Methods
3.1. Growth of Bacterial Cultures
3.2. Quantification of Photopigments and Photosynthetic Complexes
3.3. RNA Isolation and Quantificaiton
3.4. Bioinformatic Data Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roop, R.M.; Gee, J.M.; Robertson, G.T.; Richardson, J.M.; Ng, W.-L.; Winkler, M.E. Brucella stationary-phase gene expression and virulence. Annu. Rev. Microbiol. 2003, 57, 57–76. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, M.; Eisenhardt, K.; Remes, B.; Konzer, A.; Klug, G. Adaptation of the alphaproteobacterium Rhodobacter sphaeroides to stationary phase. Environ. Microbiol. 2019, 21, 4425–4445. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, R.L. Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J. Bacteriol. 2019, 201, 10–128. [Google Scholar] [CrossRef]
- Jaishankar, J.; Srivastava, P. Molecular basis of stationary phase survival and applications. Front. Microbiol. 2017, 8, 2000. [Google Scholar] [CrossRef] [PubMed]
- Bakkeren, E.; Diard, M.; Hardt, W.-D. Evolutionary causes and consequences of bacterial antibiotic persistence. Nat. Rev. Microbiol. 2020, 18, 479–490. [Google Scholar] [CrossRef]
- Helaine, S.; Cheverton, A.M.; Watson, K.G.; Faure, L.M.; Matthews, S.A.; Holden, D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science 2014, 343, 204–208. [Google Scholar] [CrossRef]
- McKay, S.L.; Portnoy, D.A. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob. Agents Chemother. 2015, 59, 6992–6999. [Google Scholar] [CrossRef]
- Jõers, A.; Tenson, T. Growth resumption from stationary phase reveals memory in Escherichia coli cultures. Sci. Rep. 2016, 6, 24055. [Google Scholar] [CrossRef]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.-M.; Tindall, B.J.; Gronow, S.; Kyrpides, N.C.; Woyke, T.; Göker, M. Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef]
- Remes, B.; Rische-Grahl, T.; Müller, K.M.H.; Förstner, K.U.; Yu, S.-H.; Weber, L.; Jäger, A.; Peuser, V.; Klug, G. An RpoHI-dependent response promotes outgrowth after extended stationary phase in the alphaproteobacterium Rhodobacter sphaeroides. J. Bacteriol. 2017, 199, 10–1128. [Google Scholar] [CrossRef]
- Nuss, A.M.; Glaeser, J.; Klug, G. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J. Bacteriol. 2009, 191, 220–230. [Google Scholar] [CrossRef]
- Berghoff, B.A.; Konzer, A.; Mank, N.N.; Looso, M.; Rische, T.; Förstner, K.U.; Krüger, M.; Klug, G. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLoS Genet. 2013, 9, e1003576. [Google Scholar] [CrossRef]
- Dufour, Y.S.; Imam, S.; Koo, B.-M.; Green, H.A.; Donohue, T.J. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet. 2012, 8, e1002929. [Google Scholar] [CrossRef]
- Donohue, T.J. Shedding light on a Group IV (ECF11) alternative σ factor. Mol. Microbiol. 2019, 112, 374–384. [Google Scholar] [CrossRef]
- Bathke, J.; Konzer, A.; Remes, B.; McIntosh, M.; Klug, G. Comparative analyses of the variation of the transcriptome and proteome of Rhodobacter sphaeroides throughout growth. BMC Genom. 2019, 20, 358. [Google Scholar] [CrossRef] [PubMed]
- Grützner, J.; Remes, B.; Eisenhardt, K.M.H.; Scheller, D.; Kretz, J.; Madhugiri, R.; McIntosh, M.; Klug, G. sRNA-mediated RNA processing regulates bacterial cell division. Nucleic Acids Res. 2021, 49, 7035–7052. [Google Scholar] [CrossRef] [PubMed]
- Weber, L.; Thoelken, C.; Volk, M.; Remes, B.; Lechner, M.; Klug, G. The conserved dcw gene cluster of R. sphaeroides is preceded by an uncommonly extended 5′ leader featuring the sRNA UpsM. PLoS ONE 2016, 11, e0165694. [Google Scholar] [CrossRef]
- Zeilstra-Ryalls, J.H.; Kaplan, S. Oxygen intervention in the regulation of gene expression: The photosynthetic bacterial paradigm. Cell. Mol. Life Sci. 2004, 61, 417–436. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Bauer, C.E. Controlling the delicate balance of tetrapyrrole biosynthesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120262. [Google Scholar] [CrossRef]
- Klug, G.; Adams, C.W.; Belasco, J.; Doerge, B.; Cohen, S.N. Biological consequences of segmental alterations in mRNA stability: Effects of deletion of the intercistronic hairpin loop region of the Rhodobacter capsulatus puf operon. EMBO J. 1987, 6, 3515–3520. [Google Scholar] [CrossRef]
- Mank, N.N.; Berghoff, B.A.; Hermanns, Y.N.; Klug, G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc. Natl. Acad. Sci. USA 2012, 109, 16306–16311. [Google Scholar] [CrossRef] [PubMed]
- Eisenhardt, K.M.H.; Reuscher, C.M.; Klug, G. PcrX, an sRNA derived from the 3′- UTR of the Rhodobacter sphaeroides puf operon modulates expression of puf genes encoding proteins of the bacterial photosynthetic apparatus. Mol. Microbiol. 2018, 110, 325–334. [Google Scholar] [CrossRef]
- Reuscher, C.M.; Klug, G. Antisense RNA asPcrL regulates expression of photosynthesis genes in Rhodobacter sphaeroides by promoting RNase III-dependent turn-over of puf mRNA. RNA Biol. 2021, 18, 1445–1457. [Google Scholar] [CrossRef]
- Eisenhardt, K.M.H.; Remes, B.; Grützner, J.; Spanka, D.-T.; Jäger, A.; Klug, G. A complex network of sigma factors and sRNA StsR regulates stress responses in R. sphaeroides. Int. J. Mol. Sci. 2021, 22, 7557. [Google Scholar] [CrossRef] [PubMed]
- Berghoff, B.A.; Glaeser, J.; Sharma, C.M.; Zobawa, M.; Lottspeich, F.; Vogel, J.; Klug, G. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol. Microbiol. 2011, 80, 1479–1495. [Google Scholar] [CrossRef] [PubMed]
- Grützner, J.; Börner, J.; Jäger, A.; Klug, G. The small RNA-binding protein CcaF1 promotes formation of photosynthetic complexes in Rhodobacter sphaeroides. Int. J. Mol. Sci. 2023, 24, 9515. [Google Scholar] [CrossRef]
- Förstner, K.U.; Reuscher, C.M.; Haberzettl, K.; Weber, L.; Klug, G. RNase E cleavage shapes the transcriptome of Rhodobacter sphaeroides and strongly impacts phototrophic growth. Life Sci. Alliance 2018, 1, e201800080. [Google Scholar] [CrossRef]
- Börner, J.; Friedrich, T.; Bartkuhn, M.; Klug, G. Ribonuclease E strongly impacts bacterial adaptation to different growth conditions. RNA Biol. 2023, 20, 120–135. [Google Scholar] [CrossRef]
- Klug, G.; Kaufmann, N.; Drews, G. Gene expression of pigment-binding proteins of the bacterial photosynthetic apparatus: Transcription and assembly in the membrane of Rhodopseudomonas capsulata. Proc. Natl. Acad. Sci. USA 1985, 82, 6485–6489. [Google Scholar] [CrossRef]
- Börner, J.; Friedrich, T.; Klug, G. RNase III participates in control of quorum sensing, pigmentation and oxidative stress resistance in Rhodobacter sphaeroides. Mol. Microbiol. 2023, 120, 874–892. [Google Scholar] [CrossRef]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A web-based tool for Gene Ontology searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.W.; Forrest, M.E.; Cohen, S.N.; Beatty, J.T. Structural and functional analysis of transcriptional control of the Rhodobacter capsulatus puf operon. J. Bacteriol. 1989, 171, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Wellington, C.L.; Beatty, J.T. Overlapping mRNA transcripts of photosynthesis gene operons in Rhodobacter capsulatus. J. Bacteriol. 1991, 173, 1432–1443. [Google Scholar] [CrossRef]
- Bauer, C.E.; Buggy, J.J.; Yang, Z.M.; Marrs, B.L. The superoperonal organization of genes for pigment biosynthesis and reaction center proteins is a conserved feature in Rhodobacter capsulatus: Analysis of overlapping bchB and puhA transcripts. Mol. Gen. Genet. 1991, 228, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Sikora, F.; Budja, L.V.P.; Milojevic, O.; Ziemniewicz, A.; Dudys, P.; Görke, B. Multiple regulatory inputs including cell envelope stress orchestrate expression of the Escherichia coli rpoN operon. Mol. Microbiol. 2024, 122, 11–28. [Google Scholar] [CrossRef]
- Lejars, M.; Hajnsdorf, E. RNase III participates in the adaptation to temperature shock and oxidative stress in Escherichia coli. Microorganisms 2022, 10, 699. [Google Scholar] [CrossRef]
- Bechhofer, D.H.; Deutscher, M.P. Bacterial ribonucleases and their roles in RNA metabolism. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 242–300. [Google Scholar] [CrossRef]
- Massé, E.; Escorcia, F.E.; Gottesman, S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003, 17, 2374–2383. [Google Scholar] [CrossRef]
- Holmqvist, E.; Vogel, J. RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 2018, 16, 601–615. [Google Scholar] [CrossRef]
- Sinha, D.; de Lay, N.R. Target recognition by RNase E RNA-binding domain AR2 drives sRNA decay in the absence of PNPase. Proc. Natl. Acad. Sci. USA 2022, 119, e2208022119. [Google Scholar] [CrossRef]
- Ponath, F.; Hör, J.; Vogel, J. An overview of gene regulation in bacteria by small RNAs derived from mRNA 3′ ends. FEMS Microbiol. Rev. 2022, 46, fuac017. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chao, Y.; Matera, G.; Gao, Q.; Vogel, J. The conserved 3′ UTR-derived small RNA NarS mediates mRNA crossregulation during nitrate respiration. Nucleic Acids Res. 2020, 48, 2126–2143. [Google Scholar] [CrossRef]
- Iosub, I.A.; Marchioretto, M.; van Nues, R.W.; McKellar, S.; Viero, G.; Granneman, S. The mRNA derived MalH sRNA contributes to alternative carbon source utilization by tuning maltoporin expression in E. coli. RNA Biol. 2021, 18, 914–931. [Google Scholar] [CrossRef]
- Desgranges, E.; Barrientos, L.; Herrgott, L.; Marzi, S.; Toledo-Arana, A.; Moreau, K.; Vandenesch, F.; Romby, P.; Caldelari, I. The 3′UTR-derived sRNA RsaG coordinates redox homeostasis and metabolism adaptation in response to glucose-6-phosphate uptake in Staphylococcus aureus. Mol. Microbiol. 2022, 117, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Hui, M.P.; Foley, P.L.; Belasco, J.G. Messenger RNA degradation in bacterial cells. Annu. Rev. Genet. 2014, 48, 537–559. [Google Scholar] [CrossRef]
- Jain, C.; Deana, A.; Belasco, J.G. Consequences of RNase E scarcity in Escherichia coli. Mol. Microbiol. 2002, 43, 1053–1064. [Google Scholar] [CrossRef]
- Richards, J.; Belasco, J.G. Obstacles to scanning by RNase E govern bacterial mRNA lifetimes by hindering access to distal cleavage sites. Molecular Cell 2019, 74, 284–295.e5. [Google Scholar] [CrossRef] [PubMed]
- Lejars, M.; Hajnsdorf, E. Bacterial RNase III: Targets and physiology. Biochimie 2024, 217, 54–65. [Google Scholar] [CrossRef]
- Altuvia, Y.; Bar, A.; Reiss, N.; Karavani, E.; Argaman, L.; Margalit, H. In vivo cleavage rules and target repertoire of RNase III in Escherichia coli. Nucleic Acids Res. 2018, 46, 10380–10394. [Google Scholar] [CrossRef]
- Lejars, M.; Kobayashi, A.; Hajnsdorf, E. RNase III, Ribosome Biogenesis and Beyond. Microorganisms 2021, 9, 2608. [Google Scholar] [CrossRef]
- Saramago, M.; Robledo, M.; Matos, R.G.; Jiménez-Zurdo, J.I.; Arraiano, C.M. Sinorhizobium meliloti RNase III: Catalytic features and impact on symbiosis. Front. Genet. 2018, 9, 350. [Google Scholar] [CrossRef]
- Snow, S.; Bacon, E.; Bergeron, J.; Katzman, D.; Wilhelm, A.; Lewis, O.; Syangtan, D.; Calkins, A.; Archambault, L.; Anacker, M.L.; et al. Transcript decay mediated by RNase III in Borrelia burgdorferi. Biochem. Biophys. Res. Commun. 2020, 529, 386–391. [Google Scholar] [CrossRef]
- Ifill, G.; Blimkie, T.; Lee, A.H.-Y.; Mackie, G.A.; Chen, Q.; Stibitz, S.; Hancock, R.E.W.; Fernandez, R.C. RNase III and RNase E influence posttranscriptional regulatory networks involved in virulence factor production, metabolism, and regulatory RNA processing in Bordetella pertussis. mSphere 2021, 6, e0065021. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.C.; Cameron, J.C.; Pfleger, B.F. RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. mBio 2017, 8, 10–1128. [Google Scholar] [CrossRef]
- McKellar, S.W.; Ivanova, I.; Arede, P.; Zapf, R.L.; Mercier, N.; Chu, L.-C.; Mediati, D.G.; Pickering, A.C.; Briaud, P.; Foster, R.G.; et al. RNase III CLASH in MRSA uncovers sRNA regulatory networks coupling metabolism to toxin expression. Nat. Commun. 2022, 13, 3560. [Google Scholar] [CrossRef]
- Mediati, D.G.; Wong, J.L.; Gao, W.; McKellar, S.; Pang, C.N.I.; Wu, S.; Wu, W.; Sy, B.; Monk, I.R.; Biazik, J.M.; et al. RNase III-CLASH of multi-drug resistant Staphylococcus aureus reveals a regulatory mRNA 3′UTR required for intermediate vancomycin resistance. Nat. Commun. 2022, 13, 3558. [Google Scholar] [CrossRef] [PubMed]
- Karls, R.K.; Brooks, J.; Rossmeissl, P.; Luedke, J.; Donohue, T.J. Metabolic roles of a Rhodobacter sphaeroides member of the sigma32 family. J. Bacteriol. 1998, 180, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Nuss, A.M.; Glaeser, J.; Berghoff, B.A.; Klug, G. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J. Bacteriol. 2010, 192, 2613–2623. [Google Scholar] [CrossRef] [PubMed]
- Grützner, J.; Billenkamp, F.; Spanka, D.-T.; Rick, T.; Monzon, V.; Förstner, K.U.; Klug, G. The small DUF1127 protein CcaF1 from Rhodobacter sphaeroides is an RNA-binding protein involved in sRNA maturation and RNA turnover. Nucleic Acids Res. 2021, 49, 3003–3019. [Google Scholar] [CrossRef]
- Quendera, A.P.; Seixas, A.F.; Dos Santos, R.F.; Santos, I.; Silva, J.P.N.; Arraiano, C.M.; Andrade, J.M. RNA-binding proteins driving the regulatory activity of small non-coding RNAs in bacteria. Front. Mol. Biosci. 2020, 7, 78. [Google Scholar] [CrossRef]
- Holmqvist, E.; Berggren, S.; Rizvanovic, A. RNA-binding activity and regulatory functions of the emerging sRNA-binding protein ProQ. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194596. [Google Scholar] [CrossRef]
- Ng Kwan Lim, E.; Sasseville, C.; Carrier, M.-C.; Massé, E. Keeping up with RNA-based regulation in bacteria: New roles for RNA binding proteins. Trends Genet. 2021, 37, 86–97. [Google Scholar] [CrossRef]
- Santiago-Frangos, A.; Woodson, S.A. Hfq chaperone brings speed dating to bacterial sRNA. Wiley Interdiscip. Rev. RNA 2018, 9, e1475. [Google Scholar] [CrossRef] [PubMed]
- Westermann, A.J.; Venturini, E.; Sellin, M.E.; Förstner, K.U.; Hardt, W.-D.; Vogel, J. The major RNA-binding protein ProQ impacts virulence gene expression in Salmonella enterica Serovar Typhimurium. mBio 2019, 10, 10–128. [Google Scholar] [CrossRef]
- Eraso, J.M.; Kaplan, S. prrA, a putative response regulator involved in oxygen regulation of photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 1994, 176, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Eraso, J.M.; Kaplan, S. Complex regulatory activities associated with the histidine kinase PrrB in expression of photosynthesis genes in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1996, 178, 7037–7046. [Google Scholar] [CrossRef]
- Gomelsky, M.; Kaplan, S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 1997, 179, 128–134. [Google Scholar] [CrossRef]
- Braatsch, S.; Gomelsky, M.; Kuphal, S.; Klug, G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 2002, 45, 827–836. [Google Scholar] [CrossRef]
- Masuda, S.; Bauer, C.E. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 2002, 110, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Moskvin, O.V.; Kaplan, S.; Gilles-Gonzalez, M.-A.; Gomelsky, M. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 2007, 282, 28740–28748. [Google Scholar] [CrossRef]
- Spanka, D.-T.; Reuscher, C.M.; Klug, G. Impact of PNPase on the transcriptome of Rhodobacter sphaeroides and its cooperation with RNase III and RNase E. BMC Genom. 2021, 22, 106. [Google Scholar] [CrossRef] [PubMed]
- Spanka, D.-T.; Klug, G. Maturation of UTR-derived sRNAs is modulated during adaptation to different growth conditions. Int. J. Mol. Sci. 2021, 22, 12260. [Google Scholar] [CrossRef]
- Jordan, B.; Weidenbach, K.; Schmitz, R.A. The power of the small: The underestimated role of small proteins in bacterial and archaeal physiology. Curr. Opin. Microbiol. 2023, 76, 102384. [Google Scholar] [CrossRef]
- Van Niel, C.B. The culture, general physiology, morphology, and classification of the non-sulfur purple and brown bacteria. Bacteriol. Rev. 1944, 8, 1–118. [Google Scholar] [CrossRef]
- Remes, B.; Berghoff, B.A.; Förstner, K.U.; Klug, G. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genom. 2014, 15, 794. [Google Scholar] [CrossRef]
- Apirion, D. Isolation, genetic mapping and some characterization of a mutation in Escherichia coli that affects the processing of ribonuleic acid. Genetics 1978, 90, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. 1989, 219, 480–485. [Google Scholar] [CrossRef]
- Blumenkamp, P.; Pfister, M.; Diedrich, S.; Brinkrolf, K.; Jaenicke, S.; Goesmann, A. Curare and GenExVis: A versatile toolkit for analyzing and visualizing RNA-Seq data. BMC Bioinform. 2024, 25, 138. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dündar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160-5. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Hadley, W. Ggplot2: Elegrant Graphics for Data Analysis; Springer: Cham, Switzerland, 2016; ISBN 9783319242774. [Google Scholar]
- Jørgensen, M.G.; Pettersen, J.S.; Kallipolitis, B.H. sRNA-mediated control in bacteria: An increasing diversity of regulatory mechanisms. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194504. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Melamed, S. Small RNAs, large networks: Posttranscriptional regulons in Gram-negative bacteria. Annu. Rev. Microbiol. 2023, 77, 23–43. [Google Scholar] [CrossRef] [PubMed]

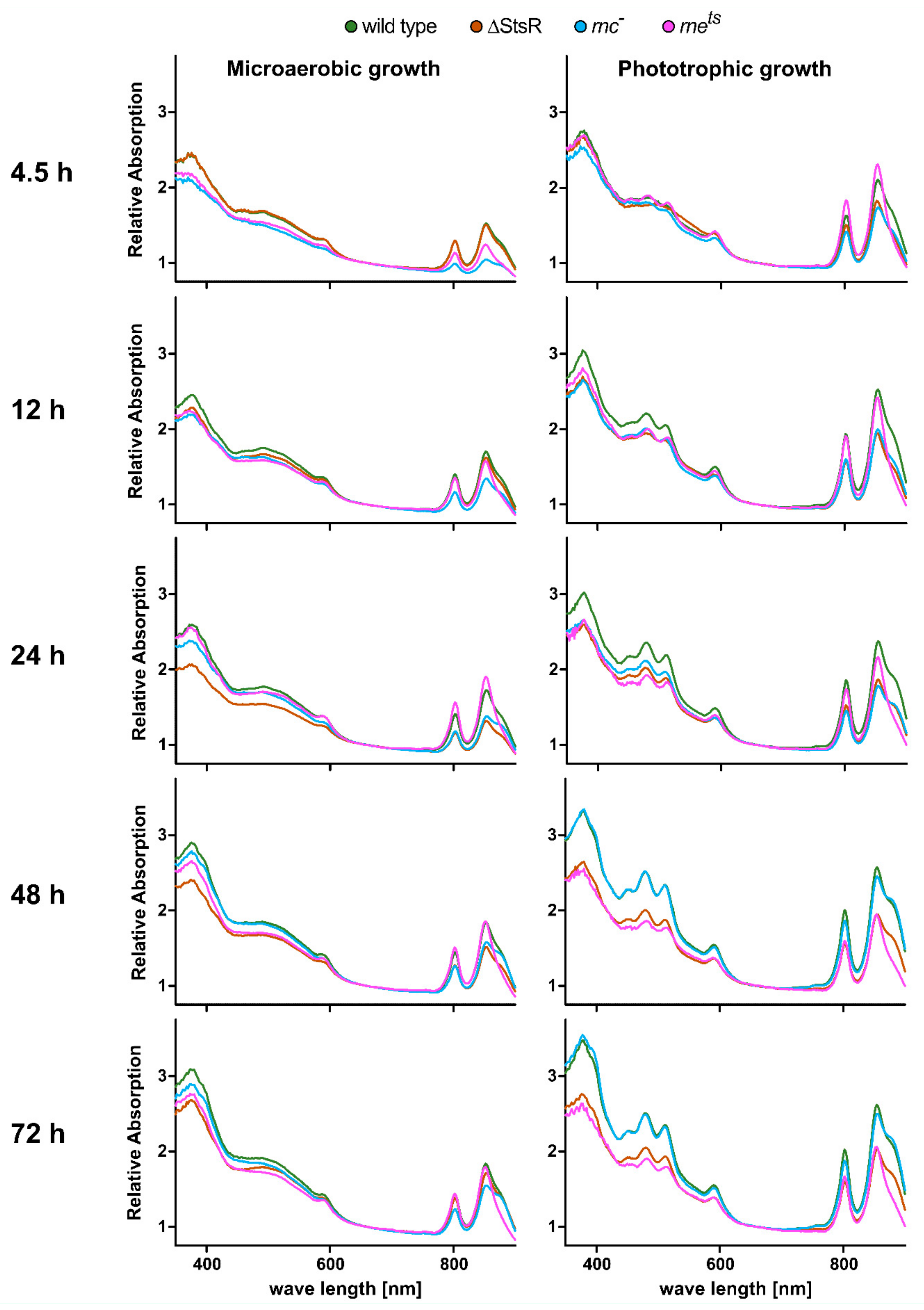
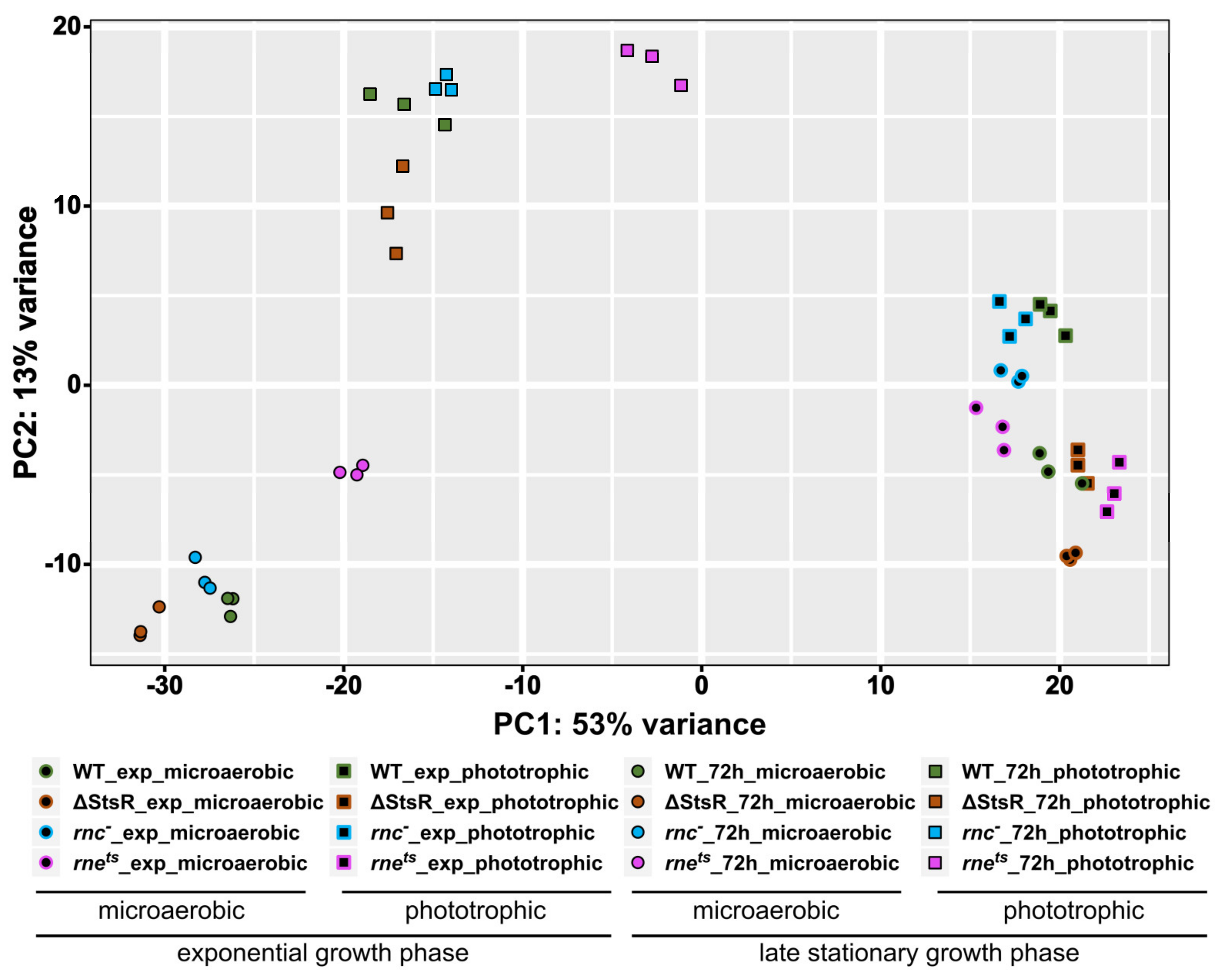
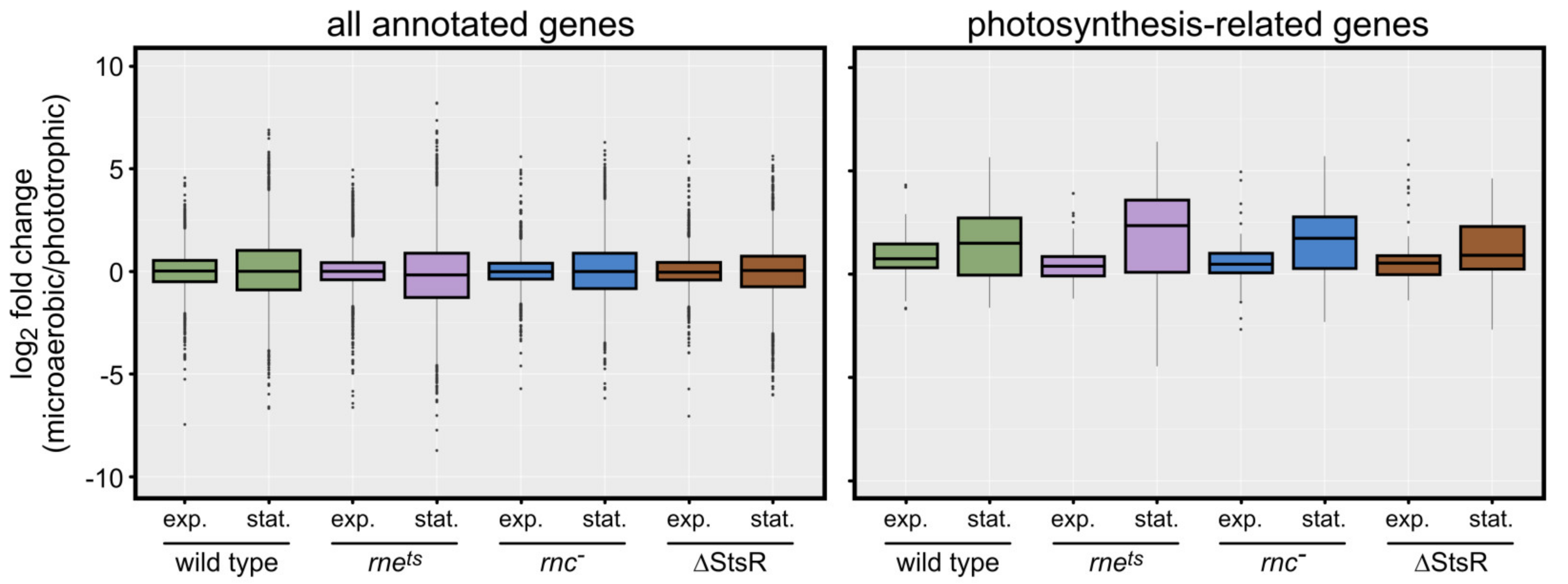
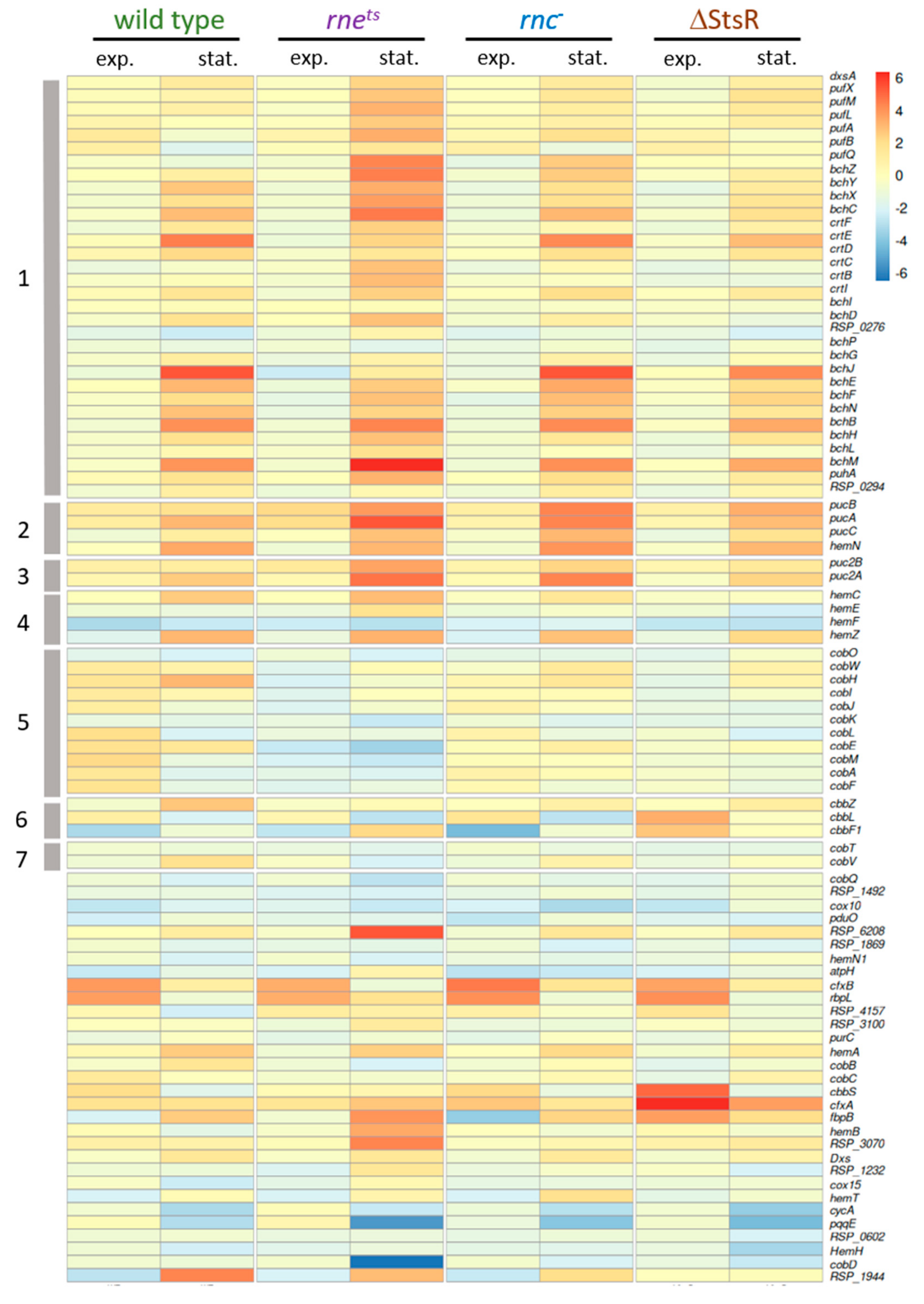
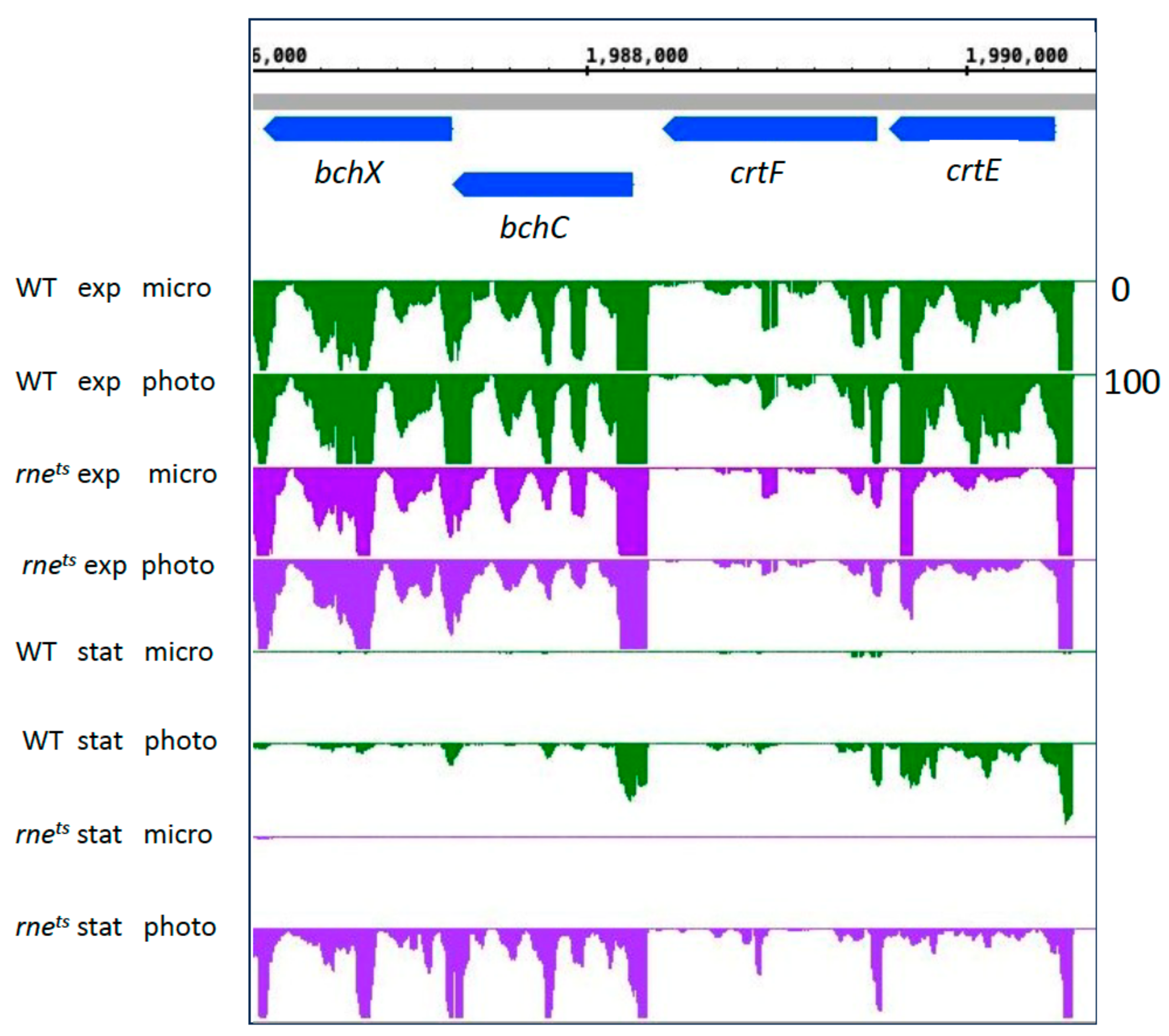
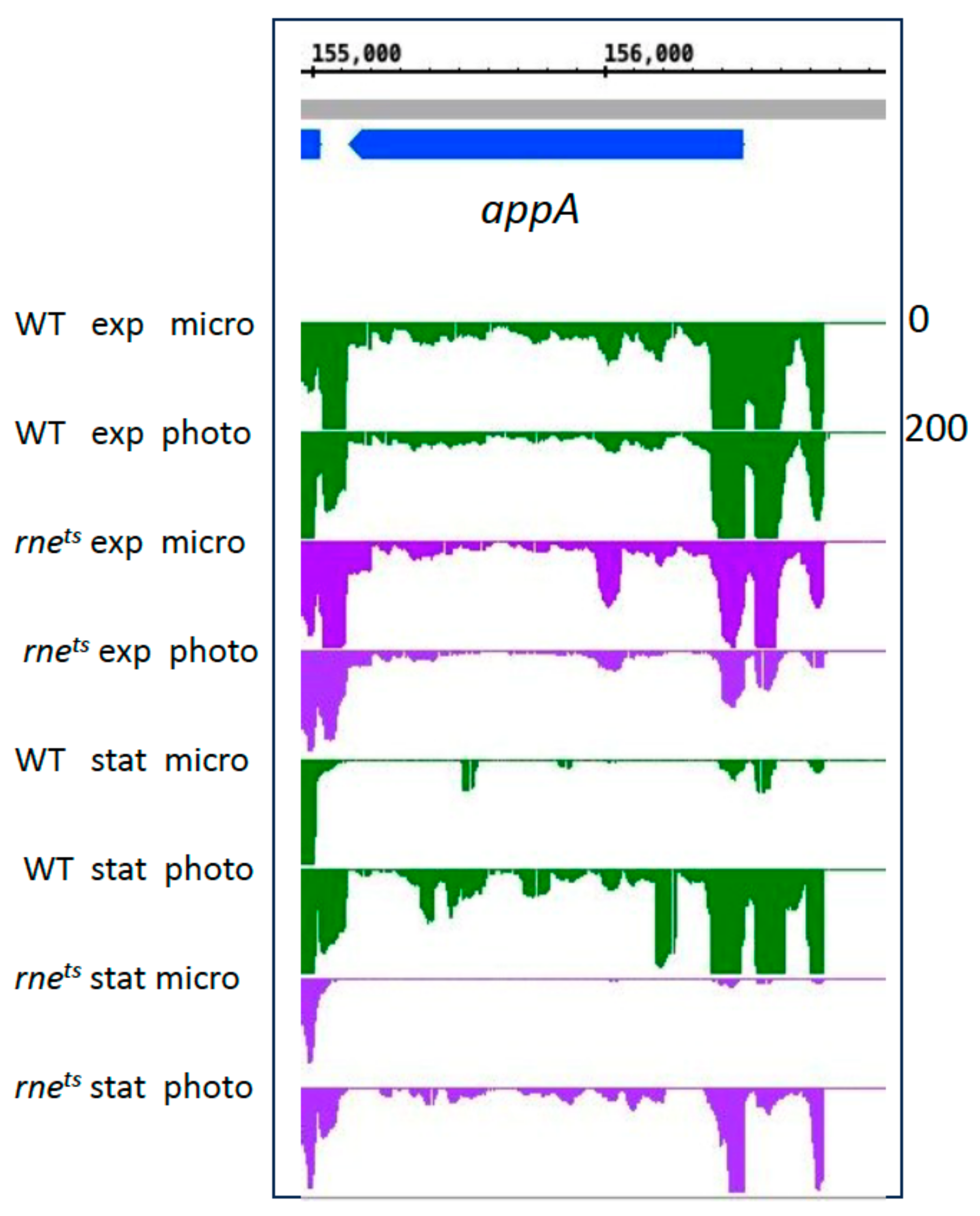
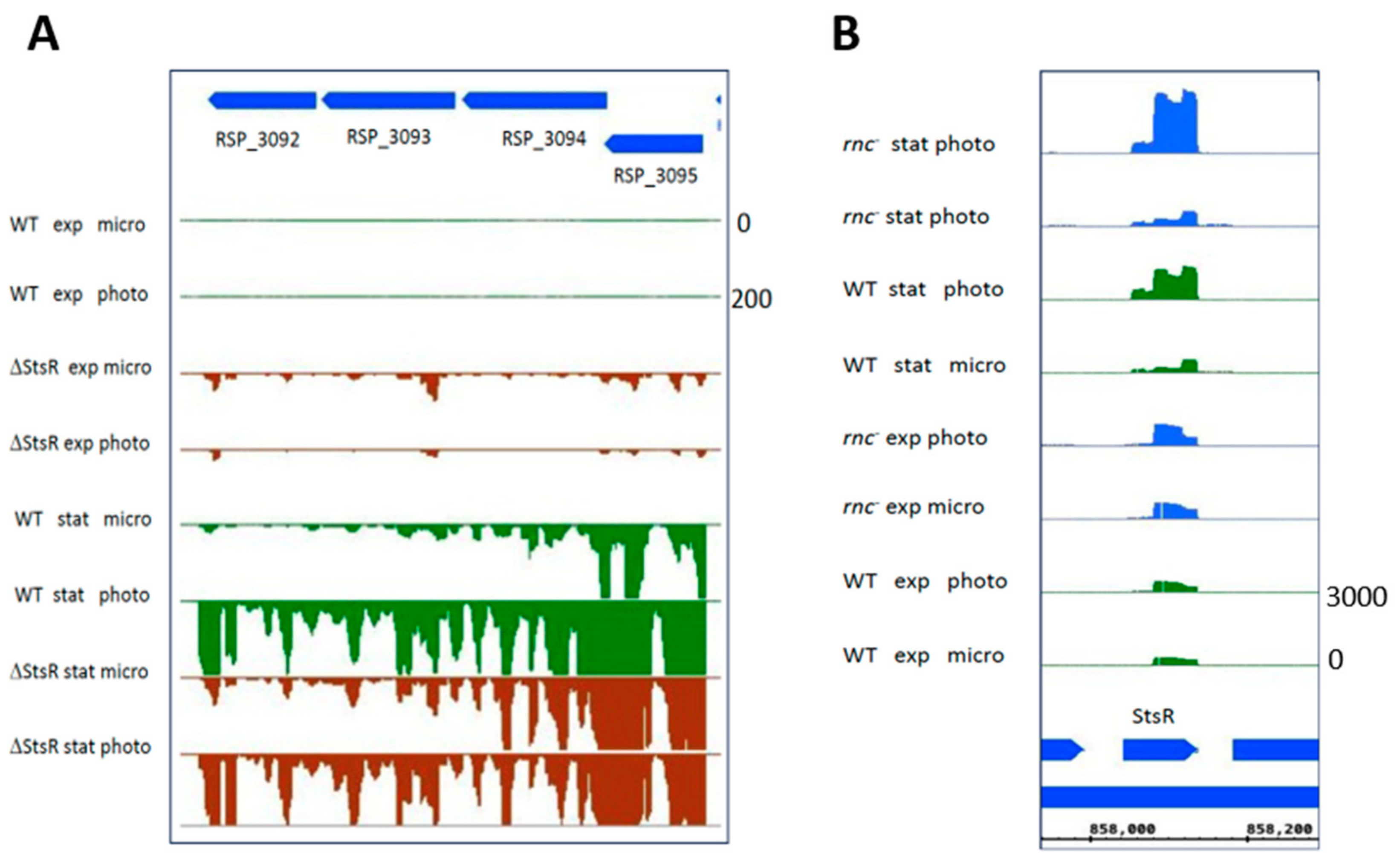
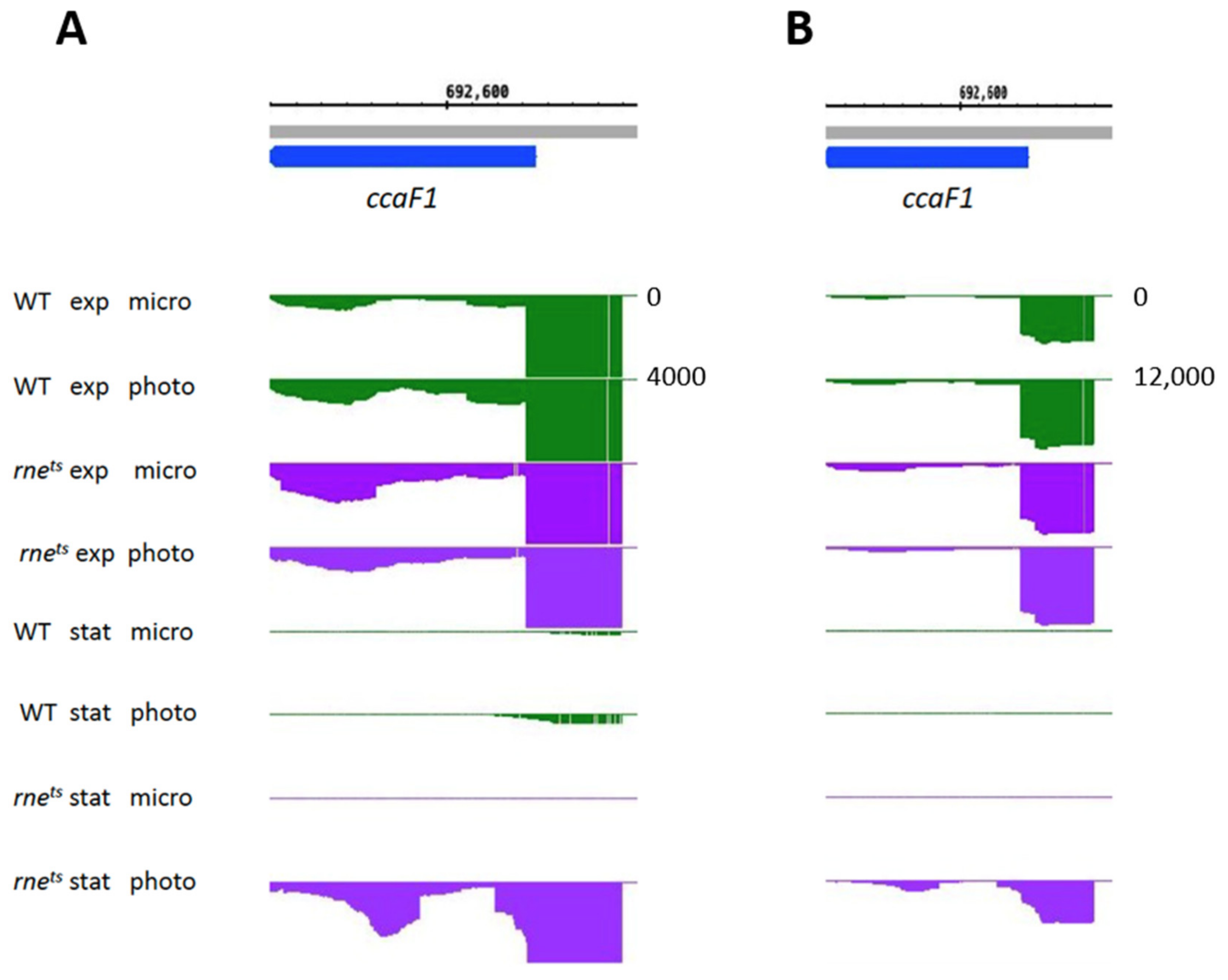
| Gene Expression Change from Microaerobic to Phototrophic Given as log2 Fold Change | ||||||||
|---|---|---|---|---|---|---|---|---|
| Wild Type | rnets | rnc− | ΔStsR | |||||
| Exp. | Stat. | Exp. | Stat. | Exp. | Stat. | Exp. | Stat. | |
| PS gene cluster (n = 38) | 0.7 | 2.0 | 0.5 | 2.7 | 0.5 | 2.2 | 0.5 | 1.6 |
| motility (n = 63) | −1.7 | −0.7 | 2.8 | −2.2 | −0.5 | −0.4 | 0.3 | −1.1 |
| dcw gene cluster (n = 24) | −0.2 | 0.5 | 0.1 | 0.3 | −0.3 | 0.8 | −0.3 | 0.6 |
| ribosomal gene cluster (n = 37) | −0.4 | 1.3 | 0 | 0.9 | −0.4 | 1.1 | 0 | 0.7 |
| nuo gene cluster (n = 22) | −0.5 | 0.1 | −0.5 | −0.7 | −0.5 | −0.2 | 0.7 | 0.1 |
| Gene Expression Change from Microaerobic to Phototrophic Given as log2 fold Change | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exponential Phase | Stationary Phase | ||||||||
| WT | rnets | rnc− | ΔStsR | WT | rnets | rnc− | ΔStsR | ||
| A | pucBAC | 1.5 | 2.2 * | 1.4 | 1.1 | 2.8 * | 4.5 * | 4.4 * | 3.4 * |
| pufQ-X | 1.6 * | 0.8 * | 1.0 | 1.2 * | 0.8 | 3.5 * | 2.0 | 1.6 * | |
| hemZ | −0.4 | 0.1 | −0.7 * | 0.0 | 3.7 * | 3.7 * | 3.4 * | 2.9 * | |
| hemA | 1.4 * | 0.3 | 0.9 * | 0.6 * | 3.3 * | 3.2 * | 3.0 * | 2.1 * | |
| crtE | 1.2 * | −0.2 | 0.7 * | 0.7 * | 4.8 * | 3.0 * | 4.6 * | 3.5 * | |
| crtIB | 1.1 * | −0.4 | 0.8 * | 0.5 | 1.8 * | 3.4 * | 1.8 * | 1.1 * | |
| crtD | 1.5 * | 0.6 * | 1.0 * | 0.6 * | 3.0 * | 2.4 * | 2.7 * | 2.4 * | |
| bchCXYZ | 0.7 | 0.5 | 0.3 | 0.4 | 2.9 * | 4.5 * | 3.1 * | 2.3 * | |
| bchEJ | 0.6 | −0.4 | 0.4 | 0.9 * | 4.6 * | 2.6 * | 4.8 * | 3.7 * | |
| crtA-bchI | 1.3 * | 1.1 * | 0.9 * | 0.9 * | 2.3 * | 3.3 * | 2.4 * | 1.7 * | |
| RSP_0290 | 1.1 * | 0.9 * | 0.9 * | 0.8 * | 2.3 * | 3.3 * | 2.9 * | 1.8 * | |
| fbcFBC | 0.8 | 0.6 * | 0.5 | 0.6 | 1.8 * | 4.1 * | 2.0 * | 2.3 * | |
| B | prrA | 0.4 | −0.1 | −0.1 | 0.0 | 0.3 | 1.4 * | 0.8 * | 0.2 |
| prrB | 0.2 | 0.1 | 0.1 | −0.3 * | 0.3 | −0.5 * | 0.3 | 1.0 * | |
| fnrL | −1.0 * | −0.3 | −0.4 * | −0.6 * | −1.1 * | −0.3 | −0.7 * | −0.1 | |
| appA | −0.2 | −0.6 * | −0.7 * | −0.1 | 3.4 * | 2.4 * | 2.8 * | 1.9 * | |
| ppsR | −0.9 * | −0.3 | −0.4 * | −0.7 * | 0.1 | 0.6 * | 0.5 * | 0.0 | |
| C | rne | −1.0 * | n.a. | −0.5 * | −1.0 * | 0.4 * | n.a. | 0.7 * | 0.7 * |
| rnc | −0.7 * | 0.2 | n.a. | −0.4 * | 1.8 * | 0.0 | n.a. | 1.0 * | |
| pnp | 0.1 | 0.8 * | 0.1 | 0.4 * | −0.5 * | 0.3 | 0.0 | −0.8 * | |
| D | rpoHI | 2.2 * | 1.2 * | 1.5 * | 1.1 * | 0.1 | −2.1 * | −0.8 * | 0.1 |
| rpoHII | 0.3 | −0.6 * | 0.2 | 1.3 * | 0.7 * | 0.8 * | 0.6 * | 2.9 | |
| rpoE1 | 0.7 * | 0.2 | 0.5 * | 0.7 * | 2.1 * | 0.7 * | 1.7 * | 2.5 * | |
| RSP_3095 | 0.1 | −0.2 | −0.1 | −0.2 | 2.3 * | 0.6 * | 0.3 | −0.4 | |
| E | pcrZ | 0.0 | 0.4 * | 0.4 | 2.6 * | 0.8 * | −1.7 * | 0.0 | −0.4 * |
| stsR | 0.6 * | 0.9 * | 0.0 | n.a. | 1.4 * | 1.4 | 1.7 * | n.a. | |
| upsM | −1.1 * | −1.0 * | −0.5 | −0.3 | −1.6 * | −0.6 | −0.2 | 0.3 | |
| ccsR1 | 1.7 * | 0.7 * | 0.5 | 1.6 * | 3.1 * | 5.2 * | 2.2 * | 5.2 * | |
| F | hfq | 0.2 | 0.2 | 0.2 | 0.2 | -0.1 | 0.4 * | 0.3 * | 0.4 * |
| ccaF1 | 0.7 * | 0.7 | −0.3 | −0.1 | 2.0 * | 6.3 * | 2.9 * | 4.2 * | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Börner, J.; Grützner, J.; Gerken, F.; Klug, G. The Impact of the Major Endoribonucleases RNase E and RNase III and of the sRNA StsR on Photosynthesis Gene Expression in Rhodobacter sphaeroides Is Growth-Phase-Dependent. Int. J. Mol. Sci. 2024, 25, 9123. https://doi.org/10.3390/ijms25169123
Börner J, Grützner J, Gerken F, Klug G. The Impact of the Major Endoribonucleases RNase E and RNase III and of the sRNA StsR on Photosynthesis Gene Expression in Rhodobacter sphaeroides Is Growth-Phase-Dependent. International Journal of Molecular Sciences. 2024; 25(16):9123. https://doi.org/10.3390/ijms25169123
Chicago/Turabian StyleBörner, Janek, Julian Grützner, Florian Gerken, and Gabriele Klug. 2024. "The Impact of the Major Endoribonucleases RNase E and RNase III and of the sRNA StsR on Photosynthesis Gene Expression in Rhodobacter sphaeroides Is Growth-Phase-Dependent" International Journal of Molecular Sciences 25, no. 16: 9123. https://doi.org/10.3390/ijms25169123
APA StyleBörner, J., Grützner, J., Gerken, F., & Klug, G. (2024). The Impact of the Major Endoribonucleases RNase E and RNase III and of the sRNA StsR on Photosynthesis Gene Expression in Rhodobacter sphaeroides Is Growth-Phase-Dependent. International Journal of Molecular Sciences, 25(16), 9123. https://doi.org/10.3390/ijms25169123






