Metabolomics-Assisted Breeding in Oil Palm: Potential and Current Perspectives
Abstract
:1. Introduction
2. Brief History of Oil Palm Breeding
3. Overview of Current Breeding Strategies
4. The Potential of Metabolomics in Oil Palm
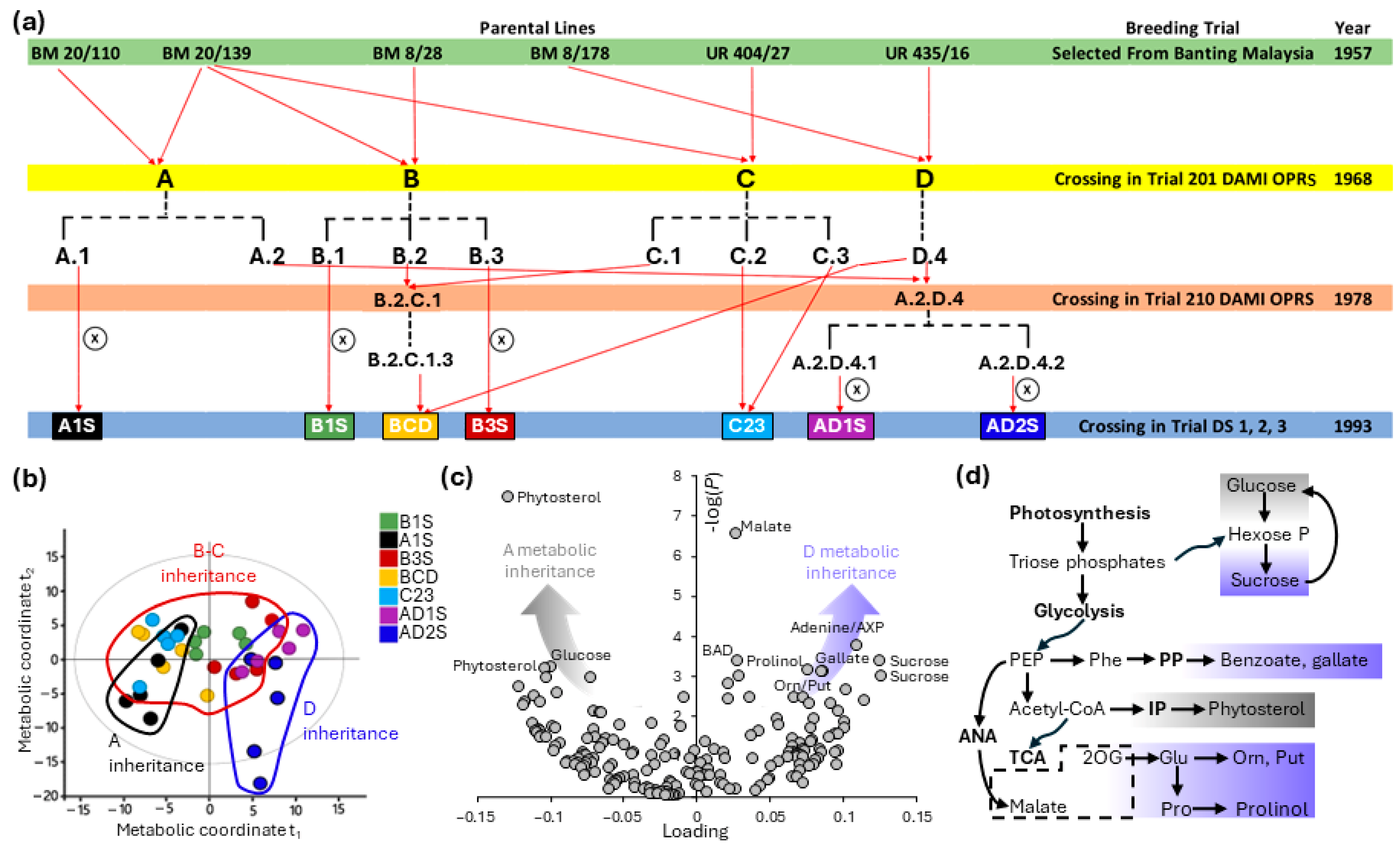
5. Practical Imperatives for Metabolomics-Assisted Breeding of Oil Palm
- -
- First, there should be good candidate metabolome-based descriptors of traits of interest. The case of K use efficiency is illustrated above in Section 4 and suggests that building a quantitative multivariate variable that can be used to monitor traits is possible (Figure 2b,c). In the case of nutrient utilization, this represents a remarkable opportunity, since it provides access to a complex trait. In the case of potassium nutrition, assessing potassium utilization by trees is a long and resource-consuming process, generally involving agronomic trials at some stages to define a line- and -environment-dependent critical %K in leaflets [37], differential decomposition of growth response curves [50], or plantation K balance [51]. In other words, there could be a universal metabolic signature that can be used to anticipate physiological palm K sufficiency without the need to conduct long and costly agronomic trials required by other methods. However, to date, uncertainty remains as to whether reliable metabolic descriptors will be accessible for traits other than K use efficiency. Additionally, it is worth noting that a candidate metabolic marker is reliable when the factor of interest (drought, nutrient availability, etc.) varies while other parameters are controlled to avoid bias due to the involvement of common metabolites in several responses to environmental cues (i.e., the influence of confounding factors).
- -
- Second, there must be natural variations amongst oil palm families and genotypes in metabolic pathways, making possible associations between genetic markers and metabolites. That is, under the assumption that the traits of interest are determined by metabolic properties, differences in metabolic content can be used to select oil palm lines and perform breeding.
- -
- Third, metabolomics analysis and interpretation themselves must be relatively fast. In fact, should metabolomics be a slow step, it would compensate for the time gained by bypassing long agronomic trials and data recording. That is, in the breeding process (illustrated step by step in Figure 4), uncertainty remains as to whether metabolomics themselves might represent a bottleneck. This is an important question because metabolomics-assisted breeding requires metabolomics analysis, adding steps associated with both identification of candidate metabolome signatures (dark-blue steps, Figure 4a) and validation (light-blue steps, Figure 4a). It is unlikely that metabolomics analyses per se would be highly time-consuming. In particular, rapid methods like 1H-NMR can be used, allowing acquisition of well-resolved spectra within 10 min and identification and quantification of many metabolites, including sugars, amino acids, catecholamines, and polyamines, in oil palm [11,45]. Also, automated exact mass spectrometry by GC–MS can perform derivatization prior to injection, facilitating sample processing and acquisition [61]. LC–MS analyses can also be performed relatively rapidly (about 25 min per sample) including on crude extracts. For a recent example, see [46]. However, data treatment and extraction can be relatively long (Figure 4b, dark-blue bar) for LC–MS datasets, due to spectral cleaning (elimination of adducts, noise m/z signals, etc.) and compound annotations. Recent methods have been developed for routine LC–MS data trimming, such as MS-CleanR [62], that facilitate the simplification and integration of relevant metabolomics features. However, annotation is still challenging.
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ramli, U.S.; Othman, A.; Tahir, N.I.M.; Lau, B.Y.C.; Shahwan, S.; Hassan, H.; Nurazah, Z.; Dzulkafli, S.B.; Rozali, N.L.; Ishak, N.A. Omics—A potential tool for oil palm improvement and productivity. In The Oil Palm Genome; Ithnin, M., Kushairi, A., Eds.; Springer: Cham, Switzerland, 2020; pp. 141–157. [Google Scholar]
- Martin, J.; Jeyakumar, J.; Yarra, R.; Wei, L.; Cao, H. Oil palm breeding in the modern era: Challenges and opportunities. Plants 2022, 11, 1395. [Google Scholar] [CrossRef] [PubMed]
- Subhi, S.; Tahir, N.I.; Abd Rasid, O.; Ramli, U.S.; Othman, A.; Masani, M.Y.A.; Parveez, G.K.A.; Kushairi, A. Post-genomic technologies for the advancement of oil palm research. J. Oil Palm Res. 2017, 29, 469–486. [Google Scholar]
- Santiago, K.A.A.; Wong, W.C.; Goh, Y.K.; Tey, S.H.; Ting, A.S.Y. Metabolomics approach in identifying biomarkers from pathogenic Ganoderma boninense involved in early interactions with oil palm host. Physiol. Mol. Plant Pathol. 2023, 125, 101980. [Google Scholar] [CrossRef]
- Othman, A.; Lau, B.Y.C.; Nurazah, Z.; Shahwan, S.; Rusli, M.H.; Singh, R.; Abdullah, M.O.; Marjuni, M.; Yaakub, Z.; Sundram, S. Comparative proteomic and metabolomic studies between partial resistant and susceptible oil palm reveal the molecular mechanism associated with Ganoderma boninense infection. Physiol. Mol. Plant Pathol. 2024, 129, 102198. [Google Scholar] [CrossRef]
- Baharum, S.N.; Ahmad, M.F.; Ajeng, A.A.; Abdullah, R. Metabolites profiling of Ganoderma-infected oil palms rachis grown on tropical soils reveals choline phosphate and 2-oxoglutaramate as potential biomarkers in the disease detection. Physiol. Mol. Plant Pathol. 2023, 125, 102001. [Google Scholar] [CrossRef]
- Neto, J.C.R.; Vieira, L.R.; de Aquino Ribeiro, J.A.; de Sousa, C.A.F.; Júnior, M.T.S.; Abdelnur, P.V. Metabolic effect of drought stress on the leaves of young oil palm (Elaeis guineensis) plants using UHPLC–MS and multivariate analysis. Sci. Rep. 2021, 11, 18271. [Google Scholar] [CrossRef]
- Cui, J.; Chao de la Barca, J.M.; Lamade, E.; Tcherkez, G. Potassium nutrition in oil palm: The potential of metabolomics as a tool for precision agriculture. Plant People Planet 2021, 3, 350–354. [Google Scholar] [CrossRef]
- Cui, J.; Davanture, M.; Lamade, E.; Zivy, M.; Tcherkez, G. Plant low-K responses are partly due to Ca prevalence and the low-K biomarker putrescine does not protect from Ca side effects but acts as a metabolic regulator. Plant Cell Environ. 2021, 44, 1565–1579. [Google Scholar] [CrossRef]
- Cui, J.; Davanture, M.; Zivy, M.; Lamade, E.; Tcherkez, G. Metabolic responses to potassium availability and waterlogging reshape respiration and carbon use efficiency in oil palm. New Phytol. 2019, 223, 310–322. [Google Scholar] [CrossRef]
- Cui, J.; Lamade, E.; Tcherkez, G. Potassium deficiency reconfigures sugar export and induces catecholamine accumulation in oil palm leaves. Plant Sci. 2020, 300, 110628. [Google Scholar] [CrossRef]
- Sahebi, M.; Hanafi, M.M.; van Wijnen, A.J.; Akmar, A.S.N.; Azizi, P.; Idris, A.S.; Taheri, S.; Foroughi, M. Profiling secondary metabolites of plant defence mechanisms and oil palm in response to Ganoderma boninense attack. Int. Biodeterior. Biodegrad. 2017, 122, 151–164. [Google Scholar] [CrossRef]
- Tahir, N.I.; Rozali, N.L.; Rahmah, A.R.S.; Amiruddin, M.D.; Hwa, L.F.; Shaari, K.; Abas, F.; Othman, A.; Parveez, G.K.A.; Ramli, U.S. Metabolome study of oil palm (Elaeis guineensis Jacq.) planted in different environment conditions. Trop. Plant Biol. 2022, 15, 211–232. [Google Scholar] [CrossRef]
- Rodrigues-Neto, J.C.; Correia, M.V.; Souto, A.L.; Ribeiro, J.A.d.A.; Vieira, L.R.; Souza, M.T.; Rodrigues, C.M.; Abdelnur, P.V. Metabolic fingerprinting analysis of oil palm reveals a set of differentially expressed metabolites in fatal yellowing symptomatic and non-symptomatic plants. Metabolomics 2018, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Corley, R.; Tinker, P. The Oil Palm, 5th ed.; Wiley-Blackwell: Chichester, UK, 2016. [Google Scholar]
- Soh, A.; Wong, G.; Hor, T.; Tan, C.; Chew, P. Oil palm genetic improvement. Plant Breed. Rev. 2003, 22, 165–220. [Google Scholar]
- Cros, D.; Sánchez, L.; Cochard, B.; Samper, P.; Denis, M.; Bouvet, J.-M.; Fernández, J. Estimation of genealogical coancestry in plant species using a pedigree reconstruction algorithm and application to an oil palm breeding population. Theor. Appl. Genet. 2014, 127, 981–994. [Google Scholar] [CrossRef]
- Durand-Gasselin, T.; Kouame, R.; Cochard, B.; Adon, B.; Amblard, P. Varietal diffusion of oil palm (Elaeis guineensis Jacq.). Oléagineux Corps Gras Lipides 2000, 7, 207–2014. [Google Scholar] [CrossRef]
- Gascon, J.P.; De Berchoux, C. Characteristics of the production of E. guineensis (Jacq.) forms of varied origin and of their crosses. Application to oil-palm breeding. Oléagineux 1964, 19, 75–84. [Google Scholar]
- Seyum, E.G.; Bille, N.H.; Abtew, W.G.; Rastas, P.; Arifianto, D.; Domonhédo, H.; Cochard, B.; Jacob, F.; Riou, V.; Pomiès, V.; et al. Genome properties of key oil palm (Elaeis guineensis Jacq.) breeding populations. J. Appl. Genet. 2022, 63, 633–650. [Google Scholar] [CrossRef]
- Rosenquist, E.A. The genetic base of oil palm breeding populations. In Workshop Proceedings of the Palm Oil Research Institute of Malaysia, Proceedings of the International Workshop on Oil Palm Germplasm and Utilization, Selangor, Malaysia, 26–27 March 1985; Institut Penyelidikan Minyak Kelapa Sawit Malaysia: Selangor, Malaysia, 1986; Volume 10, pp. 27–56. [Google Scholar]
- Soh, A.C.; Mayes, S.; Roberts, J.A. Oil Palm Breeding: Genetics and Genomics; CRC Press: New York, NY, USA, 2017. [Google Scholar]
- Nyouma, A.; Bell, J.M.; Jacob, F.; Cros, D. From mass selection to genomic selection: One century of breeding for quantitative yield components of oil palm (Elaeis guineensis Jacq.). Tree Genet. Genom. 2019, 15, 69. [Google Scholar] [CrossRef]
- Cros, D.; Denis, M.; Bouvet, J.-M.; Sanchez, L. Comparing strategies of genomic selection to increase oil palm fresh fruit bunch yield. In Proceedings of the 4th International Oil Palm Conference, Bali, Indonesia, 12–14 February 2014. Document no. 574368. [Google Scholar]
- Nyouma, A.; Bell, J.M.; Jacob, F.; Riou, V.; Manez, A.; Pomiès, V.; Nodichao, L.; Syahputra, I.; Affandi, D.; Cochard, B. Genomic predictions improve clonal selection in oil palm (Elaeis guineensis Jacq.) hybrids. Plant Sci. 2020, 299, 110547. [Google Scholar] [CrossRef]
- Cros, D.; Denis, M.; Bouvet, J.-M.; Sánchez, L. Long-term genomic selection for heterosis without dominance in multiplicative traits: Case study of bunch production in oil palm. BMC Genom. 2015, 16, 651. [Google Scholar] [CrossRef] [PubMed]
- Purba, A.R.; Flori, A.; Baudouin, L.; Hamon, S. Prediction of oil palm (Elaeis guineensis, Jacq.) agronomic performances using the best linear unbiased predictor (BLUP). Theor. Appl. Genet. 2001, 102, 787–792. [Google Scholar] [CrossRef]
- Nyouma, A.; Bell, J.M.; Jacob, F.; Riou, V.; Manez, A.; Pomiès, V.; Domonhedo, H.; Arifiyanto, D.; Cochard, B.; Durand-Gasselin, T.; et al. Improving the accuracy of genomic predictions in an outcrossing species with hybrid cultivars between heterozygote parents: A case study of oil palm (Elaeis guineensis Jacq.). Mol. Genet. Genom. 2022, 297, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Cros, D.; Denis, M.; Sánchez, L.; Cochard, B.; Flori, A.; Durand-Gasselin, T.; Nouy, B.; Omoré, A.; Pomiès, V.; Riou, V. Genomic selection prediction accuracy in a perennial crop: Case study of oil palm (Elaeis guineensis Jacq.). Theor. Appl. Genet. 2015, 128, 397–410. [Google Scholar] [CrossRef]
- Tchounke, B.; Sanchez, L.; Bell, J.M.; Cros, D. Mate selection: A useful approach to maximize genetic gain and control inbreeding in genomic and conventional oil palm (Elaeis guineensis Jacq.) hybrid breeding. PLoS Comp. Biol. 2023, 19, e1010290. [Google Scholar] [CrossRef]
- Garzón-Martínez, G.A.; Osorio-Guarín, J.A.; Moreno, L.P.; Bastidas, S.; Barrero, L.S.; Lopez-Cruz, M.; Enciso-Rodríguez, F.E. Genomic selection for morphological and yield-related traits using genome-wide SNPs in oil palm. Mol. Breed. 2022, 42, 71. [Google Scholar] [CrossRef]
- Muhammad, I.I.; Abdullah, S.N.A.; Saud, H.M.; Shaharuddin, N.A.; Isa, N.M. The dynamic responses of oil palm leaf and root metabolome to phosphorus deficiency. Metabolites 2021, 11, 217. [Google Scholar] [CrossRef]
- Mirande-Ney, C.; Tcherkez, G.; Balliau, T.; Zivy, M.; Gilard, F.; Cui, J.; Ghashghaie, J.; Lamade, E. Metabolic leaf responses to potassium availability in oil palm (Elaeis guineensis Jacq.) trees grown in the field. Environ. Exp. Bot. 2020, 175, 104062. [Google Scholar] [CrossRef]
- Mirande-Ney, C.; Tcherkez, G.; Gilard, F.; Ghashghaie, J.; Lamade, E. Effects of potassium fertilization on oil palm fruit metabolism and mesocarp lipid accumulation. J. Agric. Food Chem. 2019, 67, 9432–9440. [Google Scholar] [CrossRef]
- Martin, J.; Wu, Q.; Feng, M.; Li, R.; Zhou, L.; Zhang, S.; Yang, C.; Cao, H. Lipidomic profiles of lipid biosynthesis in oil palm during fruit development. Metabolites 2023, 13, 727. [Google Scholar] [CrossRef]
- Wei, L.; Yang, C.; John Martin, J.J.; Li, R.; Zhou, L.; Cheng, S.; Cao, H.; Liu, X. Metabonomics and transcriptomic analysis of free fatty acid synthesis in seedless and Tenera oil palm. Int. J. Mol. Sci. 2024, 25, 1686. [Google Scholar] [CrossRef] [PubMed]
- Caliman, J.-P.; Daniel, C.; Tailliez, B. La nutrition minérale du palmier à huile. Plant. Rech. Dev. 1994, 1, 36–54. [Google Scholar]
- Chapman, G.W.; Gray, H.M. Leaf analysis and the nutrition of oil palm. Ann. Bot. 1949, 13, 415–433. [Google Scholar] [CrossRef]
- Foster, H.; Chang, K. Seasonal fluctuations in oil palm leaf nutrient levels. MARDI Res. Bull. 1977, 5, 74–90. [Google Scholar]
- Astorkia, M.; Hernández, M.; Bocs, S.; Ponce, K.; León, O.; Morales, S.; Quezada, N.; Orellana, F.; Wendra, F.; Sembiring, Z.; et al. Detection of significant SNP associated with production and oil quality traits in interspecific oil palm hybrids using RARSeq. Plant Sci. 2020, 291, 110366. [Google Scholar] [CrossRef]
- Babu, B.K.; Mathur, R.; Ravichandran, G.; Anita, P.; Venu, M. Genome wide association study (GWAS) and identification of candidate genes for yield and oil yield related traits in oil palm (Eleaeis guineensis) using SNPs by genotyping-based sequencing. Genomics 2020, 112, 1011–1020. [Google Scholar] [CrossRef]
- Apriyanto, A.; Compart, J.; Zimmermann, V.; Alseekh, S.; Fernie, A.R.; Fettke, J. Indication that starch and sucrose are biomarkers for oil yield in oil palm (Elaeis guineensis Jacq.). Food Chem. 2022, 393, 133361. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, N.A. Assessment of the Potential for Metabolomics-Assisted Breeding in Oil Palm-Part 1. Ph.D. Report, University of Angers, Beaucouze, France, 2024. [Google Scholar]
- Lamade, E.; Tcherkez, G. Revisiting foliar diagnosis for oil palm potassium nutrition. Eur. J. Agron. 2023, 143, 126694. [Google Scholar] [CrossRef]
- Pancoro, A.; Karima, E.; Apriyanto, A.; Effendi, Y. 1H NMR metabolomics analysis of oil palm stem tissue infected by Ganoderma boninense based on field severity Indices. Sci. Rep. 2022, 12, 21087. [Google Scholar] [CrossRef]
- Santiago, K.A.A.; Wong, W.C.; Goh, Y.K.; Tey, S.H.; Ting, A.S.Y. Pathogenicity of monokaryotic and dikaryotic mycelia of Ganoderma boninense revealed via LC–MS-based metabolomics. Sci. Rep. 2024, 14, 5330. [Google Scholar] [CrossRef]
- Daval, A.; Pomiès, V.; Le Squin, S.; Denis, M.; Riou, V.; Breton, F.; Nopariansyah; Bink, M.; Cochard, B.; Jacob, F. In silico QTL mapping in an oil palm breeding program reveals a quantitative and complex genetic resistance to Ganoderma boninense. Mol. Breed. 2021, 41, 53. [Google Scholar] [CrossRef] [PubMed]
- Rahmadi, H.Y.; Syukur, M.; Widodo; Suwarno, W.B.; Wening, S.; Simamora, A.N.; Nugroho, S. 1H NMR analysis of metabolites from leaf tissue of resistant and susceptible oil palm breeding materials against Ganoderma boninense. Metabolomics 2024, 20, 89. [Google Scholar]
- Nusaibah, S.A.; Siti Nor Akmar, A.; Idris, A.S.; Sariah, M.; Mohamad Pauzi, Z. Involvement of metabolites in early defense mechanism of oil palm (Elaeis guineensis Jacq.) against Ganoderma disease. Plant Physiol. Biochem. 2016, 109, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Foster, H.L. Assessment of oil palm requirements. In The Oil Palm, Management for Large and Sustainable Yields; Fairhurst, T.H., Härdter, R., Eds.; Potash and Phosphate Institute of Canada (ESEAP): Singapore, 2003; pp. 231–257. [Google Scholar]
- Kee, K.K.; Goh, K.; Chew, P.S.; Tey, S.H. An integrated site specific fertilizer recommendation system (INFERS) for high productivity in mature oil palms. In Management for Enhanced Profitability in Plantations; Chee, K., Ed.; Incoporated Society of Planters: Kuala Lumpur, Malaysia, 1994; pp. 83–100. [Google Scholar]
- Jin, J.; Sun, Y.; Qu, J.; syah, R.; Lim, C.-H.; Alfiko, Y.; Rahman, N.E.B.; Suwanto, A.; Yue, G.; Wong, L.; et al. Transcriptome and functional analysis reveals hybrid vigor for oil biosynthesis in oil palm. Sci. Rep. 2017, 7, 439. [Google Scholar] [CrossRef]
- Lonien, J.; Schwender, J. Analysis of metabolic flux phenotypes for two Arabidopsis mutants with severe impairment in seed storage lipid synthesis. Plant Physiol. 2009, 151, 1617–1634. [Google Scholar] [CrossRef] [PubMed]
- Guerin, C.; Joët, T.; Serret, J.; Lashermes, P.; Vaissayre, V.; Agbessi, M.D.T.; Beulé, T.; Severac, D.; Amblard, P.; Tregear, J.; et al. Gene coexpression network analysis of oil biosynthesis in an interspecific backcross of oil palm. Plant J. 2016, 87, 423–441. [Google Scholar] [CrossRef]
- Apriyanto, A.; Compart, J.; Fettke, J. Transcriptomic analysis of mesocarp tissue during fruit development of the oil palm revealed specific isozymes related to starch metabolism that control oil yield. Front. Plant Sci. 2023, 14, 1220237. [Google Scholar] [CrossRef]
- Dussert, S.; Guerin, C.; Andersson, M.; Joët, T.; Tranbarger, T.J.; Pizot, M.; Sarah, G.; Omore, A.; Durand-Gasselin, T.; Morcillo, F. Comparative transcriptome analysis of three oil palm fruit and seed tissues that differ in oil content and fatty acid composition. Plant Physiol. 2013, 162, 1337–1358. [Google Scholar] [CrossRef]
- Zhang, A.; Jin, L.; Yarra, R.; Cao, H.; Chen, P.; John Martin, J.J. Transcriptome analysis reveals key developmental and metabolic regulatory aspects of oil palm (Elaeis guineensis Jacq.) during zygotic embryo development. BMC Plant Biol. 2022, 22, 112. [Google Scholar] [CrossRef]
- Henson, I.E. Comparative ecophysiology of oil palm and tropical rainforests. In Oil Palm and the Environment—A Malaysian Perspective; Gurmit, S., Ed.; Malaysian Oil Palm Growers Council: Kuala Lumpur, Malaysia, 1999; pp. 9–39. [Google Scholar]
- Lamade, E.; Tcherkez, G.; Darlan, N.H.; Rodrigues, R.L.; Fresneau, C.; Mauve, C.; Lamothe-Sibold, M.; Sketriené, D.; Ghashghaie, J. Natural 13C distribution in oil palm (Elaeis guineensis Jacq.) and consequences for allocation pattern. Plant Cell Environ. 2016, 39, 199–212. [Google Scholar] [CrossRef]
- Lamade, E.; Fresneau, C.; Lamothe, M.; Mauve, C.; Tcherkez, G.; Ghashghaie, J. Exploring carbon allocation patterns using natural carbon isotope abundance in oil palm in a North Sumatra environment. In Proceedings of the First National Symposium of Scientific Ecology, Montpellier, France, 2–4 September 2010; pp. 31–32. [Google Scholar]
- Abadie, C.; Lalande, J.; Tcherkez, G. Exact mass GC-MS analysis: Protocol, database, advantages and application to plant metabolic profiling. Plant Cell Environ. 2022, 45, 3171–3183. [Google Scholar] [CrossRef] [PubMed]
- Fraisier-Vannier, O.; Chervin, J.; Cabanac, G.; Puech, V.; Fournier, S.; Durand, V.; Amiel, A.; André, O.; Benamar, O.A.; Dumas, B. MS-CleanR: A feature-filtering workflow for untargeted LC–MS based metabolomics. Anal. Chem. 2020, 92, 9971–9981. [Google Scholar] [CrossRef] [PubMed]
- Chassagne, F.; Cabanac, G.; Hubert, G.; David, B.; Marti, G. The landscape of natural product diversity and their pharmacological relevance from a focus on the Dictionary of Natural Products®. Phytochem. Rev. 2019, 18, 601–622. [Google Scholar] [CrossRef]
- Yang, Z.; Liu, Z.; Xu, H.; Li, Y.; Huang, S.; Cao, G.; Shi, M.; Zhu, J.; Zhou, J.; Li, R.; et al. ArecaceaeMDB: A comprehensive multi-omics database for Arecaceae breeding and functional genomics studies. Plant Biotechnol. J. 2023, 21, 11–13. [Google Scholar] [CrossRef]
- Zhou, S.; Kremling, K.A.; Bandillo, N.; Richter, A.; Zhang, Y.K.; Ahern, K.R.; Artyukhin, A.B.; Hui, J.X.; Younkin, G.C.; Schroeder, F.C.; et al. Metabolome-scale genome-wide association studies reveal chemical diversity and genetic control of maize specialized metabolites. Plant Cell 2019, 31, 937–955. [Google Scholar] [CrossRef]
- Chen, J.; Xue, M.; Liu, H.; Fernie, A.R.; Chen, W. Exploring the genic resources underlying metabolites through mGWAS and mQTL in wheat: From large-scale gene identification and pathway elucidation to crop improvement. Plant Commun. 2021, 2, 100216. [Google Scholar] [CrossRef]
- Razzaq, A.; Wishart, D.S.; Wani, S.H.; Hameed, M.K.; Mubin, M.; Saleem, F. Advances in metabolomics-driven diagnostic breeding and crop improvement. Metabolites 2022, 12, 511. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Liu, S.; Wang, T.; Li, F.; Cheng, J.; Lai, J.; Qin, F.; Li, Z.; Wang, X.; Jiang, C. Metabolomics-driven gene mining and genetic improvement of tolerance to salt-induced osmotic stress in maize. New Phytol. 2021, 230, 2355–2370. [Google Scholar] [CrossRef]
- Rival, A. Breeding the oil palm (Elaeis guineensis Jacq.) for climate change. Oilseed Fat Crops Lipids 2017, 24, D107. [Google Scholar]
- Rival, A.; Bessou, C. Climate change is challenging oil palm (Elaeis guineensis Jacq.) production systems. In Cultivation for Climate Change Resilience; Abdul-Soad, A., Al-Khayri, J., Eds.; CRC Press: Boca-Raton, FL, USA, 2023; Volume 1, pp. 109–126. [Google Scholar]
- Bahariah, B.; Masani, M.Y.A.; Jamaludin, N.; Fizree, M.D.P.M.A.A.; Syuhada, W.S.W.N.; Rasid, O.A.; Parveez, G.K.A. Application of CRISPR/Cas9-Mediated Genome Editing for Trait Improvement in Oil Palm. In Industrial Crop Plants; Kumar, N., Ed.; Springer Nature: Singapore, 2024; pp. 201–226. [Google Scholar]
- Bahariah, B.; Masani, M.Y.A.; Fizree, M.P.M.A.A.; Rasid, O.A.; Parveez, G.K.A. Multiplex CRISPR/Cas9 gene-editing platform in oil palm targeting mutations in EgFAD2 and EgPAT genes. J. Genet. Eng. Biotechnol. 2023, 21, 3. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nugroho, R.A.P.; Zaag, I.; Lamade, E.; Lukman, R.; Caliman, J.-P.; Tcherkez, G. Metabolomics-Assisted Breeding in Oil Palm: Potential and Current Perspectives. Int. J. Mol. Sci. 2024, 25, 9833. https://doi.org/10.3390/ijms25189833
Nugroho RAP, Zaag I, Lamade E, Lukman R, Caliman J-P, Tcherkez G. Metabolomics-Assisted Breeding in Oil Palm: Potential and Current Perspectives. International Journal of Molecular Sciences. 2024; 25(18):9833. https://doi.org/10.3390/ijms25189833
Chicago/Turabian StyleNugroho, Rizki Anjal P., Ismail Zaag, Emmanuelle Lamade, Rudy Lukman, Jean-Pierre Caliman, and Guillaume Tcherkez. 2024. "Metabolomics-Assisted Breeding in Oil Palm: Potential and Current Perspectives" International Journal of Molecular Sciences 25, no. 18: 9833. https://doi.org/10.3390/ijms25189833
APA StyleNugroho, R. A. P., Zaag, I., Lamade, E., Lukman, R., Caliman, J.-P., & Tcherkez, G. (2024). Metabolomics-Assisted Breeding in Oil Palm: Potential and Current Perspectives. International Journal of Molecular Sciences, 25(18), 9833. https://doi.org/10.3390/ijms25189833





