Mining of Candidate Genes Associated with Leaf Shape Traits in Grapes
Abstract
1. Introduction
2. Results
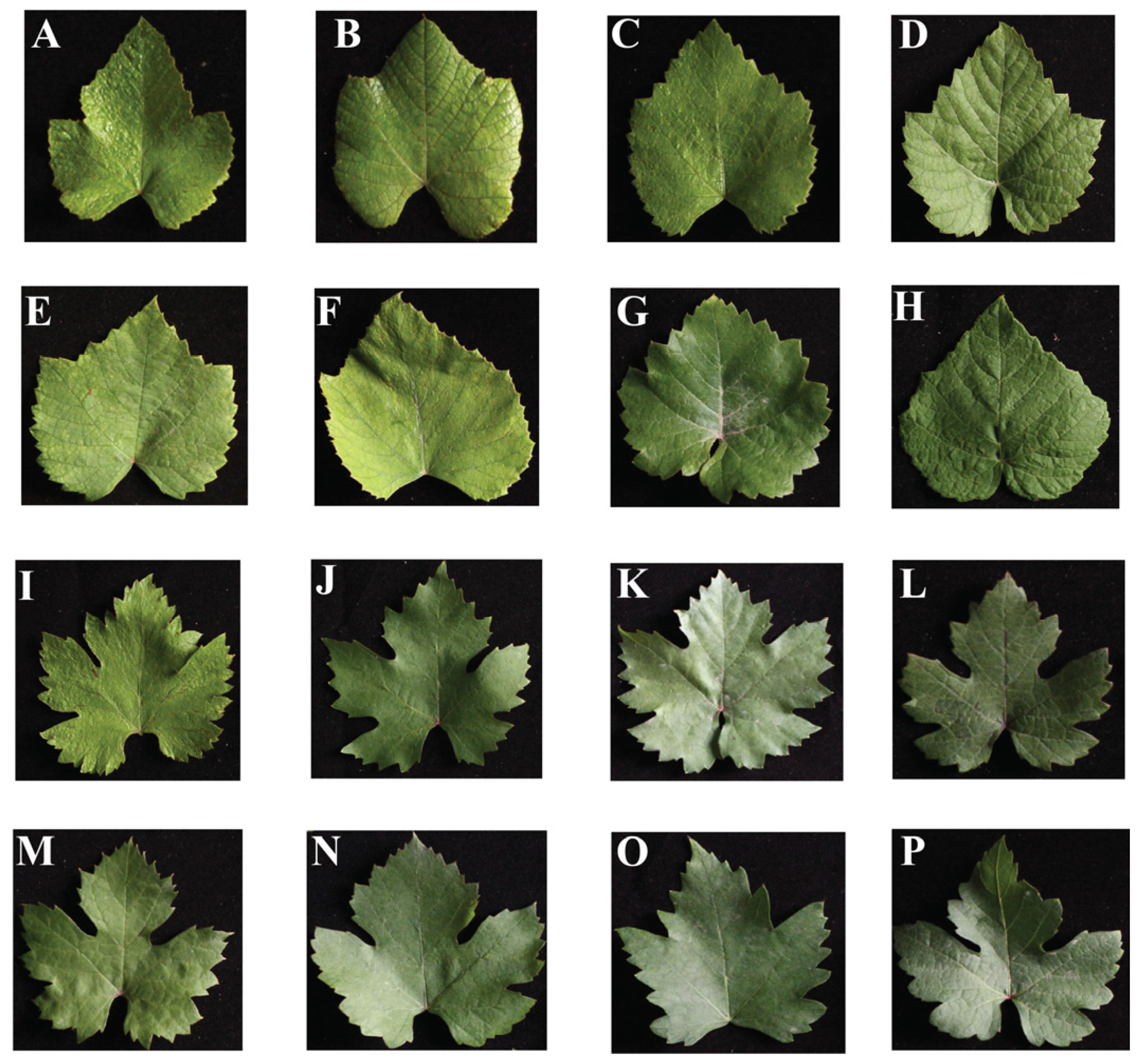
2.1. Changes in Grape Leaf Shape Character Parameters Determined Using TA
2.2. Principal Component Analysis and Correlation Analysis of Grape Leaf Shape Trait-Related Parameters
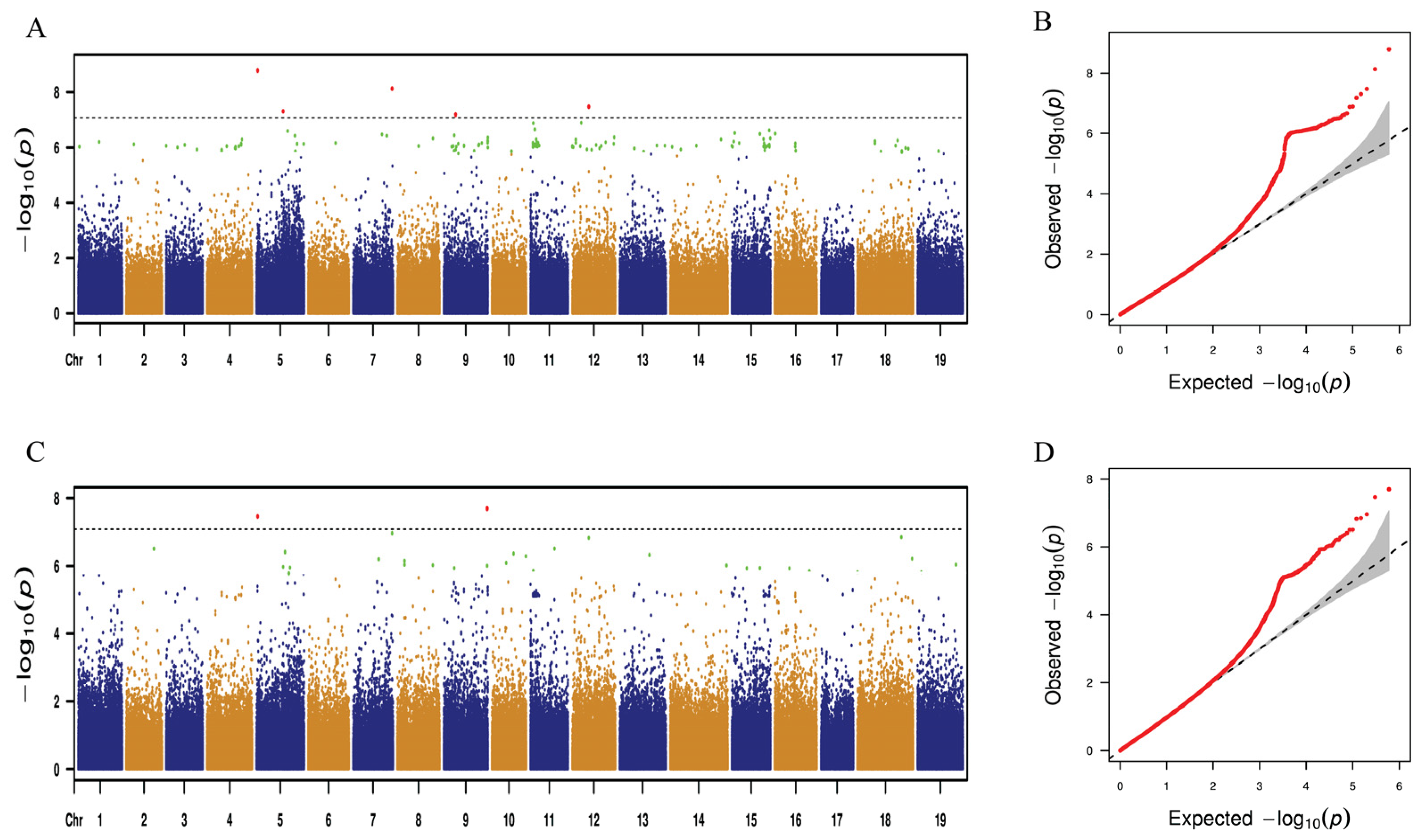
2.3. GWAS for Grape Leaf Traits
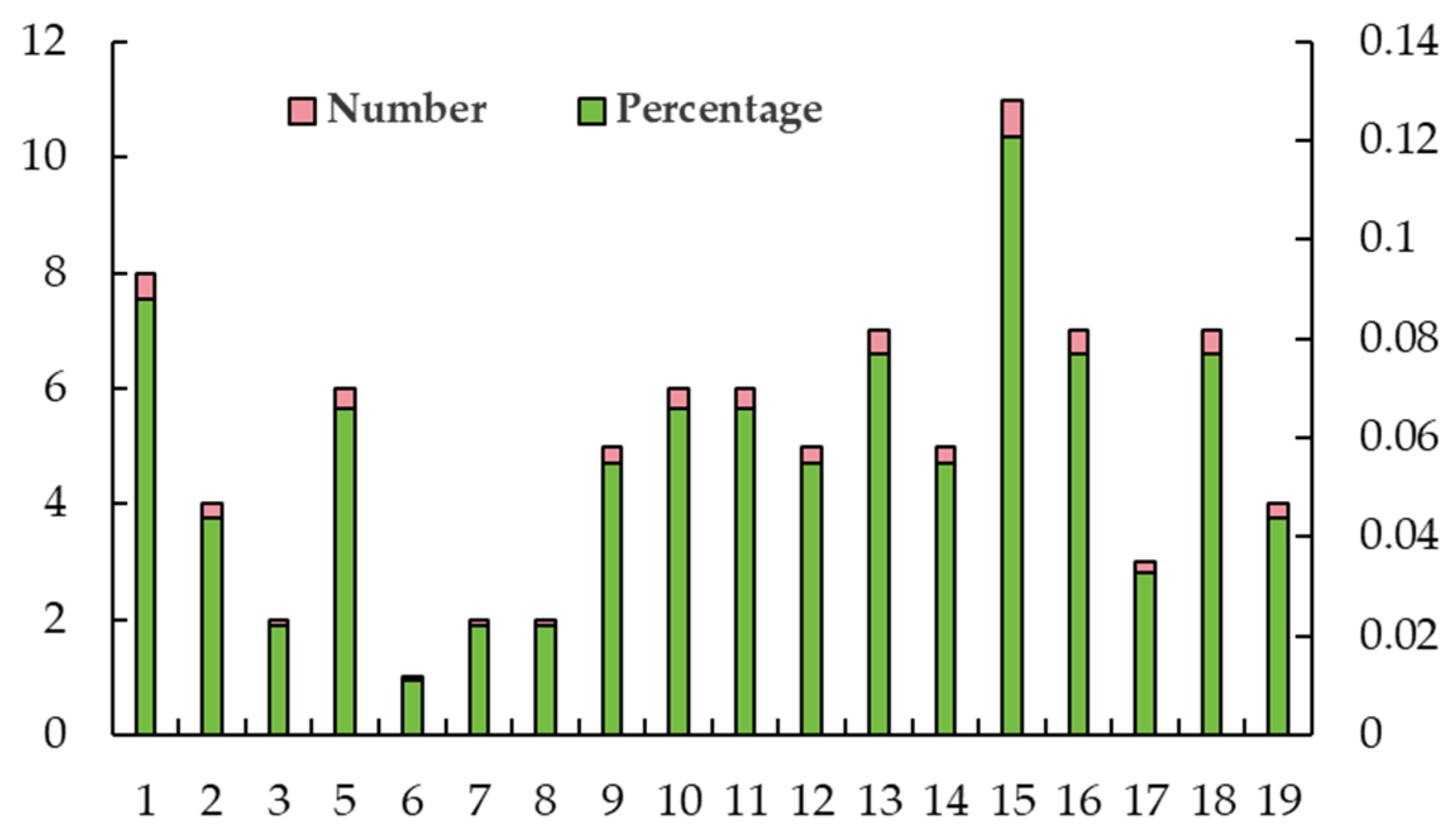
2.4. Distribution of SNP Loci Controlling Leaf Traits on Chromosomes
2.5. Candidate Genes for Leaf Traits
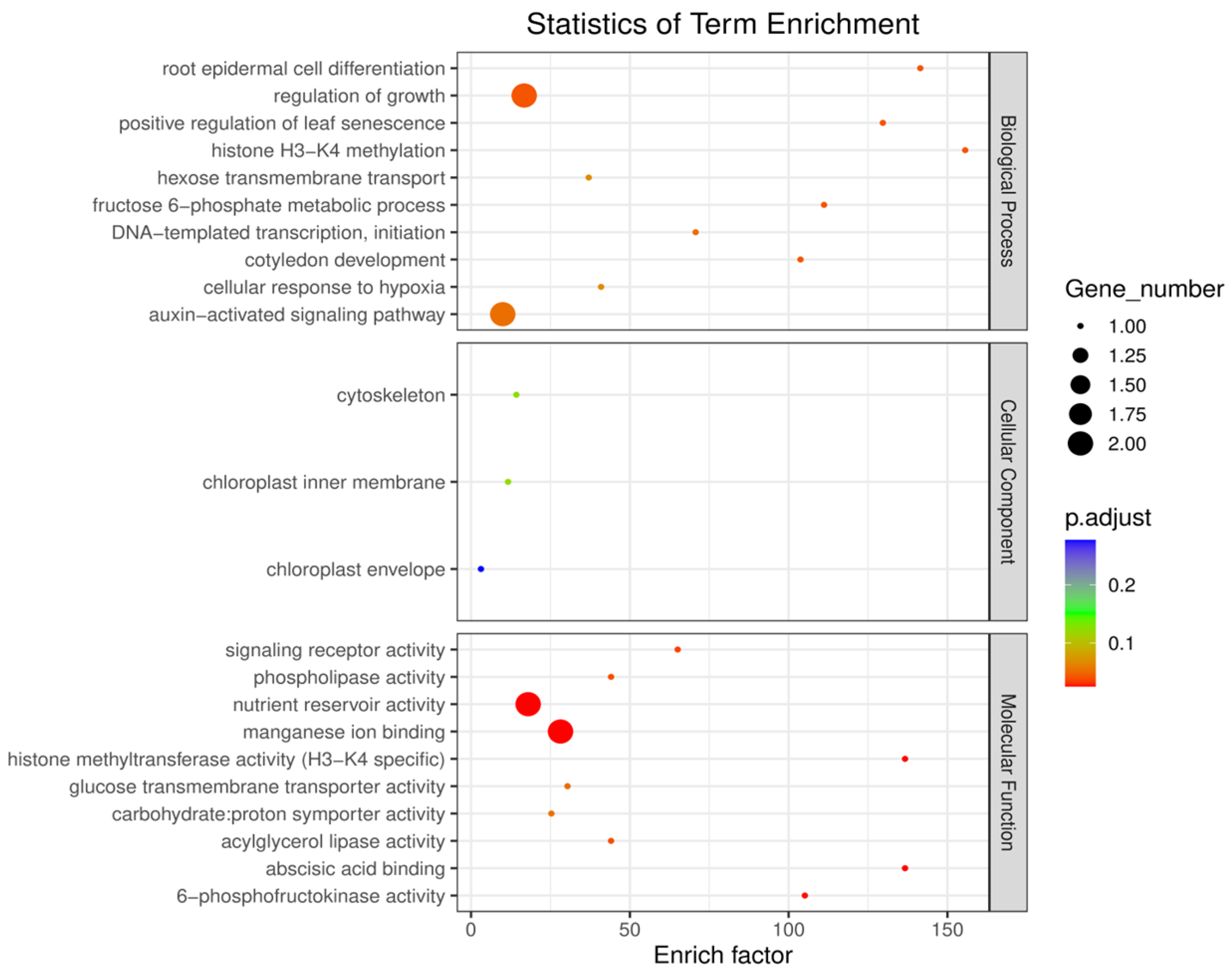
2.6. Enrichment Analysis of Candidate Genes for Grape Leaf Traits
2.7. Tissue-Specific Expression Analysis of Candidate Genes
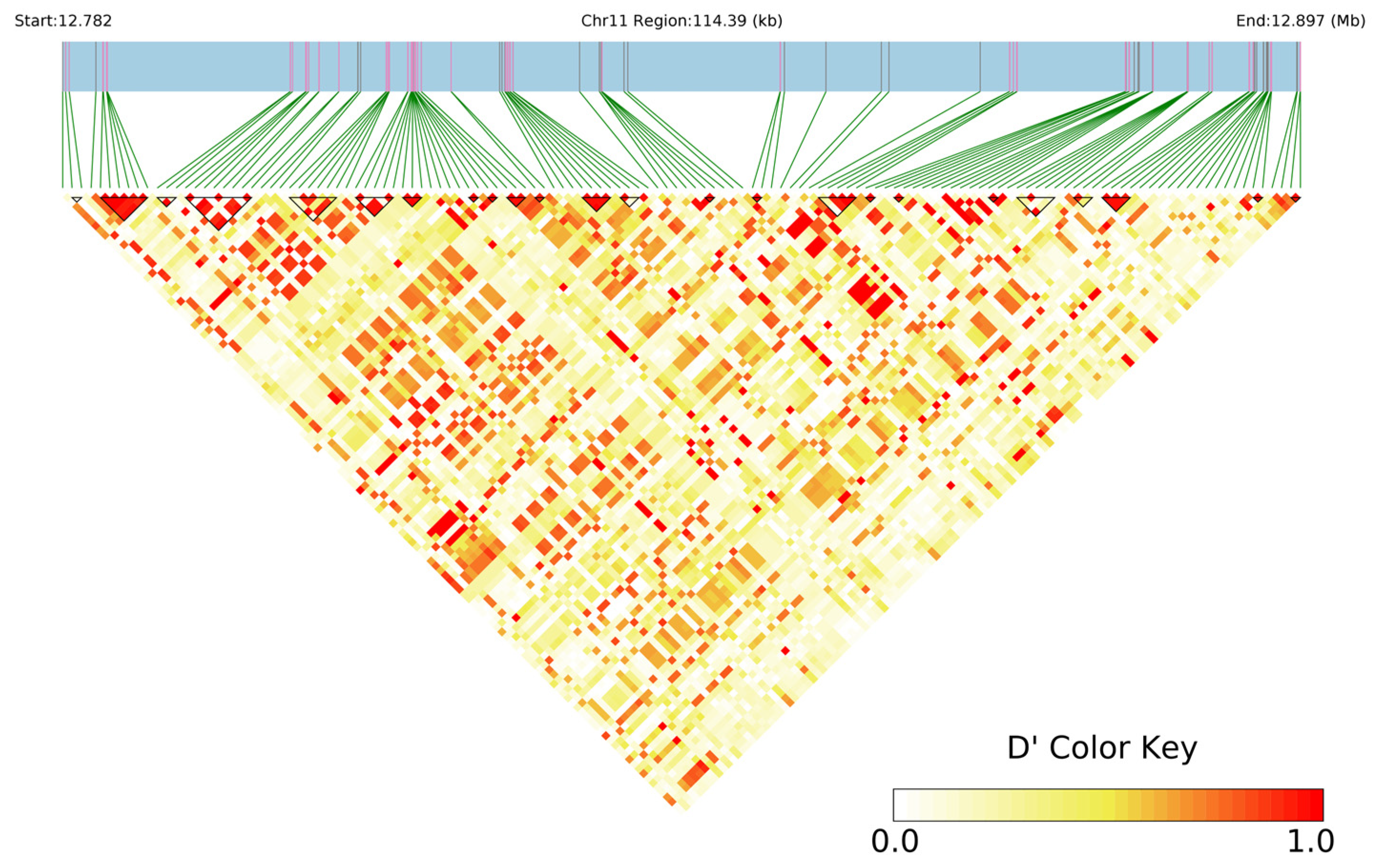
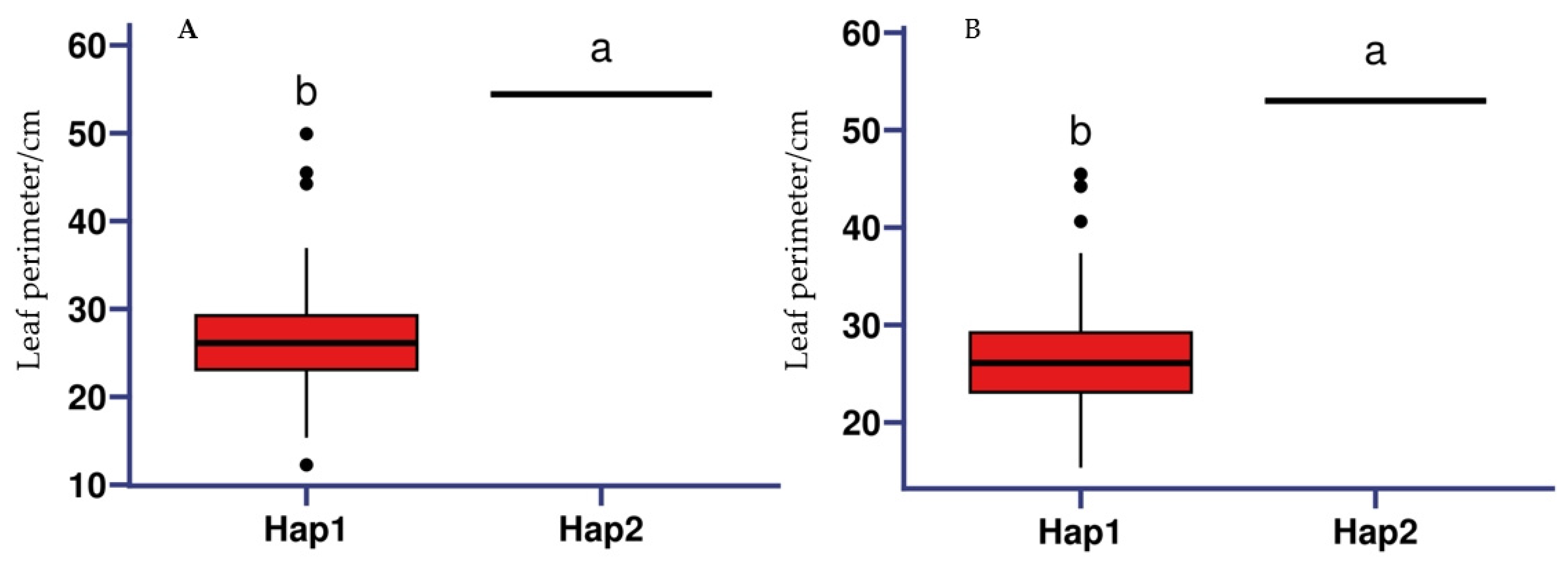
2.8. Haplotype Analysis of Candidate Genes Related to Grape Leaf Traits
3. Discussion
3.1. TA Greatly Expands Its Application Range in Measuring Plant Phenotype and Organ Morphology and Identifying Leaf Traits of Grape Germplasm Resources
3.2. GWAS of Grape Leaf Shape Trait-Related Genes
4. Materials and Methods
4.1. Grape Resources and Sample Collection
4.2. Relevant Parameters of Grape Leaf Shape Were Analyzed Using TA
4.3. Genome-Wide Association Study
4.4. Annotation and Function Prediction of Candidate Genes
4.5. Haplotype Analysis and Expression Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsukaya, H. Comparative leaf development in angiosperms. Curr. Opin. Plant Biol. 2014, 17, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Yahiaoui, I.; Mzoughi, O.; Boujemaa, N. Leaf Shape Descriptor for Tree Species Identification. In Proceedings of the 2012 IEEE International Conference on Multimedia and Expo, IEEE Computer Society, Melbourne, VIC, Australia, 9–13 July 2012; pp. 254–259. [Google Scholar]
- Conklin, P.A.; Strable, J.; Li, S.; Scanlon, M.J. On the mechanisms of development in monocot and eudicot leaves. New Phytol. 2019, 221, 706–724. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kong, F.; Zhou, C. From genes to networks: The genetic control of leaf development. J. Integr. Plant Biol. 2021, 63, 1181–1196. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, M.D. Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol. 2009, 149, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.G.; Long, S.P.; Ort, D.R. Improving photosynthetic efficiency for greater yield. Annu. Rev. Plant Biol. 2010, 61, 235–261. [Google Scholar] [CrossRef]
- Leigh, A.; Sevanto, S.; Close, J.D.; Nicotra, A.B. The influence of leaf size and shape on leaf thermal dynamics: Does theory hold up under natural conditions? Plant Cell Environ. 2017, 40, 237–248. [Google Scholar] [CrossRef]
- Galet, P. Precis d’Ampelographie Pratique. Impr. P. Dehan. Montpellier. 1952, 256. [Google Scholar]
- Chitwood, D.H.; Klein, L.L.; O’Hanlon, R.; Chacko, S.; Greg, M.; Kitchen, C.; Miller, A.J.; Londo, J.P. Latent developmental and evolutionary shapes embedded within the grapevine leaf. New Phytol. 2016, 210, 343–355. [Google Scholar] [CrossRef]
- Moore, M.O. Classification and systematics of eastern North American vitis L.(Vitaceae) north of Mexico. Sida Contrib. Bot. 1991, 14, 339–367. [Google Scholar]
- Yan, L.-H.; Qi, C.-J.; Liu, X.-X. A study on the flora of the seed vines in Central China region. Bull. Bot. Res. 2006, 26, 497. [Google Scholar]
- Mullins, M. Biology of the Grapevine; Cambridge University Press: Cambridge, UK, 1992. [Google Scholar]
- Chitwood, D.H.; Ranjan, A.; Martinez, C.C.; Headland, L.R.; Thiem, T.; Kumar, R.; Covington, M.F.; Hatcher, T.; Naylor, D.T.; Zimmerman, S. A modern ampelography: A genetic basis for leaf shape and venation patterning in grape. Plant Physiol. 2014, 164, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Barthélémy, D.; Boujemaa, N.; Mathieu, D.; Molino, J.-F.; Joly, A.; Mouysset, E. The Pl@ntnet project: Plant computational identification and collaborative information system. In Proceedings of the IBC2011, XVIII International Botanical Congress, Melbourne, Australia, 23 July 2011. [Google Scholar]
- Wäldchen, J.; Mäder, P. Plant Species Identification Using Computer Vision Techniques: A Systematic Literature Review. Arch. Comput. Methods Eng. 2018, 25, 507–543. [Google Scholar] [CrossRef] [PubMed]
- Cope, J.S.; Corney, D.; Clark, J.Y.; Remagnino, P.; Wilkin, P. Plant species identification using digital morphometrics: A review. Expert Syst. Appl. 2012, 39, 7562–7573. [Google Scholar] [CrossRef]
- Upadhyaya, H.; Gowda, C.; Sastry, D. Plant genetic resources management: Collection, characterization, conservation and utilization. J. SAT Agric. Res. 2008, 6, 16. [Google Scholar]
- Zhang, S.; Huang, W.; Huang, Y.-a.; Zhang, C. Plant species recognition methods using leaf image: Overview. Neurocomputing 2020, 408, 246–272. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Cobb, J.N.; DeClerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef]
- Németh, M. Ampelográfiai album. Termesztett borszőlőfajták 1. In Ampelographic Album, Cultivated Grapevine Cultivars I; Mezőgazdasági Kiadó: Budapest, Hungary, 1967. [Google Scholar]
- Bešlić, Z.; Todić, S.; Rakonjac, V. Inheritance of some morphological traits in hybridization of grapevine cultivars Drenak crveni and Afuz-ali. Genetika 2005, 37, 137–144. [Google Scholar] [CrossRef]
- Bodor, P.; Baranyai, L.; Ladányi, M.; Bálo, B.; Strever, A.; Bisztray, G.; Hunter, J. Stability of ampelometric characteristics of Vitis vinifera L. cv. ‘Syrah’ and ‘Sauvignon blanc’ leaves: Impact of within-vineyard variability and pruning method/bud load. S. Afr. J. Enol. Vitic. 2013, 34, 129–137. [Google Scholar]
- Wang, H.; Xu, Y.; Hong, L.; Zhang, X.; Wang, X.; Zhang, J.; Ding, Z.; Meng, Z.; Wang, Z.Y.; Long, R.; et al. HEADLESS Regulates Auxin Response and Compound Leaf Morphogenesis in Medicago truncatula. Front. Plant Sci. 2019, 10, 1024. [Google Scholar] [CrossRef]
- He, L.; Liu, Y.; He, H.; Liu, Y.; Qi, J.; Zhang, X.; Li, Y.; Mao, Y.; Zhou, S.; Zheng, X. A molecular framework underlying the compound leaf pattern of Medicago truncatula. Nat. Plants 2020, 6, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Yang, B.; Zhang, A.; Zhang, L.; Xu, X.; Li, J.; Liu, L. Screening of candidate leaf morphology genes by integration of QTL mapping and RNA sequencing technologies in oilseed rape (Brassica napus L.). PLoS ONE 2017, 12, e0169641. [Google Scholar] [CrossRef] [PubMed]
- Fasoli, M.; Dal Santo, S.; Zenoni, S.; Tornielli, G.B.; Farina, L.; Zamboni, A.; Porceddu, A.; Venturini, L.; Bicego, M.; Murino, V. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell 2012, 24, 3489–3505. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Moyseenko, J.B.; Robbins, M.D.; Morejón, N.H.; Francis, D.M.; van der Knaap, E. Tomato Analyzer: A useful software application to collect accurate and detailed morphological and colorimetric data from two-dimensional objects. JoVE (J. Vis. Exp.) 2010, 37, 1856. [Google Scholar]
- Gonzalo, M.J.; Van Der Knaap, E. A comparative analysis into the genetic bases of morphology in tomato varieties exhibiting elongated fruit shape. Theor. Appl. Genet. 2008, 116, 647–656. [Google Scholar] [CrossRef]
- Mazzucato, A.; Ficcadenti, N.; Caioni, M.; Mosconi, P.; Piccinini, E.; Sanampudi, V.R.R.; Sestili, S.; Ferrari, V. Genetic diversity and distinctiveness in tomato (Solanum lycopersicum L.) landraces: The Italian case study of ‘A pera Abruzzese’. Sci. Hortic. 2010, 125, 55–62. [Google Scholar] [CrossRef]
- Hurtado, M.; Vilanova, S.; Plazas, M.; Gramazio, P.; Herraiz, F.J.; Andújar, I.; Prohens, J. Phenomics of fruit shape in eggplant (Solanum melongena L.) using Tomato Analyzer software. Sci. Hortic. 2013, 164, 625–632. [Google Scholar] [CrossRef]
- Tripodi, P.; Greco, B. Large scale phenotyping provides insight into the diversity of vegetative and reproductive organs in a wide collection of wild and domesticated peppers (Capsicum spp.). Plants 2018, 7, 103. [Google Scholar] [CrossRef]
- Brewer, M.T.; Lang, L.; Fujimura, K.; Dujmovic, N.; Gray, S.; van der Knaap, E. Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 2006, 141, 15–25. [Google Scholar] [CrossRef]
- Brewer, M.T.; Moyseenko, J.B.; Monforte, A.J.; van der Knaap, E. Morphological variation in tomato: A comprehensive study of quantitative trait loci controlling fruit shape and development. J. Exp. Bot. 2007, 58, 1339–1349. [Google Scholar] [CrossRef]
- Gonzalo, M.J.; Brewer, M.T.; Anderson, C.; Sullivan, D.; Gray, S.; van der Knaap, E. Tomato fruit shape analysis using morphometric and morphology attributes implemented in Tomato Analyzer software program. J. Am. Soc. Hortic. Sci. 2009, 134, 77–87. [Google Scholar] [CrossRef]
- Orsi, C.H.; Tanksley, S.D. Natural variation in an ABC transporter gene associated with seed size evolution in tomato species. PLoS Genet. 2009, 5, e1000347. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Cai, X.; Yuan, W.; Vick, B.; Hu, J. Mapping Quantitative Trait Loci (QTL) Controlling Seed Morphology and Disk Diameter in Sunflower (Helianthus annuus L.)/Mapeo de Loci Para Caracteres Cuantitativos (QTL) Que Controlan Morfología de Semillas y Diámetro del Disco en Girasol (Helianthus annuus L.)/Établissement D’une Carte de Loci des Caractéristiques Quantitatives (QTL) Contrôlant la Morphologie de la Graine et le Diamètre du Capitule du Tournesol (Helianthus annuus L.). Helia 2009, 32, 17–36. [Google Scholar]
- Xiao, H.; Jiang, N.; Schaffner, E.; Stockinger, E.J.; van der Knaap, E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 2008, 319, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef]
- Scarpella, E.; Barkoulas, M.; Tsiantis, M. Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. 2010, 2, a001511. [Google Scholar] [CrossRef]
- Byrne, M.E. Making leaves. Curr. Opin. Plant Biol. 2012, 15, 24–30. [Google Scholar] [CrossRef]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inzé, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef]
- Vanhaeren, H.; Gonzalez, N.; Coppens, F.; De Milde, L.; Van Daele, T.; Vermeersch, M.; Eloy, N.B.; Storme, V.; Inzé, D. Combining growth-promoting genes leads to positive epistasis in Arabidopsis thaliana. eLife 2014, 3, e02252. [Google Scholar] [CrossRef] [PubMed]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Finkelstein, R.R.; Gampala, S.S.; Rock, C.D. Abscisic acid signaling in seeds and seedlings. Plant Cell 2002, 14 (Suppl. S1), S15–S45. [Google Scholar] [CrossRef]
- Cornforth, J.W.; Milborrow, B.V.; Ryback, G.; Wareing, P.F. Chemistry and Physiology of ‘Dormins’ In Sycamore: Identity of Sycamore ‘Dormin’ with Abscisin II. Nature 1965, 205, 1269–1270. [Google Scholar] [CrossRef]
- Finkelstein, R.; Reeves, W.; Ariizumi, T.; Steber, C. Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 2008, 59, 387–415. [Google Scholar] [CrossRef]
- Ohkuma, K.; Lyon, J.L.; Addicott, F.T.; Smith, O.E. Abscisin II, an Abscission-Accelerating Substance from Young Cotton Fruit. Science 1963, 142, 1592–1593. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef]
- Xing, L.; Zhao, Y.; Gao, J.; Xiang, C.; Zhu, J.-K. The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci. Rep. 2016, 6, 27177. [Google Scholar] [CrossRef]
- Boneh, U.; Biton, I.; Schwartz, A.; Ben-Ari, G. Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 2012, 187, 89–96. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Li, S.; Yang, L.; Liang, Z. ABA signaling pathway genes and function during abiotic stress and berry ripening in Vitis vinifera. Gene 2021, 769, 145226. [Google Scholar] [CrossRef]
- Ren, C.; Kuang, Y.; Lin, Y.; Guo, Y.; Li, H.; Fan, P.; Li, S.; Liang, Z. Overexpression of grape ABA receptor gene VaPYL4 enhances tolerance to multiple abiotic stresses in Arabidopsis. BMC Plant Biol. 2022, 22, 271. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Dumenil, J.; Lu, F.-H.; Na, L.; Vanhaeren, H.; Naumann, C.; Klecker, M.; Prior, R.; Smith, C.; McKenzie, N. Ubiquitylation activates a peptidase that promotes cleavage and destabilization of its activating E3 ligases and diverse growth regulatory proteins to limit cell proliferation in Arabidopsis. Genes Dev. 2017, 31, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, L.; Corke, F.; Smith, C.; Bevan, M.W. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008, 22, 1331–1336. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Yang, W. E3 ubiquitin ligases: Ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Zhu, L.; Bu, Q.-Y.; Huq, E. MAX2 affects multiple hormones to promote photomorphogenesis. Mol. Plant 2012, 5, 750–762. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.T.; Scaffidi, A.; Sun, Y.K.; Flematti, G.R.; Smith, S.M. The karrikin response system of Arabidopsis. Plant J. 2014, 79, 623–631. [Google Scholar] [CrossRef]
- Piao, C.; Gao, Z.; Yuan, S.; Li, F.; Cui, M.L. The R2R3-MYB gene CgMYB4 is involved in the regulation of cell differentiation and fiber development in the stamens of Chelone glabra L. Protoplasma 2022, 259, 1397–1407. [Google Scholar] [CrossRef]
- Oppenheimer, D.G.; Herman, P.L.; Sivakumaran, S.; Esch, J.; Marks, M.D. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 1991, 67, 483–493. [Google Scholar] [CrossRef]
- Kang, Y.H.; Kirik, V.; Hulskamp, M.; Nam, K.H.; Hagely, K.; Lee, M.M.; Schiefelbein, J. The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 2009, 21, 1080–1094. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Nukumizu, Y.; Sato, S.; Kato, T.; Tabata, S.; Wada, T. Functional divergence of MYB-related genes, WEREWOLF and AtMYB23 in Arabidopsis. Biosci. Biotechnol. Biochem. 2012, 76, 883–887. [Google Scholar] [CrossRef][Green Version]
- Lee, M.M.; Schiefelbein, J. WEREWOLF, a MYB-Related Protein in Arabidopsis, Is a Position-Dependent Regulator of Epidermal Cell Patterning. Cell 1999, 99, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Vannini, A.; Cramer, P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 2012, 45, 439–446. [Google Scholar] [CrossRef] [PubMed]
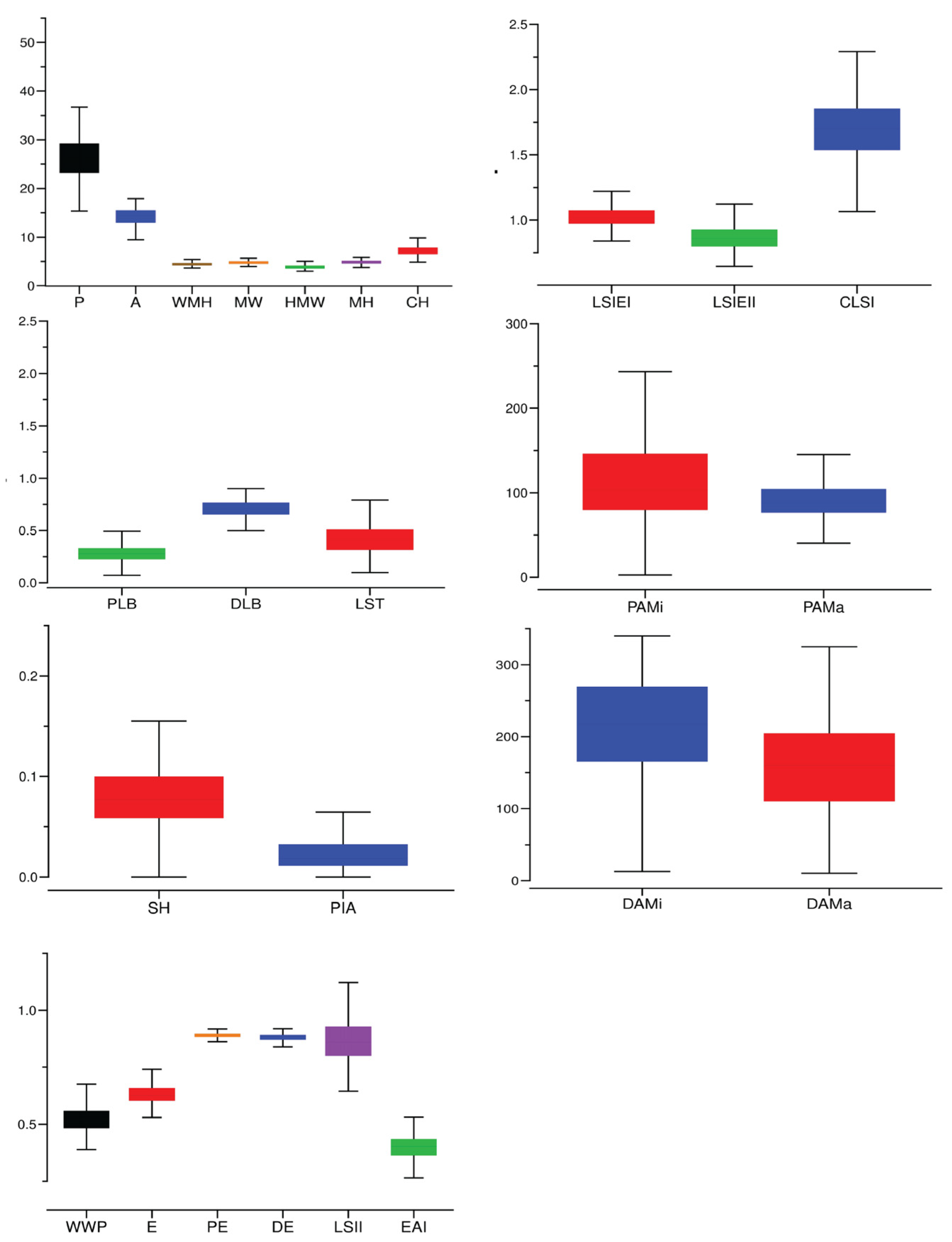
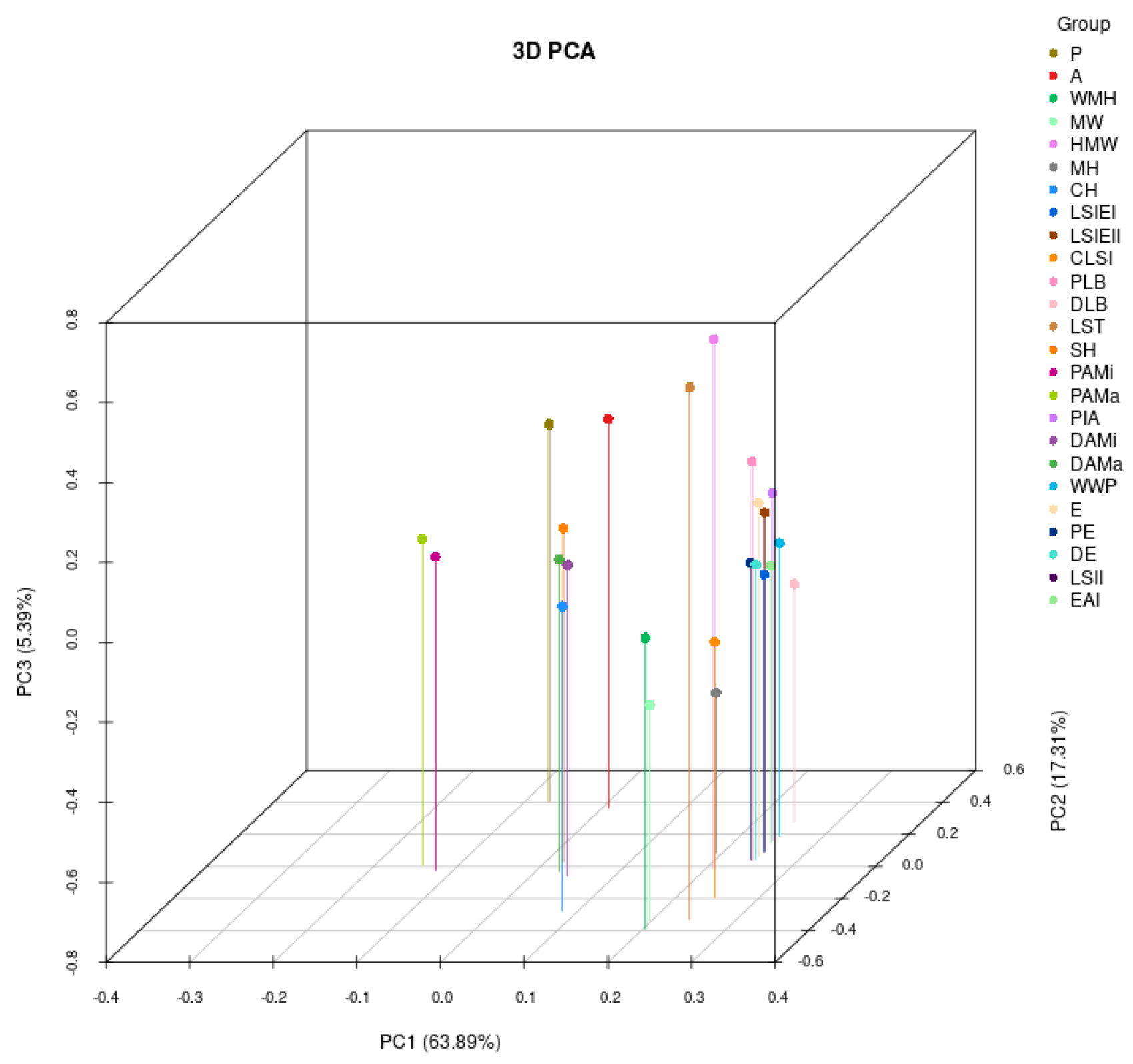
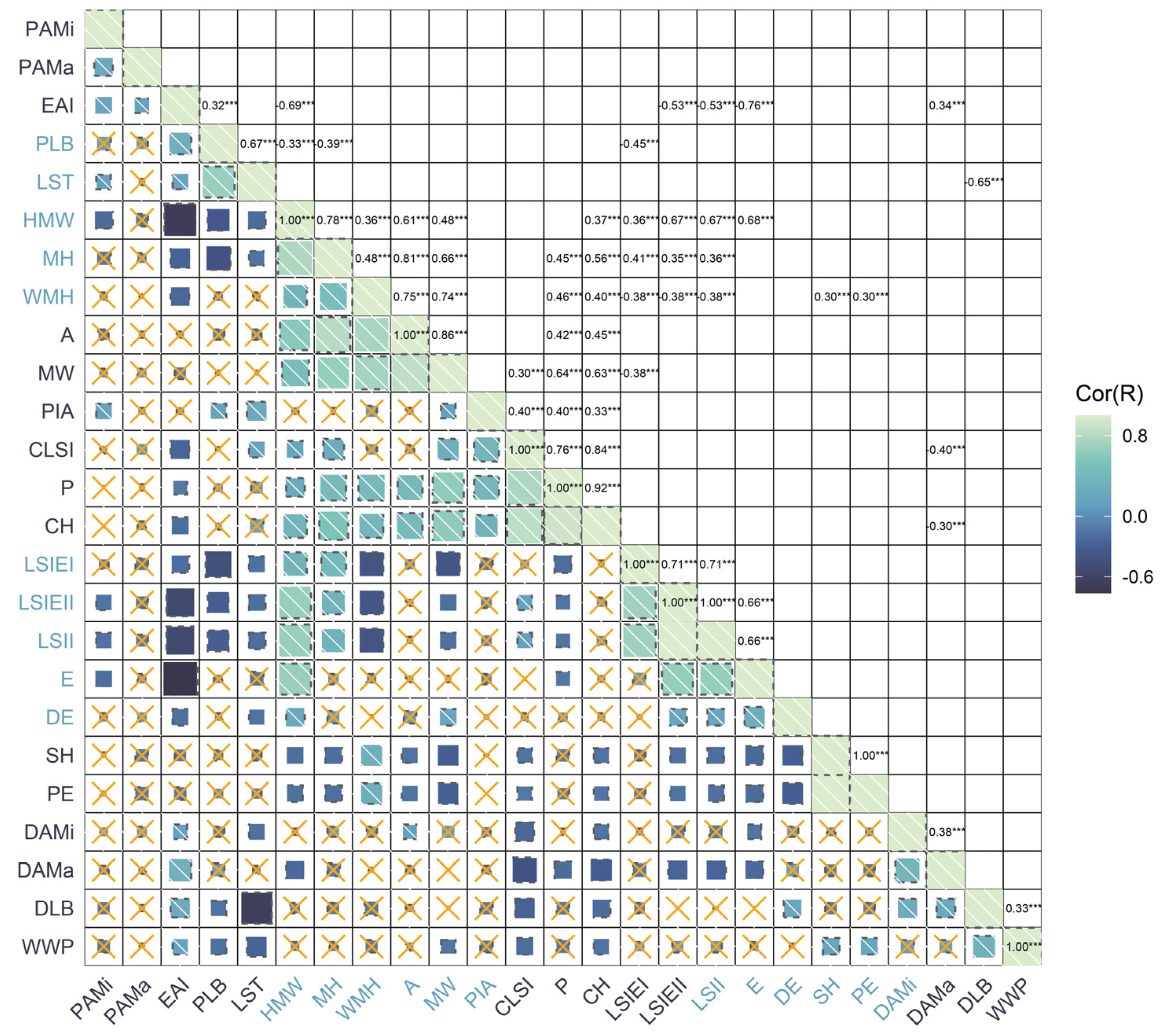


| Characteristic | Maximum | Minimum | Mean | Standard Deviation | Coefficient of Variation |
|---|---|---|---|---|---|
| Area | 53.71 | 14.10 | 26.49 | 4.86 | 18.36% |
| Curved leaf shape index | 17.91 | 5.26 | 14.25 | 1.88 | 13.19% |
| Curved height | 6.30 | 2.61 | 4.44 | 0.40 | 8.90% |
| Distal-angle macro | 5.87 | 2.71 | 4.75 | 0.42 | 8.75% |
| Distal-angle micro | 5.03 | 2.49 | 3.83 | 0.44 | 11.56% |
| Distal eccentricity | 5.84 | 3.01 | 4.85 | 0.42 | 8.71% |
| Distal leaf blockiness | 12.46 | 3.92 | 7.23 | 1.16 | 16.03% |
| Eccentricity | 1.47 | 0.84 | 1.03 | 0.08 | 7.71% |
| Eccentricity area index | 1.34 | 0.60 | 0.87 | 0.10 | 11.68% |
| Leaf shape index external I | 2.61 | 1.07 | 1.71 | 0.26 | 15.06% |
| Leaf shape index external II | 0.57 | 0.07 | 0.29 | 0.09 | 30.36% |
| Leaf shape index internal | 0.95 | 0.16 | 0.70 | 0.10 | 14.49% |
| Leaf shape triangle | 2.44 | 0.10 | 0.45 | 0.23 | 50.23% |
| Height mid-width | 0.40 | 0.00 | 0.08 | 0.03 | 40.32% |
| Maximum height | 316.20 | 2.63 | 116.41 | 55.43 | 47.61% |
| Maximum width | 230.39 | 0.95 | 90.81 | 26.81 | 29.53% |
| Proximal-angle macro | 0.27 | 0.00 | 0.03 | 0.03 | 118.37% |
| Proximal-angle micro | 339.76 | 6.03 | 208.37 | 75.68 | 36.32% |
| Proximal eccentricity | 324.73 | 10.37 | 159.97 | 68.56 | 42.85% |
| Proximal leaf blockiness | 0.73 | 0.34 | 0.52 | 0.07 | 12.81% |
| Proximal indentation area | 0.77 | 0.45 | 0.63 | 0.05 | 7.29% |
| Shoulder height | 1.16 | 0.82 | 0.89 | 0.02 | 2.80% |
| Width mid-height | 1.03 | 0.73 | 0.88 | 0.04 | 4.07% |
| Width widest position | 1.34 | 0.57 | 0.87 | 0.10 | 11.74% |
| Perimeter | 0.55 | 0.18 | 0.40 | 0.06 | 14.47% |
| Gene ID | Location | Nr Annotation |
|---|---|---|
| VIT_01s0182g00160 | 1:13463652–13467087 | PREDICTED: galactoside 2-alpha-L-fucosyltransferase [Vitis vinifera] |
| VIT_03s0088g01090 | 3:9340147–9341749 | PREDICTED: RING finger protein 44 [Vitis vinifera] |
| VIT_05s0020g00420 | 5:2340655–2343157 | PREDICTED: polygalacturonase At1g48100 [Vitis vinifera] |
| VIT_05s0029g00040 | 5:14450243–14453228 | PREDICTED: cyclin-dependent kinase inhibitor 5 [Vitis vinifera] |
| VIT_05s0124g00250 | 5:21154095–21220251 | PREDICTED: histone-lysine N-methyltransferase ATX1 [Vitis vinifera] |
| VIT_09s0002g02020 | 9:1789739–1791953 | PREDICTED: putative F-box/LRR-repeat protein At5g02700 [Vitis vinifera] |
| VIT_10s0003g01920 | 10:6997420–7015431 | PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X1 [Vitis vinifera] |
| VIT_11s0078g00480 | 11:15480609–15481984 | PREDICTED: myb-related protein Myb4 [Vitis vinifera] |
| VIT_12s0178g00200 | 12:11429553–11433532 | PREDICTED: actin-101 isoform X1 [Vitis vinifera] |
| VIT_13s0047g00320 | 13:16190297–16192565 | PREDICTED: cell division cycle protein 123 homolog [Vitis vinifera] |
| VIT_14s0006g02400 | 14:19942477–19949902 | PREDICTED: putative germin-like protein 2-1 [Vitis vinifera] |
| VIT_14s0006g02420 | 14:19962832–19963625 | PREDICTED: putative germin-like protein 2-1 [Vitis vinifera] |
| VIT_14s0006g02720 | 14:20731846–20739925 | PREDICTED: plastid hexose transporter isoform X1 [Vitis vinifera] |
| VIT_15s0048g00530 | 15:14642820–14643902 | PREDICTED: auxin-responsive protein SAUR36 [Vitis vinifera] |
| VIT_15s0046g01050 | 15:18133272–18136212 | PREDICTED: abscisic acid receptor PYL9 [Vitis vinifera] |
| VIT_17s0053g00990 | 17:17733901–17735829 | PREDICTED: expansin-A10 [Vitis vinifera] |
| VIT_17s0053g01010 | 17:17937970–17944266 | PREDICTED: transcription initiation factor IIB-2 [Vitis vinifera] |
| VIT_19s0027g00020 | 19:18803078–18804735 | PREDICTED: serine/threonine-protein kinase WAG1 [Vitis vinifera] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Yadav, V.; Cui, L. Mining of Candidate Genes Associated with Leaf Shape Traits in Grapes. Int. J. Mol. Sci. 2024, 25, 12101. https://doi.org/10.3390/ijms252212101
Zhang C, Yadav V, Cui L. Mining of Candidate Genes Associated with Leaf Shape Traits in Grapes. International Journal of Molecular Sciences. 2024; 25(22):12101. https://doi.org/10.3390/ijms252212101
Chicago/Turabian StyleZhang, Chuan, Vivek Yadav, and Liwen Cui. 2024. "Mining of Candidate Genes Associated with Leaf Shape Traits in Grapes" International Journal of Molecular Sciences 25, no. 22: 12101. https://doi.org/10.3390/ijms252212101
APA StyleZhang, C., Yadav, V., & Cui, L. (2024). Mining of Candidate Genes Associated with Leaf Shape Traits in Grapes. International Journal of Molecular Sciences, 25(22), 12101. https://doi.org/10.3390/ijms252212101







