Identification of IGF-1 Effects on White Adipose Tissue and Hippocampus in Alzheimer’s Disease Mice via Transcriptomic and Cellular Analysis
Abstract
:1. Introduction
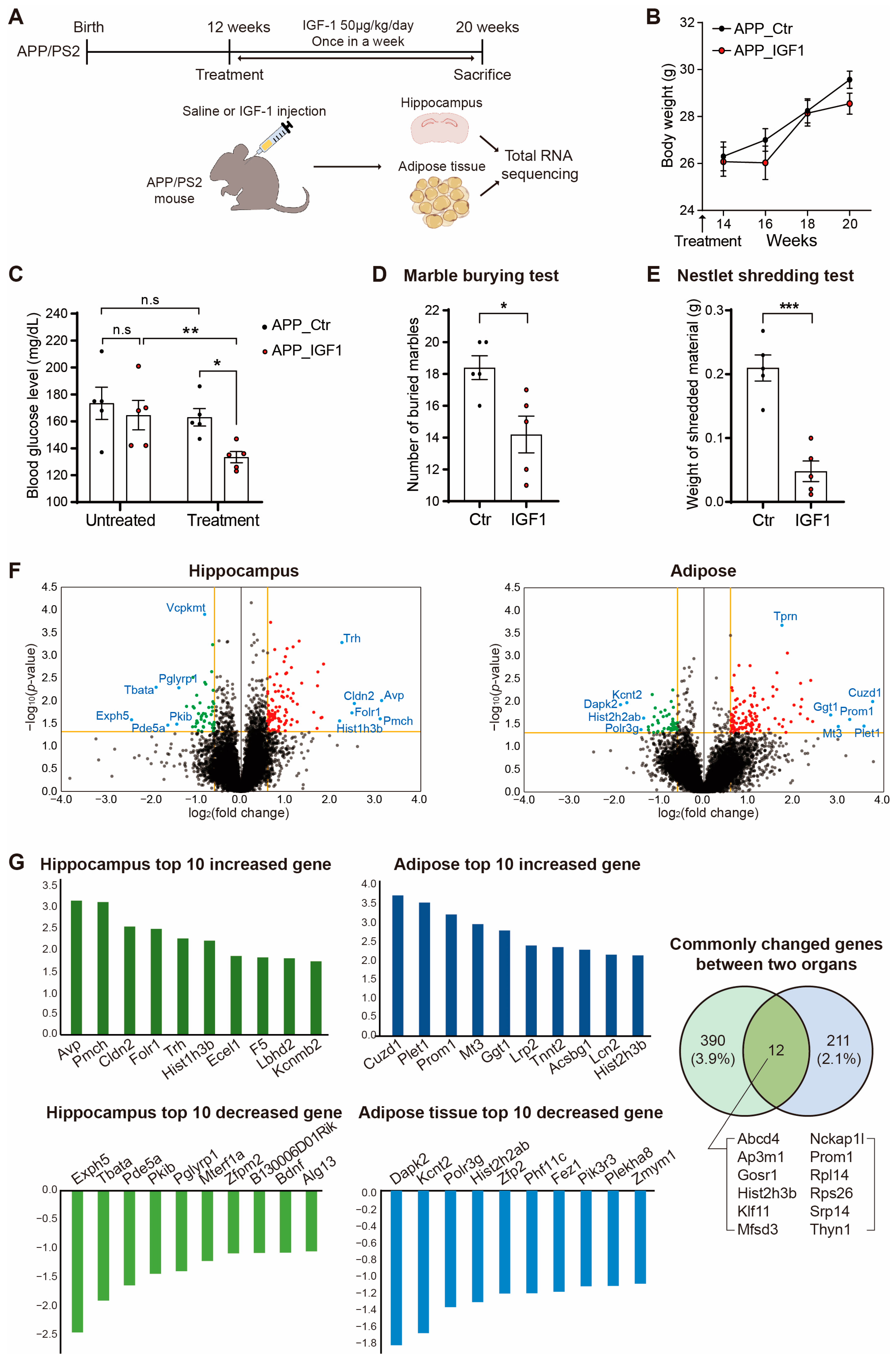
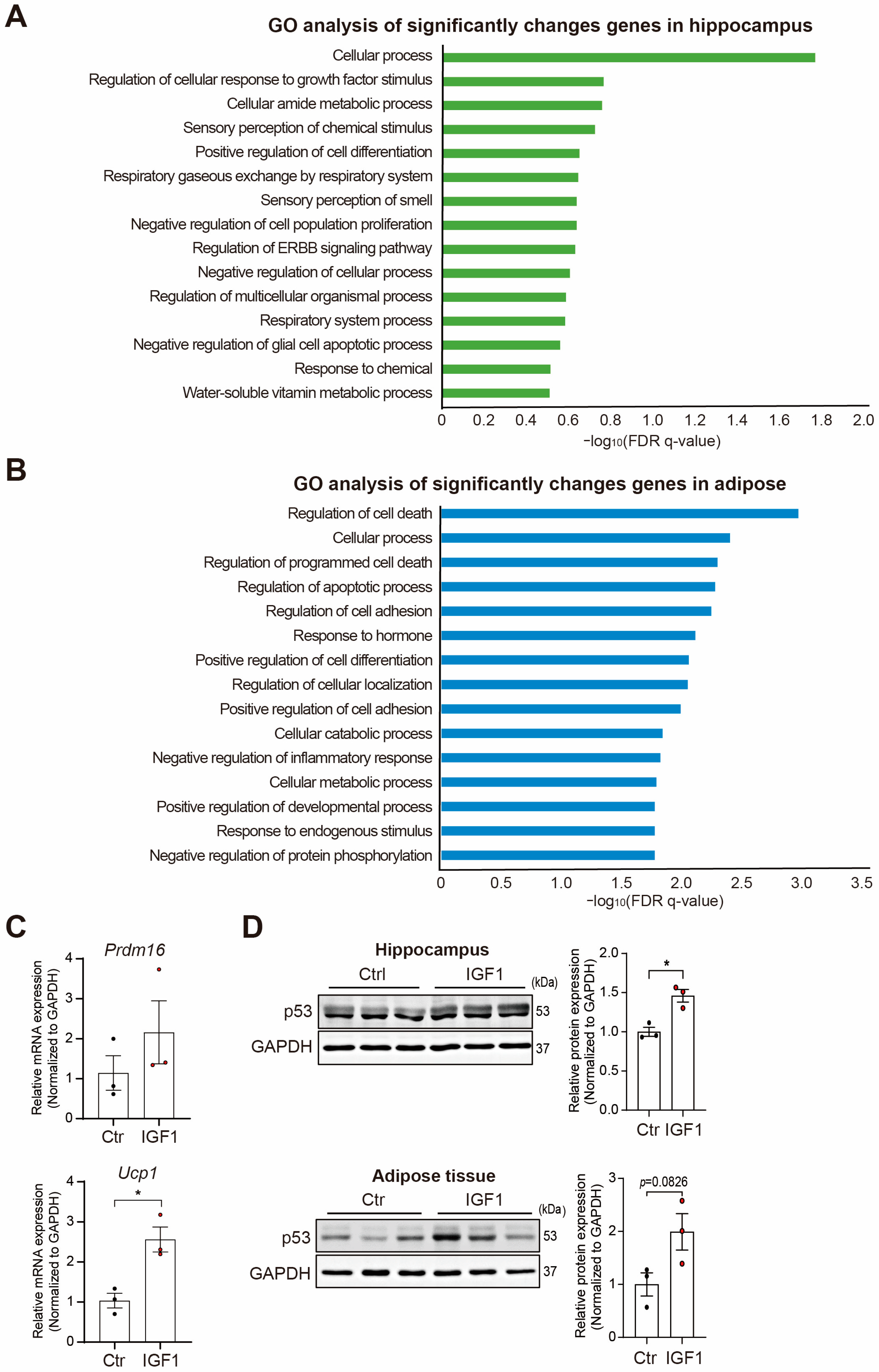
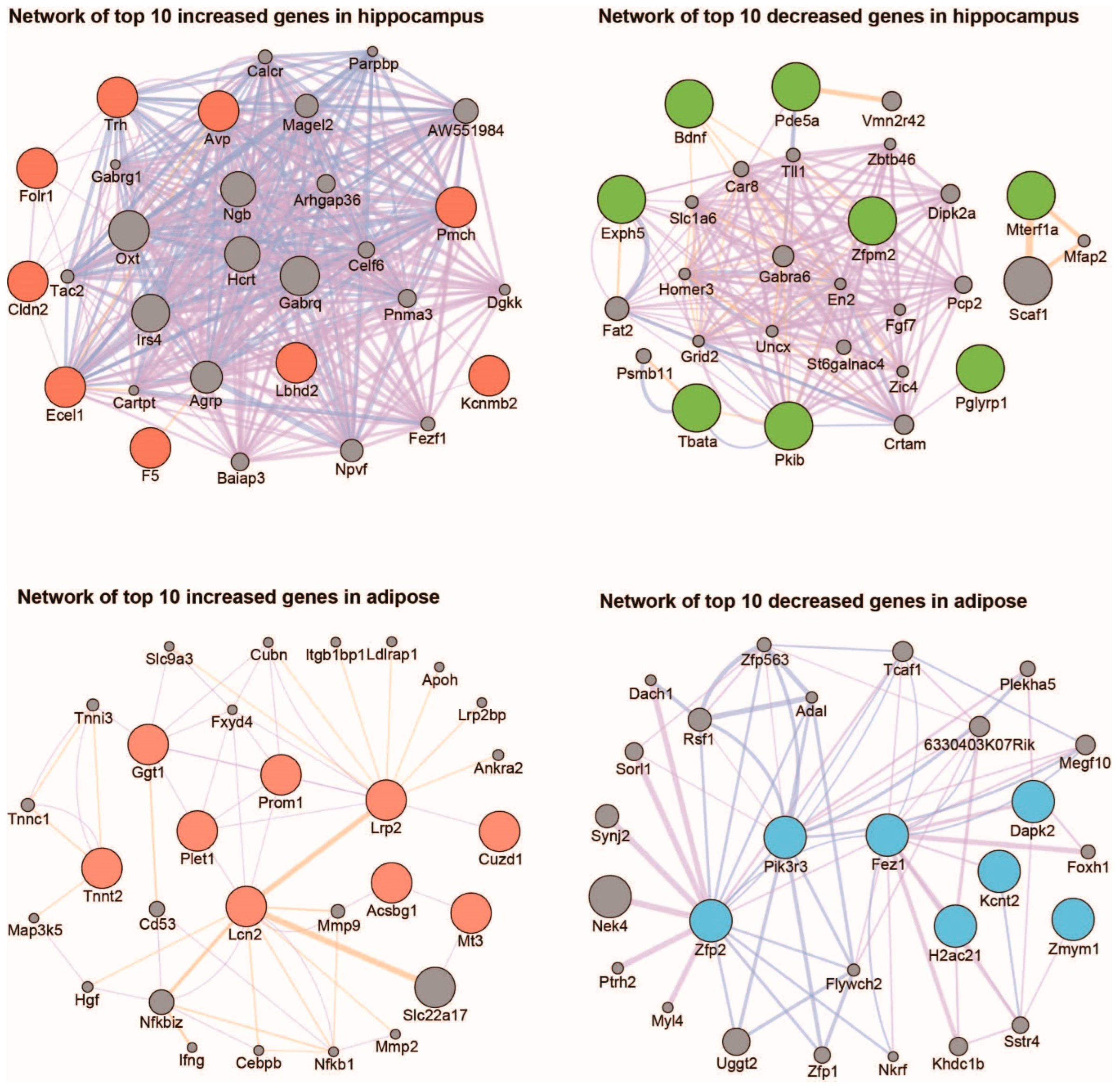
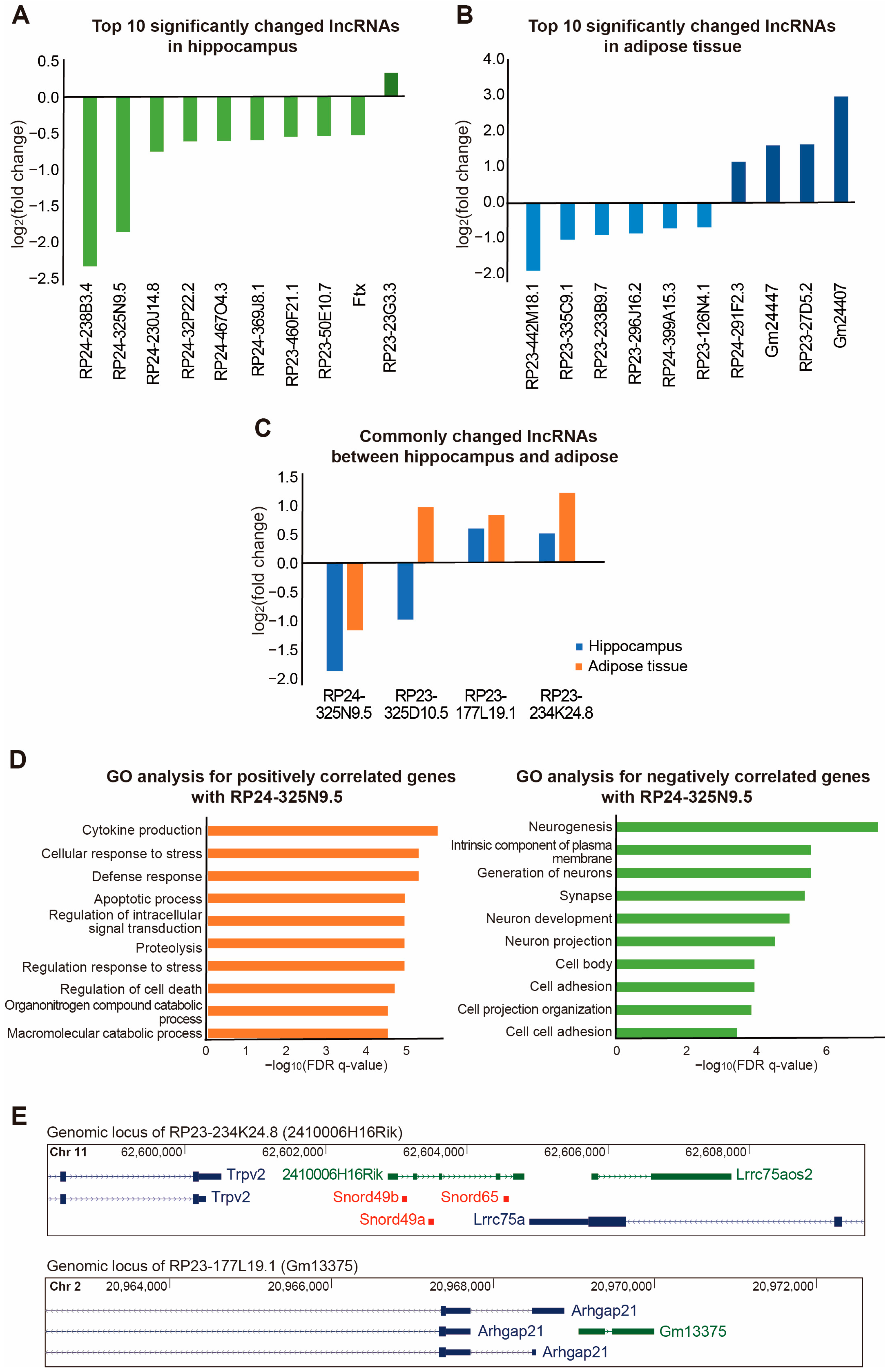
2. Results
3. Discussion
4. Materials and Methods
4.1. APP/PS2 Mouse Care and IGF-1 Injections
4.2. Marble-Burying Behavior Test
4.3. Nestlet-Shredding Behavior Test
4.4. RNA Sequencing Analysis
4.5. Analysis of RNA Sequencing Data
4.6. Functional Analysis of Changed Genes
4.7. Functional Analysis of Changed Non-Coding RNAs
4.8. RT-PCR
4.9. Western Blotting
4.10. Brain Osmotic Pump Implantation
4.11. Mouse Cytokine and Adipokine Array
4.12. Confirmation of the Circular Structure of circKsr2
4.13. Cell Fractionation
4.14. siRNA Construction
4.15. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shin, J.H. Dementia Epidemiology Fact Sheet 2022. Ann. Rehabil. Med. 2022, 46, 53–59. [Google Scholar] [CrossRef]
- Winblad, B.; Amouyel, P.; Andrieu, S.; Ballard, C.; Brayne, C.; Brodaty, H.; Cedazo-Minguez, A.; Dubois, B.; Edvardsson, D.; Feldman, H.; et al. Defeating Alzheimer’s disease and other dementias: A priority for European science and society. Lancet Neurol. 2016, 15, 455–532. [Google Scholar] [CrossRef] [PubMed]
- Scheltens, P.; Blennow, K.; Breteler, M.M.; de Strooper, B.; Frisoni, G.B.; Salloway, S.; Van der Flier, W.M. Alzheimer’s disease. Lancet 2016, 388, 505–517. [Google Scholar] [CrossRef]
- Hyman, B.T.; Phelps, C.H.; Beach, T.G.; Bigio, E.H.; Cairns, N.J.; Carrillo, M.C.; Dickson, D.W.; Duyckaerts, C.; Frosch, M.P.; Masliah, E.; et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Flores-Cordero, J.A.; Perez-Perez, A.; Jimenez-Cortegana, C.; Alba, G.; Flores-Barragan, A.; Sanchez-Margalet, V. Obesity as a Risk Factor for Dementia and Alzheimer’s Disease: The Role of Leptin. Int. J. Mol. Sci. 2022, 23, 5202. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Li, N.; Wei, W.; Zhang, L. Detection of molecular signatures and pathways shared by Alzheimer’s disease and type 2 diabetes. Gene 2022, 810, 146070. [Google Scholar] [CrossRef]
- Shinohara, M.; Kikuchi, M.; Onishi-Takeya, M.; Tashiro, Y.; Suzuki, K.; Noda, Y.; Takeda, S.; Mukouzono, M.; Nagano, S.; Fukumori, A.; et al. Upregulated expression of a subset of genes in APP;ob/ob mice: Evidence of an interaction between diabetes-linked obesity and Alzheimer’s disease. FASEB Bioadv. 2021, 3, 323–333. [Google Scholar] [CrossRef]
- Stampanoni Bassi, M.; Iezzi, E.; Buttari, F.; Gilio, L.; Simonelli, I.; Carbone, F.; Micillo, T.; De Rosa, V.; Sica, F.; Furlan, R.; et al. Obesity worsens central inflammation and disability in multiple sclerosis. Mult. Scler. 2020, 26, 1237–1246. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, G.; Hong, D.; Chen, F.; Ji, X.; Cao, G. White matter injury in ischemic stroke. Prog. Neurobiol. 2016, 141, 45–60. [Google Scholar] [CrossRef]
- Panza, F.; Frisardi, V.; Capurso, C.; Imbimbo, B.P.; Vendemiale, G.; Santamato, A.; D’Onofrio, G.; Seripa, D.; Sancarlo, D.; Pilotto, A.; et al. Metabolic syndrome and cognitive impairment: Current epidemiology and possible underlying mechanisms. J. Alzheimers Dis. 2010, 21, 691–724. [Google Scholar] [CrossRef]
- Guo, D.H.; Yamamoto, M.; Hernandez, C.M.; Khodadadi, H.; Baban, B.; Stranahan, A.M. Visceral adipose NLRP3 impairs cognition in obesity via IL-1R1 on CX3CR1+ cells. J. Clin. Investig. 2020, 130, 1961–1976. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Yi, H.A.; Won, K.S.; Lee, J.S.; Kim, H.W. Association between Visceral Adipose Tissue Metabolism and Alzheimer’s Disease Pathology. Metabolites 2022, 12, 258. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Lee, H.; Seo, H.S.; Kim, R.E.Y.; Lee, S.K.; Lee, Y.H.; Shin, C. Obesity and muscle may have synergic effect more than independent effects on brain volume in community-based elderly. Eur. Radiol. 2021, 31, 2956–2966. [Google Scholar] [CrossRef]
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Sweeney, M.D.; Montagne, A.; Sagare, A.P.; Nation, D.A.; Schneider, L.S.; Chui, H.C.; Harrington, M.G.; Pa, J.; Law, M.; Wang, D.J.J.; et al. Vascular dysfunction-The disregarded partner of Alzheimer’s disease. Alzheimers Dement. 2019, 15, 158–167. [Google Scholar] [CrossRef]
- Furiya, Y.; Ryo, M.; Kawahara, M.; Kiriyama, T.; Morikawa, M.; Ueno, S. Renin-angiotensin system blockers affect cognitive decline and serum adipocytokines in Alzheimer’s disease. Alzheimers Dement. 2013, 9, 512–518. [Google Scholar] [CrossRef]
- Kiliaan, A.J.; Arnoldussen, I.A.; Gustafson, D.R. Adipokines: A link between obesity and dementia? Lancet Neurol. 2014, 13, 913–923. [Google Scholar] [CrossRef]
- Pichiah, P.B.T.; Sankarganesh, D.; Arunachalam, S.; Achiraman, S. Adipose-Derived Molecules-Untouched Horizons in Alzheimer’s Disease Biology. Front. Aging Neurosci. 2020, 12, 17. [Google Scholar] [CrossRef]
- Nijssen, K.M.R.; Mensink, R.P.; Joris, P.J. Effects of Intranasal Insulin Administration on Cerebral Blood Flow and Cognitive Performance in Adults: A Systematic Review of Randomized, Placebo-Controlled Intervention Studies. Neuroendocrinology 2022, 113, 1–13. [Google Scholar] [CrossRef]
- Haas, S.S.; Myoraku, A.; Watson, K.; Robakis, T.; Frangou, S.; Abbasi, F.; Rasgon, N. Lower functional hippocampal connectivity in healthy adults is jointly associated with higher levels of leptin and insulin resistance. Eur. Psychiatry 2022, 65, e29. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tong, L.; Guo, F.; Zhao, S.; Zhang, J.; Guo, X.; Tao, Y.; Lin, X.; Jin, L. Depression status and insulin resistance in adults with obesity: A cross-sectional study. J. Psychosom. Res. 2022, 163, 111049. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.; Bullich, S.; Martinat, M.; Chataigner, M.; Di Miceli, M.; Simon, V.; Clark, S.; Butler, J.; Schell, M.; Chopra, S.; et al. Insulin modulates emotional behavior through a serotonin-dependent mechanism. Mol. Psychiatry 2022, 1–10. [Google Scholar] [CrossRef]
- Zhang, X.; Yee, D. Tyrosine kinase signalling in breast cancer: Insulin-like growth factors and their receptors in breast cancer. Breast Cancer Res. 2000, 2, 170–175. [Google Scholar] [CrossRef]
- Kineman, R.D.; Del Rio-Moreno, M.; Sarmento-Cabral, A. 40 YEARS of IGF1: Understanding the tissue-specific roles of IGF1/IGF1R in regulating metabolism using the Cre/loxP system. J. Mol. Endocrinol. 2018, 61, T187–T198. [Google Scholar] [CrossRef] [PubMed]
- Vitale, G.; Pellegrino, G.; Vollery, M.; Hofland, L.J. ROLE of IGF-1 System in the Modulation of Longevity: Controversies and New Insights From a Centenarians’ Perspective. Front. Endocrinol. 2019, 10, 27. [Google Scholar] [CrossRef]
- Ikemoto-Uezumi, M.; Uezumi, A.; Tsuchida, K.; Fukada, S.; Yamamoto, H.; Yamamoto, N.; Shiomi, K.; Hashimoto, N. Pro-Insulin-Like Growth Factor-II Ameliorates Age-Related Inefficient Regenerative Response by Orchestrating Self-Reinforcement Mechanism of Muscle Regeneration. Stem Cells 2015, 33, 2456–2468. [Google Scholar] [CrossRef]
- Clemmons, D.R. Involvement of insulin-like growth factor-I in the control of glucose homeostasis. Curr. Opin. Pharmacol. 2006, 6, 620–625. [Google Scholar] [CrossRef]
- Aguirre, G.A.; De Ita, J.R.; de la Garza, R.G.; Castilla-Cortazar, I. Insulin-like growth factor-1 deficiency and metabolic syndrome. J. Transl. Med. 2016, 14, 3. [Google Scholar] [CrossRef]
- Saukkonen, T.; Amin, R.; Williams, R.M.; Fox, C.; Yuen, K.C.; White, M.A.; Umpleby, A.M.; Acerini, C.L.; Dunger, D.B. Dose-dependent effects of recombinant human insulin-like growth factor (IGF)-I/IGF binding protein-3 complex on overnight growth hormone secretion and insulin sensitivity in type 1 diabetes. J. Clin. Endocrinol. Metab. 2004, 89, 4634–4641. [Google Scholar] [CrossRef]
- Fernandez, A.M.; Santi, A.; Torres Aleman, I. Insulin Peptides as Mediators of the Impact of Life Style in Alzheimer’s disease. Brain Plast. 2018, 4, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Westwood, A.J.; Beiser, A.; Decarli, C.; Harris, T.B.; Chen, T.C.; He, X.M.; Roubenoff, R.; Pikula, A.; Au, R.; Braverman, L.E.; et al. Insulin-like growth factor-1 and risk of Alzheimer dementia and brain atrophy. Neurology 2014, 82, 1613–1619. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Bai, J.; Zhong, S.; Zhang, R.; Zhang, X.; Xu, Y.; Zhao, M.; Zhao, C.; Zhou, Z. Down-Regulation of Insulin Like Growth Factor 1 Involved in Alzheimer’s Disease via MAPK, Ras, and FoxO Signaling Pathways. Oxid. Med. Cell. Longev. 2022, 2022, 8169981. [Google Scholar] [CrossRef]
- Gannon, O.J.; Robison, L.S.; Salinero, A.E.; Abi-Ghanem, C.; Mansour, F.M.; Kelly, R.D.; Tyagi, A.; Brawley, R.R.; Ogg, J.D.; Zuloaga, K.L. High-fat diet exacerbates cognitive decline in mouse models of Alzheimer’s disease and mixed dementia in a sex-dependent manner. J. Neuroinflamm. 2022, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Terzo, S.; Amato, A.; Mule, F. From obesity to Alzheimer’s disease through insulin resistance. J. Diabetes Complicat. 2021, 35, 108026. [Google Scholar] [CrossRef]
- Wittfeld, K.; Raman, M.R.; Conner, S.C.; Aslam, A.; Teumer, A.; Nauck, M.; Hosten, N.; Habes, M.; DeCarli, C.; Vasan, R.S.; et al. Insulin-Like Growth Factor, Inflammation, and MRI Markers of Alzheimer’s Disease in Predominantly Middle-Aged Adults. J. Alzheimers Dis. 2022, 88, 311–322. [Google Scholar] [CrossRef]
- Jafferali, S.; Dumont, Y.; Sotty, F.; Robitaille, Y.; Quirion, R.; Kar, S. Insulin-like growth factor-I and its receptor in the frontal cortex, hippocampus, and cerebellum of normal human and alzheimer disease brains. Synapse 2000, 38, 450–459. [Google Scholar] [CrossRef]
- Nishijima, T.; Piriz, J.; Duflot, S.; Fernandez, A.M.; Gaitan, G.; Gomez-Pinedo, U.; Verdugo, J.M.; Leroy, F.; Soya, H.; Nunez, A.; et al. Neuronal activity drives localized blood-brain-barrier transport of serum insulin-like growth factor-I into the CNS. Neuron 2010, 67, 834–846. [Google Scholar] [CrossRef]
- Morel, G.R.; Leon, M.L.; Uriarte, M.; Reggiani, P.C.; Goya, R.G. Therapeutic potential of IGF-I on hippocampal neurogenesis and function during aging. Neurogenesis 2017, 4, e1259709. [Google Scholar] [CrossRef]
- Herrero-Labrador, R.; Trueba-Saiz, A.; Martinez-Rachadell, L.; Fernandez de Sevilla, M.E.; Zegarra-Valdivia, J.A.; Pignatelli, J.; Diaz-Pacheco, S.; Fernandez, A.M.; Torres Aleman, I. Circulating Insulin-Like Growth Factor I is Involved in the Effect of High Fat Diet on Peripheral Amyloid beta Clearance. Int. J. Mol. Sci. 2020, 21, 9675. [Google Scholar] [CrossRef]
- Horvath, A.; Quinlan, P.; Eckerstrom, C.; Aberg, N.D.; Wallin, A.; Svensson, J. Low Serum Insulin-like Growth Factor-I Is Associated with Decline in Hippocampal Volume in Stable Mild Cognitive Impairment but not in Alzheimer’s Disease. J. Alzheimers Dis. 2022, 88, 1007–1016. [Google Scholar] [CrossRef]
- Walenkamp, M.J.; Karperien, M.; Pereira, A.M.; Hilhorst-Hofstee, Y.; van Doorn, J.; Chen, J.W.; Mohan, S.; Denley, A.; Forbes, B.; van Duyvenvoorde, H.A.; et al. Homozygous and heterozygous expression of a novel insulin-like growth factor-I mutation. J. Clin. Endocrinol. Metab. 2005, 90, 2855–2864. [Google Scholar] [CrossRef]
- Bonapace, G.; Concolino, D.; Formicola, S.; Strisciuglio, P. A novel mutation in a patient with insulin-like growth factor 1 (IGF1) deficiency. J. Med. Genet. 2003, 40, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Lichtenwalner, R.J.; Forbes, M.E.; Bennett, S.A.; Lynch, C.D.; Sonntag, W.E.; Riddle, D.R. Intracerebroventricular infusion of insulin-like growth factor-I ameliorates the age-related decline in hippocampal neurogenesis. Neuroscience 2001, 107, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Lopez, C.; LeRoith, D.; Torres-Aleman, I. Insulin-like growth factor I is required for vessel remodeling in the adult brain. Proc. Natl. Acad. Sci. USA 2004, 101, 9833–9838. [Google Scholar] [CrossRef] [PubMed]
- Maass, A.; Duzel, S.; Brigadski, T.; Goerke, M.; Becke, A.; Sobieray, U.; Neumann, K.; Lovden, M.; Lindenberger, U.; Backman, L.; et al. Relationships of peripheral IGF-1, VEGF and BDNF levels to exercise-related changes in memory, hippocampal perfusion and volumes in older adults. Neuroimage 2016, 131, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Svensson, J.; Diez, M.; Engel, J.; Wass, C.; Tivesten, A.; Jansson, J.O.; Isaksson, O.; Archer, T.; Hokfelt, T.; Ohlsson, C. Endocrine, liver-derived IGF-I is of importance for spatial learning and memory in old mice. J. Endocrinol. 2006, 189, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Trejo, J.L.; Llorens-Martin, M.V.; Torres-Aleman, I. The effects of exercise on spatial learning and anxiety-like behavior are mediated by an IGF-I-dependent mechanism related to hippocampal neurogenesis. Mol. Cell Neurosci. 2008, 37, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.M.; Tseng, V.; Wang, J.; Wang, D.; Matyakhina, L.; Bondy, C.A. Tau is hyperphosphorylated in the insulin-like growth factor-I null brain. Endocrinology 2005, 146, 5086–5091. [Google Scholar] [CrossRef] [PubMed]
- Carro, E.; Trejo, J.L.; Gomez-Isla, T.; LeRoith, D.; Torres-Aleman, I. Serum insulin-like growth factor I regulates brain amyloid-beta levels. Nat. Med. 2002, 8, 1390–1397. [Google Scholar] [CrossRef]
- Rivera, E.J.; Goldin, A.; Fulmer, N.; Tavares, R.; Wands, J.R.; de la Monte, S.M. Insulin and insulin-like growth factor expression and function deteriorate with progression of Alzheimer’s disease: Link to brain reductions in acetylcholine. J. Alzheimers Dis. 2005, 8, 247–268. [Google Scholar] [CrossRef] [PubMed]
- Machann, J.; Stefan, N.; Wagner, R.; Fritsche, A.; Bell, J.D.; Whitcher, B.; Haring, H.U.; Birkenfeld, A.L.; Nikolaou, K.; Schick, F.; et al. Normalized Indices Derived from Visceral Adipose Mass Assessed by Magnetic Resonance Imaging and Their Correlation with Markers for Insulin Resistance and Prediabetes. Nutrients 2020, 12, 2064. [Google Scholar] [CrossRef] [PubMed]
- Sasaguri, H.; Nilsson, P.; Hashimoto, S.; Nagata, K.; Saito, T.; De Strooper, B.; Hardy, J.; Vassar, R.; Winblad, B.; Saido, T.C. APP mouse models for Alzheimer’s disease preclinical studies. EMBO J. 2017, 36, 2473–2487. [Google Scholar] [CrossRef] [PubMed]
- Carro, E.; Trejo, J.L.; Gerber, A.; Loetscher, H.; Torrado, J.; Metzger, F.; Torres-Aleman, I. Therapeutic actions of insulin-like growth factor I on APP/PS2 mice with severe brain amyloidosis. Neurobiol. Aging 2006, 27, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Deng, L.; Chen, Q.; Xie, J.; Wei, W.; Hui, H. circPUM1 promotes polycystic ovary syndrome progression by sponging to miR-760. Gene 2020, 754, 144903. [Google Scholar] [CrossRef]
- Huang, Y.; Liao, X.; Luo, J.; Liu, H.; Zhong, S.; Chen, J. Expression of circular RNAs in the vascular dementia rats. Neurosci. Lett. 2020, 735, 135087. [Google Scholar] [CrossRef]
- Hollensen, A.K.; Thomsen, H.S.; Lloret-Llinares, M.; Kamstrup, A.B.; Jensen, J.M.; Luckmann, M.; Birkmose, N.; Palmfeldt, J.; Jensen, T.H.; Hansen, T.B.; et al. circZNF827 nucleates a transcription inhibitory complex to balance neuronal differentiation. Elife 2020, 9, e58478. [Google Scholar] [CrossRef]
- Chen, Y.; Li, X.; Meng, S.; Huang, S.; Chang, S.; Shi, J. Identification of Functional CircRNA-miRNA-mRNA Regulatory Network in Dorsolateral Prefrontal Cortex Neurons of Patients with Cocaine Use Disorder. Front. Mol. Neurosci. 2022, 15, 839233. [Google Scholar] [CrossRef]
- Song, J.; Kim, Y.K. Targeting non-coding RNAs for the treatment of retinal diseases. Mol. Ther. Nucleic Acids 2021, 24, 284–293. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Macklin, L.; Griffith, C.M.; Cai, Y.; Rose, G.M.; Yan, X.X.; Patrylo, P.R. Glucose tolerance and insulin sensitivity are impaired in APP/PS1 transgenic mice prior to amyloid plaque pathogenesis and cognitive decline. Exp. Gerontol. 2017, 88, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.C.; Lee, S.H.; Lee, D.W.; Kim, S.Y.; Kim, G.Y.; Rhim, H.S.; Choi, C.B.; Kim, H.Y.; Lee, C.U.; Choe, B.Y. Regional metabolic alteration of Alzheimer’s disease in mouse brain expressing mutant human APP-PS1 by 1H HR-MAS. Behav. Brain Res. 2010, 211, 125–131. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zheng, R.; Li, J.; Cao, Q.; Hou, T.; Zhao, Z.; Xu, M.; Chen, Y.; Lu, J.; Wang, T.; et al. Individual and Combined Associations of Glucose Metabolic Components with Cognitive Function Modified by Obesity. Front. Endocrinol. 2021, 12, 769120. [Google Scholar] [CrossRef]
- Grau-Rivera, O.; Navalpotro-Gomez, I.; Sanchez-Benavides, G.; Suarez-Calvet, M.; Mila-Aloma, M.; Arenaza-Urquijo, E.M.; Salvado, G.; Sala-Vila, A.; Shekari, M.; Gonzalez-de-Echavarri, J.M.; et al. Association of weight change with cerebrospinal fluid biomarkers and amyloid positron emission tomography in preclinical Alzheimer’s disease. Alzheimers Res. Ther. 2021, 13, 46. [Google Scholar] [CrossRef]
- Jeyabalan, S.; Bala, L.; Subramanian, K.; Jabaris, S.L.; Sekar, M.; Wong, L.S.; Subramaniyan, V.; Chidambaram, K.; Gan, S.H.; Mat Rani, N.N.I.; et al. Potential effects of noni (Morinda citrifolia L.) fruits extract against obsessive-compulsive disorder in marble burying and nestlet shredding behavior mice models. Front. Pharmacol. 2022, 13, 993927. [Google Scholar] [CrossRef]
- Thomas, A.; Burant, A.; Bui, N.; Graham, D.; Yuva-Paylor, L.A.; Paylor, R. Marble burying reflects a repetitive and perseverative behavior more than novelty-induced anxiety. Psychopharmacology 2009, 204, 361–373. [Google Scholar] [CrossRef]
- Dorninger, F.; Zeitler, G.; Berger, J. Nestlet Shredding and Nest Building Tests to Assess Features of Psychiatric Disorders in Mice. Bio Protoc. 2020, 10, e3863. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Kueon, C.; Kim, J. Influence of lead on repetitive behavior and dopamine metabolism in a mouse model of iron overload. Toxicol. Res. 2014, 30, 267–276. [Google Scholar] [CrossRef]
- Garner, J.B.; Marshall, L.S.; Boyer, N.M.; Alapatt, V.; Miller, L.L. Effects of Ketoprofen and Morphine on Pain-Related Depression of Nestlet Shredding in Male and Female Mice. Front. Pain Res. 2021, 2, 673940. [Google Scholar] [CrossRef] [PubMed]
- Santi, A.; Bot, M.; Aleman, A.; Penninx, B.; Aleman, I.T. Circulating insulin-like growth factor I modulates mood and is a biomarker of vulnerability to stress: From mouse to man. Transl. Psychiatry 2018, 8, 142. [Google Scholar] [CrossRef] [PubMed]
- Savaheli, S.; Ahmadiani, A. Obsessive-compulsive disorder and growth factors: A comparative review. Behav. Brain Res. 2019, 372, 111967. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Cheng, C.M.; Tsai, S.J.; Tsai, C.F.; Su, T.P.; Li, C.T.; Lin, W.C.; Chen, T.J.; Bai, Y.M. Obsessive-Compulsive Disorder and Dementia Risk: A Nationwide Longitudinal Study. J. Clin. Psychiatry 2021, 82, 32996. [Google Scholar] [CrossRef] [PubMed]
- Ruggeri, M.; Ricci, M.; Gerace, C.; Blundo, C. Late-onset obsessive-compulsive disorder as the initial manifestation of possible behavioural variant Alzheimer’s disease. Cogn. Neuropsychiatry 2022, 27, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.G.; Cai, P.J.; Hu, J.H.; Jiang, J.X.; Zhang, J.J.; Liu, K.F.; Yang, L.; Long, C. Altered corticostriatal synchronization associated with compulsive-like behavior in APP/PS1 mice. Exp. Neurol. 2021, 344, 113805. [Google Scholar] [CrossRef]
- Papazacharias, A.; Nardini, M. The relationship between depression and cognitive deficits. Psychiatr. Danub. 2012, 24 (Suppl. 1), S179–S182. [Google Scholar]
- Bora, E.; Harrison, B.J.; Yucel, M.; Pantelis, C. Cognitive impairment in euthymic major depressive disorder: A meta-analysis. Psychol. Med. 2013, 43, 2017–2026. [Google Scholar] [CrossRef]
- Bortolato, B.; Carvalho, A.F.; McIntyre, R.S. Cognitive dysfunction in major depressive disorder: A state-of-the-art clinical review. CNS Neurol. Disord. Drug Targets 2014, 13, 1804–1818. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Nunes, P.V.; Oliveira, K.C.; Young, L.T.; Lafer, B. Neuropathological relationship between major depression and dementia: A hypothetical model and review. Prog. Neuropsychopharmacol. Biol. Psychiatry 2016, 67, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Rial, D.; Lemos, C.; Pinheiro, H.; Duarte, J.M.; Goncalves, F.Q.; Real, J.I.; Prediger, R.D.; Goncalves, N.; Gomes, C.A.; Canas, P.M.; et al. Depression as a Glial-Based Synaptic Dysfunction. Front. Cell Neurosci. 2015, 9, 521. [Google Scholar] [CrossRef] [PubMed]
- Suh, H.S.; Zhao, M.L.; Derico, L.; Choi, N.; Lee, S.C. Insulin-like growth factor 1 and 2 (IGF1, IGF2) expression in human microglia: Differential regulation by inflammatory mediators. J. Neuroinflamm. 2013, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Deng, P.Y.; Porter, J.E.; Shin, H.S.; Lei, S. Thyrotropin-releasing hormone increases GABA release in rat hippocampus. J. Physiol. 2006, 577 Pt 2, 497–511. [Google Scholar] [CrossRef]
- Waniek, A.; Hartlage-Rubsamen, M.; Hofling, C.; Kehlen, A.; Schilling, S.; Demuth, H.U.; Rossner, S. Identification of thyrotropin-releasing hormone as hippocampal glutaminyl cyclase substrate in neurons and reactive astrocytes. Biochim. Biophys. Acta 2015, 1852, 146–155. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, F.; Wang, C.; Zhang, J.; Bai, Y.; Zhou, F.; Wang, Z.; Wu, M.; Yang, W.; Guo, J.; et al. AVP(4-8) Improves Cognitive Behaviors and Hippocampal Synaptic Plasticity in the APP/PS1 Mouse Model of Alzheimer’s Disease. Neurosci. Bull. 2020, 36, 254–262. [Google Scholar] [CrossRef]
- Tong, W.H.; Abdulai-Saiku, S.; Vyas, A. Arginine vasopressin in the medial amygdala causes greater post-stress recruitment of hypothalamic vasopressin neurons. Mol. Brain 2021, 14, 141. [Google Scholar] [CrossRef] [PubMed]
- Diniz, G.B.; Bittencourt, J.C. The Melanin-Concentrating Hormone as an Integrative Peptide Driving Motivated Behaviors. Front. Syst. Neurosci. 2017, 11, 32. [Google Scholar] [CrossRef]
- Vawter, M.P.; Schulmann, A.; Alhassen, L.; Alhassen, W.; Hamzeh, A.R.; Sakr, J.; Pauluk, L.; Yoshimura, R.; Wang, X.; Dai, Q.; et al. Melanin Concentrating Hormone Signaling Deficits in Schizophrenia: Association with Memory and Social Impairments and Abnormal Sensorimotor Gating. Int. J. Neuropsychopharmacol. 2020, 23, 53–65. [Google Scholar] [CrossRef]
- Bahjaoui-Bouhaddi, M.; Fellmann, D.; Griffond, B.; Bugnon, C. Insulin treatment stimulates the rat melanin-concentrating hormone-producing neurons. Neuropeptides 1994, 27, 251–258. [Google Scholar] [CrossRef]
- Soussi, H.; Reggio, S.; Alili, R.; Prado, C.; Mutel, S.; Pini, M.; Rouault, C.; Clement, K.; Dugail, I. DAPK2 Downregulation Associates with Attenuated Adipocyte Autophagic Clearance in Human Obesity. Diabetes 2015, 64, 3452–3463. [Google Scholar] [CrossRef]
- Shiloh, R.; Gilad, Y.; Ber, Y.; Eisenstein, M.; Aweida, D.; Bialik, S.; Cohen, S.; Kimchi, A. Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy. Nat. Commun. 2018, 9, 1759. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Guo, L.; Li, N.; Henzel, K.S.; Gu, H.; Ran, X.; Sun, W.; Liu, S.; Lu, Y.; Ehninger, D.; et al. Overexpression of Kcnmb2 in Dorsal CA1 of Offspring Mice Rescues Hippocampal Dysfunction Caused by a Methyl Donor-Rich Paternal Diet. Front. Cell Neurosci. 2018, 12, 360. [Google Scholar] [CrossRef] [PubMed]
- Katsumata, Y.; Nelson, P.T.; Ellingson, S.R.; Fardo, D.W. Gene-based association study of genes linked to hippocampal sclerosis of aging neuropathology: GRN, TMEM106B, ABCC9, and KCNMB2. Neurobiol. Aging 2017, 53, 193.e17–193.e25. [Google Scholar] [CrossRef]
- Gennet, N.; Tamburini, C.; Nan, X.; Li, M. FolR1: A novel cell surface marker for isolating midbrain dopamine neural progenitors and nascent dopamine neurons. Sci. Rep. 2016, 6, 32488. [Google Scholar] [CrossRef] [PubMed]
- Dieterich, K.; Quijano-Roy, S.; Monnier, N.; Zhou, J.; Faure, J.; Smirnow, D.A.; Carlier, R.; Laroche, C.; Marcorelles, P.; Mercier, S.; et al. The neuronal endopeptidase ECEL1 is associated with a distinct form of recessive distal arthrogryposis. Hum. Mol. Genet. 2013, 22, 1483–1492. [Google Scholar] [CrossRef] [PubMed]
- Osanai, A.; Sashinami, H.; Asano, K.; Li, S.J.; Hu, D.L.; Nakane, A. Mouse peptidoglycan recognition protein PGLYRP-1 plays a role in the host innate immune response against Listeria monocytogenes infection. Infect. Immun. 2011, 79, 858–866. [Google Scholar] [CrossRef] [PubMed]
- Huo, J.; Ren, S.; Gao, P.; Wan, D.; Rong, S.; Li, X.; Liu, S.; Xu, S.; Sun, K.; Guo, B.; et al. ALG13 participates in epileptogenesis via regulation of GABAA receptors in mouse models. Cell Death Discov. 2020, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Deis, J.A.; Guo, H.; Wu, Y.; Liu, C.; Bernlohr, D.A.; Chen, X. Adipose Lipocalin 2 overexpression protects against age-related decline in thermogenic function of adipose tissue and metabolic deterioration. Mol. Metab. 2019, 24, 18–29. [Google Scholar] [CrossRef]
- Meyers, K.; Lopez, M.; Ho, J.; Wills, S.; Rayalam, S.; Taval, S. Lipocalin-2 deficiency may predispose to the progression of spontaneous age-related adiposity in mice. Sci. Rep. 2020, 10, 14589. [Google Scholar] [CrossRef]
- Brooker, G.J.; Kalloniatis, M.; Russo, V.C.; Murphy, M.; Werther, G.A.; Bartlett, P.F. Endogenous IGF-1 regulates the neuronal differentiation of adult stem cells. J. Neurosci. Res. 2000, 59, 332–341. [Google Scholar] [CrossRef]
- Huat, T.J.; Khan, A.A.; Pati, S.; Mustafa, Z.; Abdullah, J.M.; Jaafar, H. IGF-1 enhances cell proliferation and survival during early differentiation of mesenchymal stem cells to neural progenitor-like cells. BMC Neurosci. 2014, 15, 91. [Google Scholar] [CrossRef]
- Dang, R.; Cai, H.; Zhang, L.; Liang, D.; Lv, C.; Guo, Y.; Yang, R.; Zhu, Y.; Jiang, P. Dysregulation of Neuregulin-1/ErbB signaling in the prefrontal cortex and hippocampus of rats exposed to chronic unpredictable mild stress. Physiol. Behav. 2016, 154, 145–150. [Google Scholar] [CrossRef]
- Ledonne, A.; Mercuri, N.B. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. Int. J. Mol. Sci. 2019, 21, 275. [Google Scholar] [CrossRef]
- Ledonne, A.; Mango, D.; Latagliata, E.C.; Chiacchierini, G.; Nobili, A.; Nistico, R.; D’Amelio, M.; Puglisi-Allegra, S.; Mercuri, N.B. Neuregulin 1/ErbB signalling modulates hippocampal mGluRI-dependent LTD and object recognition memory. Pharmacol. Res. 2018, 130, 12–24. [Google Scholar] [CrossRef]
- Logan, S.; Pharaoh, G.A.; Marlin, M.C.; Masser, D.R.; Matsuzaki, S.; Wronowski, B.; Yeganeh, A.; Parks, E.E.; Premkumar, P.; Farley, J.A.; et al. Insulin-like growth factor receptor signaling regulates working memory, mitochondrial metabolism, and amyloid-beta uptake in astrocytes. Mol. Metab. 2018, 9, 141–155. [Google Scholar] [CrossRef]
- Carlson, S.W.; Saatman, K.E. Central Infusion of Insulin-Like Growth Factor-1 Increases Hippocampal Neurogenesis and Improves Neurobehavioral Function after Traumatic Brain Injury. J. Neurotrauma 2018, 35, 1467–1480. [Google Scholar] [CrossRef]
- Das, S.K.; Barhwal, K.; Hota, S.K.; Thakur, M.K.; Srivastava, R.B. Disrupting monotony during social isolation stress prevents early development of anxiety and depression like traits in male rats. BMC Neurosci. 2015, 16, 2. [Google Scholar] [CrossRef]
- Ostrowski, P.P.; Barszczyk, A.; Forstenpointner, J.; Zheng, W.; Feng, Z.P. Meta-Analysis of Serum Insulin-Like Growth Factor 1 in Alzheimer’s Disease. PLoS ONE 2016, 11, e0155733. [Google Scholar] [CrossRef]
- Galle, S.A.; van der Spek, A.; Drent, M.L.; Brugts, M.P.; Scherder, E.J.A.; Janssen, J.; Ikram, M.A.; van Duijn, C.M. Revisiting the Role of Insulin-Like Growth Factor-I Receptor Stimulating Activity and the Apolipoprotein E in Alzheimer’s Disease. Front. Aging Neurosci. 2019, 11, 20. [Google Scholar] [CrossRef]
- Aghajanov, M.; Matinyan, S.; Chavushyan, V.; Danielyan, M.; Karapetyan, G.; Mirumyan, M.; Fereshetyan, K.; Harutyunyan, H.; Yenkoyan, K. The Involvement of Insulin-Like Growth Factor 1 and Nerve Growth Factor in Alzheimer’s Disease-Like Pathology and Survival Role of the Mix of Embryonic Proteoglycans: Electrophysiological Fingerprint, Structural Changes and Regulatory Effects on Neurotrophins. Int. J. Mol. Sci. 2021, 22, 7084. [Google Scholar] [CrossRef]
- Littlejohn, E.L.; Scott, D.; Saatman, K.E. Insulin-like growth factor-1 overexpression increases long-term survival of posttrauma-born hippocampal neurons while inhibiting ectopic migration following traumatic brain injury. Acta Neuropathol. Commun. 2020, 8, 46. [Google Scholar] [CrossRef]
- Dyer, A.H.; Vahdatpour, C.; Sanfeliu, A.; Tropea, D. The role of Insulin-Like Growth Factor 1 (IGF-1) in brain development, maturation and neuroplasticity. Neuroscience 2016, 325, 89–99. [Google Scholar] [CrossRef]
- Scavo, L.M.; Karas, M.; Murray, M.; Leroith, D. Insulin-like growth factor-I stimulates both cell growth and lipogenesis during differentiation of human mesenchymal stem cells into adipocytes. J. Clin. Endocrinol. Metab. 2004, 89, 3543–3553. [Google Scholar] [CrossRef]
- Boucher, J.; Mori, M.A.; Lee, K.Y.; Smyth, G.; Liew, C.W.; Macotela, Y.; Rourk, M.; Bluher, M.; Russell, S.J.; Kahn, C.R. Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nat. Commun. 2012, 3, 902. [Google Scholar] [CrossRef]
- Fischer-Posovszky, P.; Tornqvist, H.; Debatin, K.M.; Wabitsch, M. Inhibition of death-receptor mediated apoptosis in human adipocytes by the insulin-like growth factor I (IGF-I)/IGF-I receptor autocrine circuit. Endocrinology 2004, 145, 1849–1859. [Google Scholar] [CrossRef]
- Al-Massadi, O.; Porteiro, B.; Kuhlow, D.; Kohler, M.; Gonzalez-Rellan, M.J.; Garcia-Lavandeira, M.; Diaz-Rodriguez, E.; Quinones, M.; Senra, A.; Alvarez, C.V.; et al. Pharmacological and Genetic Manipulation of p53 in Brown Fat at Adult But Not Embryonic Stages Regulates Thermogenesis and Body Weight in Male Mice. Endocrinology 2016, 157, 2735–2749. [Google Scholar] [CrossRef]
- Molchadsky, A.; Ezra, O.; Amendola, P.G.; Krantz, D.; Kogan-Sakin, I.; Buganim, Y.; Rivlin, N.; Goldfinger, N.; Folgiero, V.; Falcioni, R.; et al. p53 is required for brown adipogenic differentiation and has a protective role against diet-induced obesity. Cell Death Differ. 2013, 20, 774–783. [Google Scholar] [CrossRef]
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Sompol, P.; Ittarat, W.; Tangpong, J.; Chen, Y.; Doubinskaia, I.; Batinic-Haberle, I.; Abdul, H.M.; Butterfield, D.A.; St Clair, D.K. A neuronal model of Alzheimer’s disease: An insight into the mechanisms of oxidative stress-mediated mitochondrial injury. Neuroscience 2008, 153, 120–130. [Google Scholar] [CrossRef]
- Mandal, P.K.; Saharan, S.; Tripathi, M.; Murari, G. Brain glutathione levels--a novel biomarker for mild cognitive impairment and Alzheimer’s disease. Biol. Psychiatry 2015, 78, 702–710. [Google Scholar] [CrossRef]
- Rosa, L.R.O.; Soares, G.M.; Silveira, L.R.; Boschero, A.C.; Barbosa-Sampaio, H.C.L. ARHGAP21 as a master regulator of multiple cellular processes. J. Cell. Physiol. 2018, 233, 8477–8481. [Google Scholar] [CrossRef]
- Ferreira, S.M.; Costa-Junior, J.M.; Kurauti, M.A.; Leite, N.C.; Ortis, F.; Rezende, L.F.; Barbosa, H.C.; Boschero, A.C.; Santos, G.J. ARHGAP21 Acts as an Inhibitor of the Glucose-Stimulated Insulin Secretion Process. Front. Endocrinol. 2020, 11, 599165. [Google Scholar] [CrossRef]
- Jeong, G.; Kwon, D.H.; Shin, S.; Choe, N.; Ryu, J.; Lim, Y.H.; Kim, J.; Park, W.J.; Kook, H.; Kim, Y.K. Long noncoding RNAs in vascular smooth muscle cells regulate vascular calcification. Sci. Rep. 2019, 9, 5848. [Google Scholar] [CrossRef]
- Song, J.; Kim, Y.K. Discovery and Functional Prediction of Long Non-Coding RNAs Common to Ischemic Stroke and Myocardial Infarction. J. Lipid Atheroscler. 2020, 9, 449–459. [Google Scholar] [CrossRef]
- Lawrence, J.M.; Schardien, K.; Wigdahl, B.; Nonnemacher, M.R. Roles of neuropathology-associated reactive astrocytes: A systematic review. Acta Neuropathol. Commun. 2023, 11, 42. [Google Scholar] [CrossRef]
- Muramori, F.; Kobayashi, K.; Nakamura, I. A quantitative study of neurofibrillary tangles, senile plaques and astrocytes in the hippocampal subdivisions and entorhinal cortex in Alzheimer’s disease, normal controls and non-Alzheimer neuropsychiatric diseases. Psychiatry Clin. Neurosci. 1998, 52, 593–599. [Google Scholar] [CrossRef]
- Marttinen, M.; Kurkinen, K.M.; Soininen, H.; Haapasalo, A.; Hiltunen, M. Synaptic dysfunction and septin protein family members in neurodegenerative diseases. Mol. Neurodegener. 2015, 10, 16. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, P.; Liu, H.; Zhan, J.; Wang, J.; Li, M.; Zeng, L.; Xu, P. Changes and Significance of SYP and GAP-43 Expression in the Hippocampus of CIH Rats. Int. J. Med. Sci. 2019, 16, 394–402. [Google Scholar] [CrossRef]
- Tarsa, L.; Goda, Y. Synaptophysin regulates activity-dependent synapse formation in cultured hippocampal neurons. Proc. Natl. Acad. Sci. USA 2002, 99, 1012–1016. [Google Scholar] [CrossRef]
- Kwon, S.E.; Chapman, E.R. Synaptophysin regulates the kinetics of synaptic vesicle endocytosis in central neurons. Neuron 2011, 70, 847–854. [Google Scholar] [CrossRef]
- Evgen’ev, M.B.; Krasnov, G.S.; Nesterova, I.V.; Garbuz, D.G.; Karpov, V.L.; Morozov, A.V.; Snezhkina, A.V.; Samokhin, A.N.; Sergeev, A.; Kulikov, A.M.; et al. Molecular Mechanisms Underlying Neuroprotective Effect of Intranasal Administration of Human Hsp70 in Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2017, 59, 1415–1426. [Google Scholar] [CrossRef]
- Bobkova, N.V.; Lyabin, D.N.; Medvinskaya, N.I.; Samokhin, A.N.; Nekrasov, P.V.; Nesterova, I.V.; Aleksandrova, I.Y.; Tatarnikova, O.G.; Bobylev, A.G.; Vikhlyantsev, I.M.; et al. The Y-Box Binding Protein 1 Suppresses Alzheimer’s Disease Progression in Two Animal Models. PLoS ONE 2015, 10, e0138867. [Google Scholar] [CrossRef]
- Angoa-Perez, M.; Kane, M.J.; Briggs, D.I.; Francescutti, D.M.; Kuhn, D.M. Marble burying and nestlet shredding as tests of repetitive, compulsive-like behaviors in mice. J. Vis. Exp. 2013, 82, 50978. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
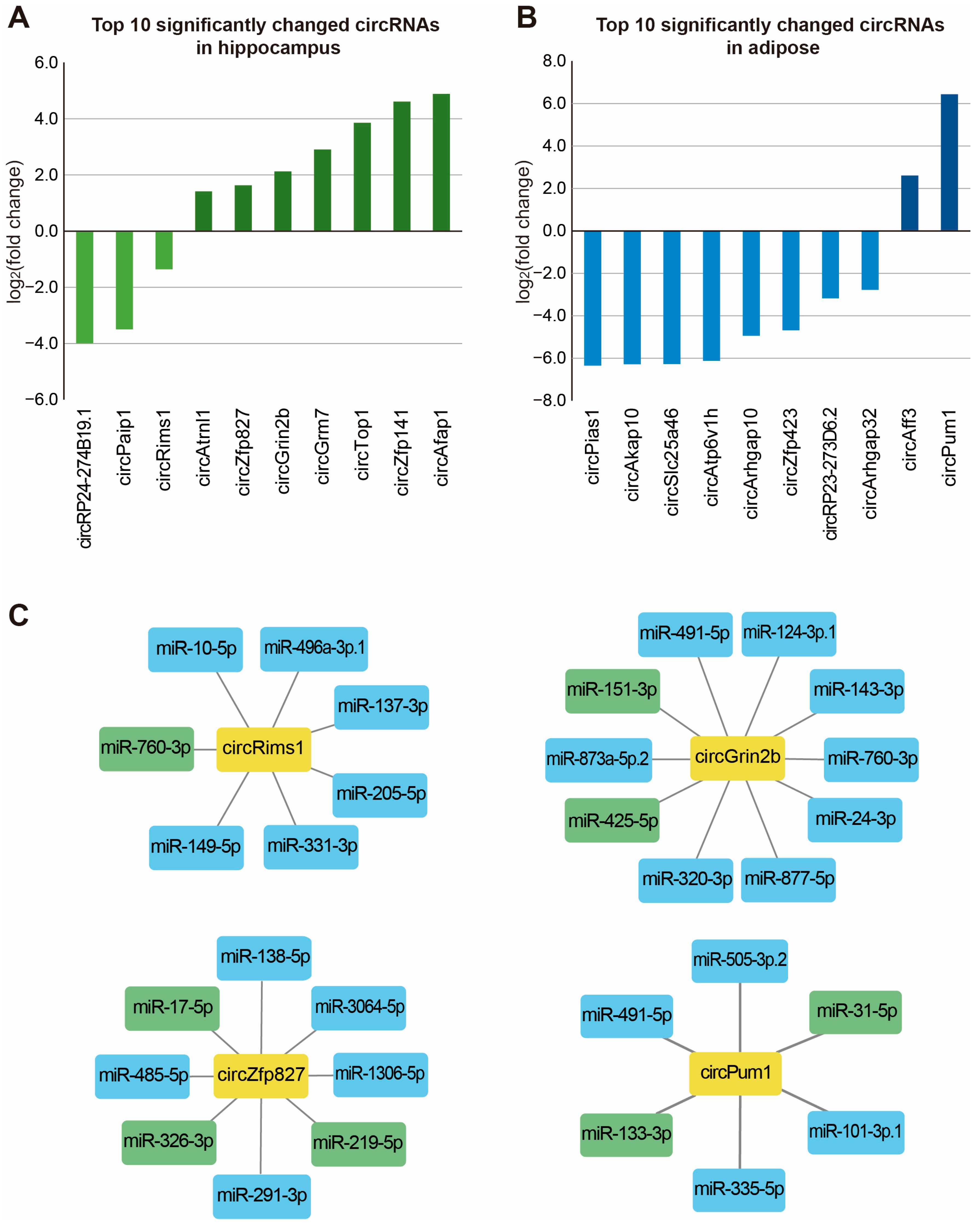
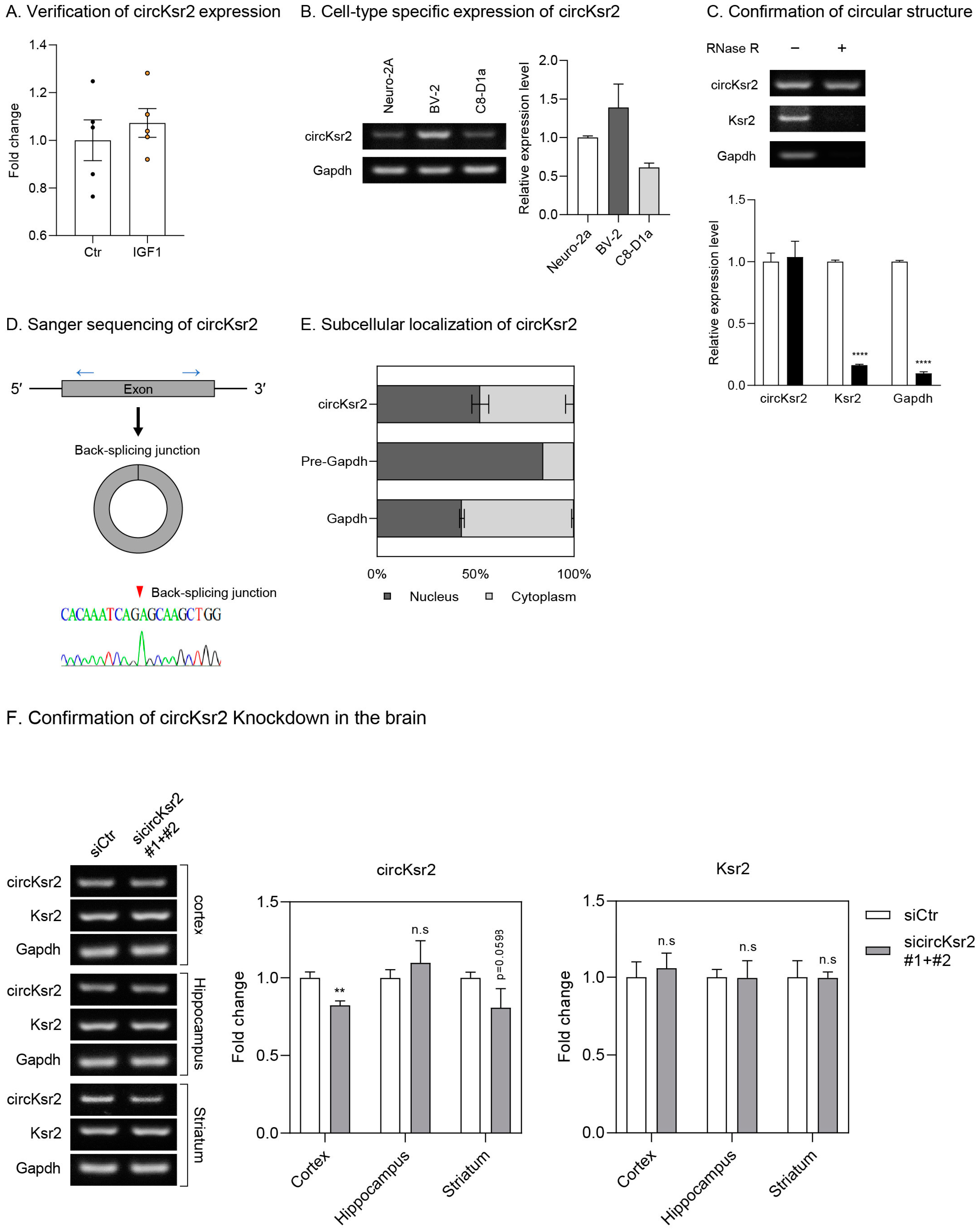

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-K.; Jo, D.; Arjunan, A.; Ryu, Y.; Lim, Y.-H.; Choi, S.Y.; Kim, H.K.; Song, J. Identification of IGF-1 Effects on White Adipose Tissue and Hippocampus in Alzheimer’s Disease Mice via Transcriptomic and Cellular Analysis. Int. J. Mol. Sci. 2024, 25, 2567. https://doi.org/10.3390/ijms25052567
Kim Y-K, Jo D, Arjunan A, Ryu Y, Lim Y-H, Choi SY, Kim HK, Song J. Identification of IGF-1 Effects on White Adipose Tissue and Hippocampus in Alzheimer’s Disease Mice via Transcriptomic and Cellular Analysis. International Journal of Molecular Sciences. 2024; 25(5):2567. https://doi.org/10.3390/ijms25052567
Chicago/Turabian StyleKim, Young-Kook, Danbi Jo, Archana Arjunan, Yeongseo Ryu, Yeong-Hwan Lim, Seo Yoon Choi, Hee Kyung Kim, and Juhyun Song. 2024. "Identification of IGF-1 Effects on White Adipose Tissue and Hippocampus in Alzheimer’s Disease Mice via Transcriptomic and Cellular Analysis" International Journal of Molecular Sciences 25, no. 5: 2567. https://doi.org/10.3390/ijms25052567






