Regulation of Disease-Resistance Genes against CWMV Infection by NbHAG1-Mediated H3K36ac
Abstract
:1. Introduction
2. Results
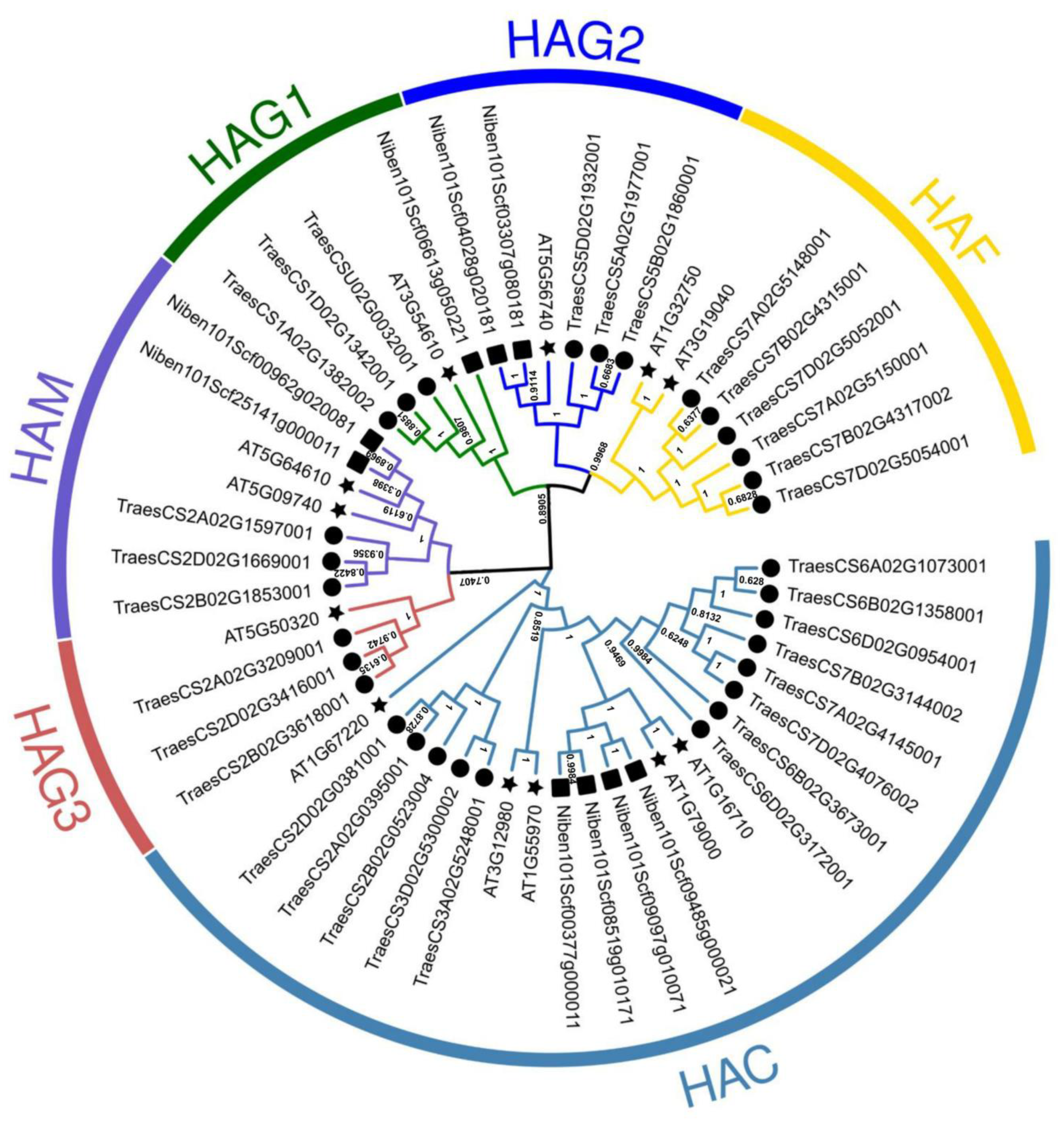
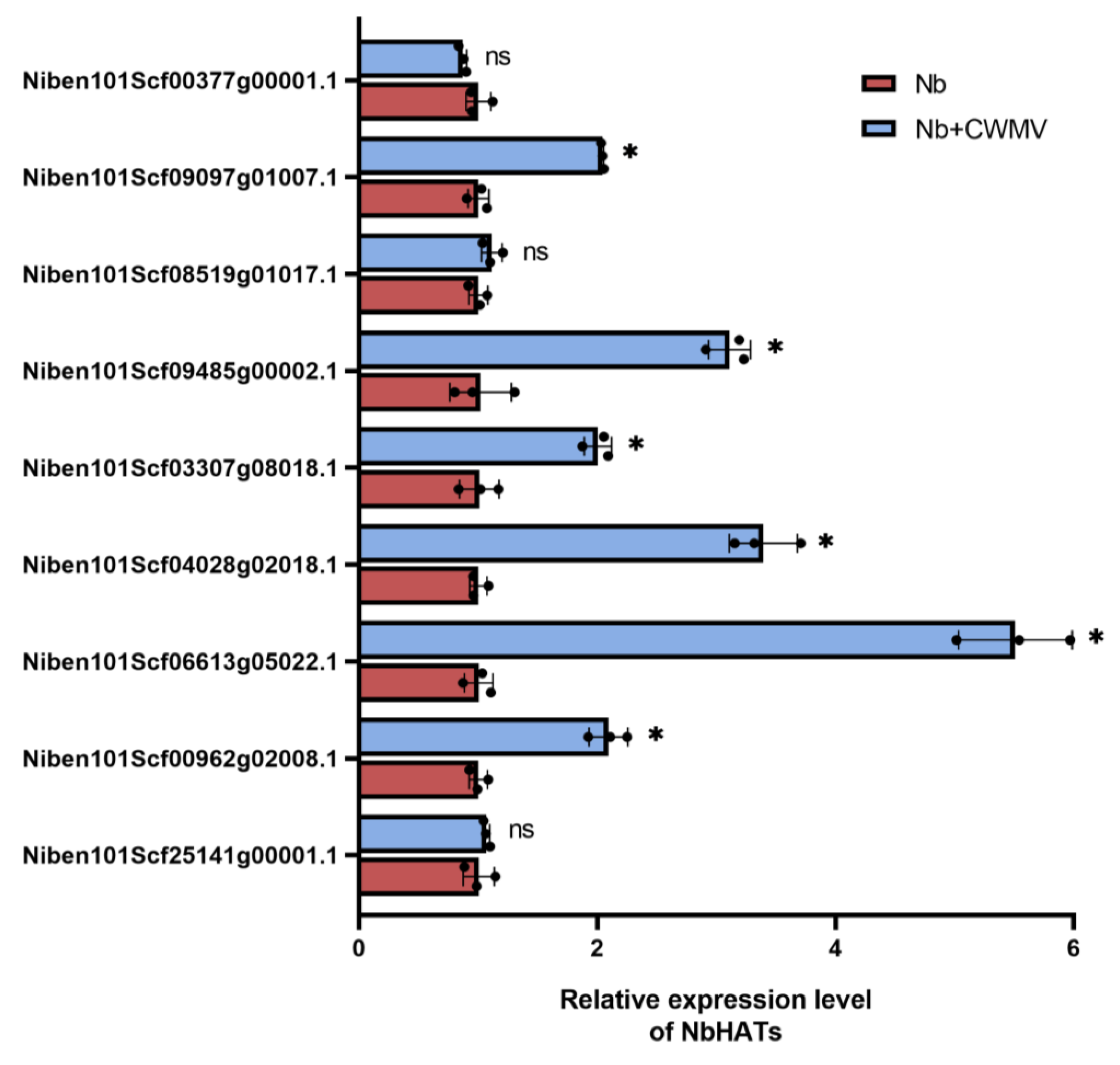
2.1. CWMV Infection Induces HAG1 Expression in N. benthamiana
2.2. NbHAG1 Positively Regulates N. benthamiana Resistance to CWMV
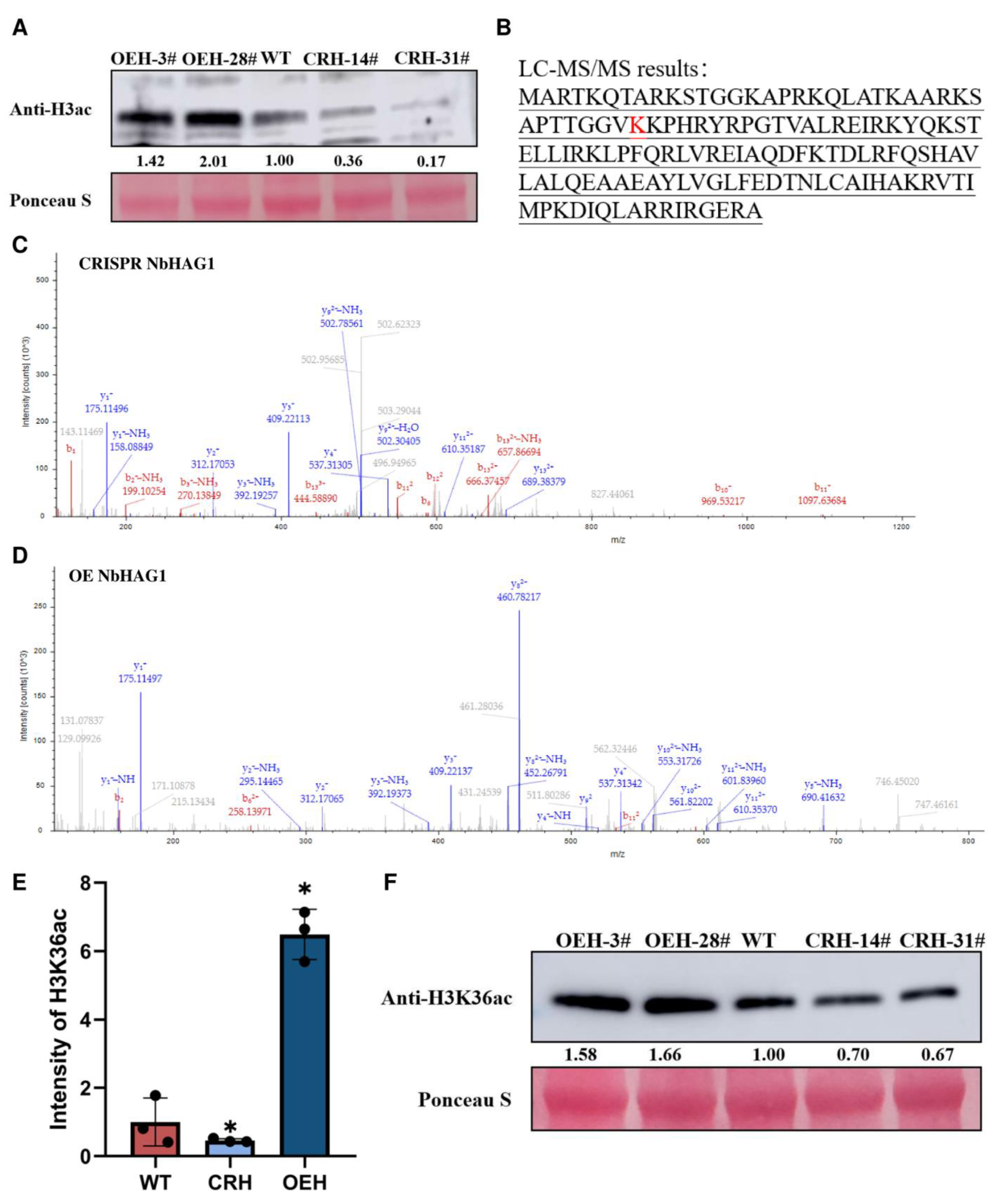
2.3. NbHAG1 Affects the Acetylation Level of H3K36
2.4. NbHAG1 May Regulate the Expression Levels of a Series of Genes
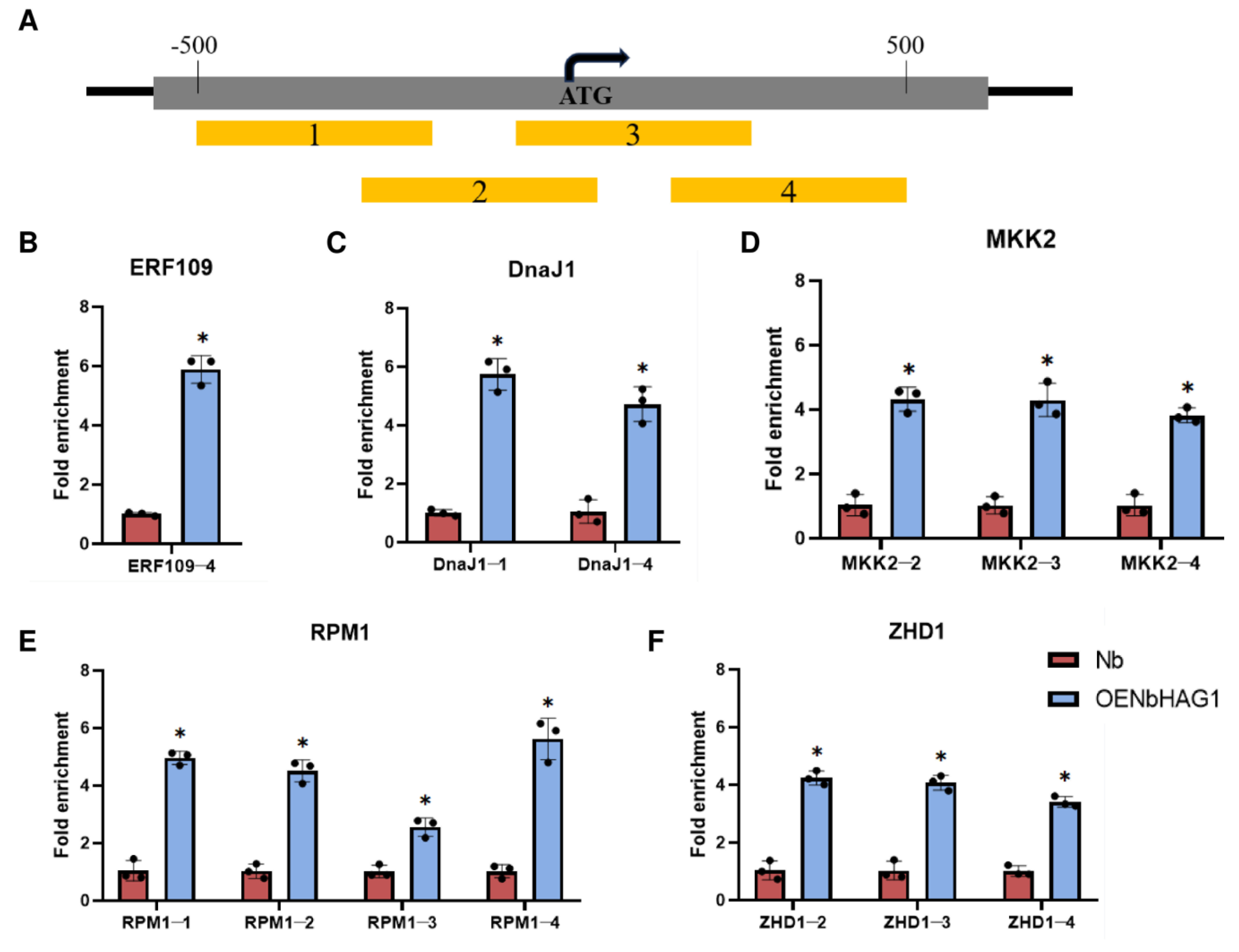
2.5. NbHAG1 Mediates H3K36ac to Regulate Downstream Gene Expression
2.6. Silencing of NbERF109 and NbMKK2 Expression Inhibits CWMV Infection
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth, and Virus Inoculation
4.2. Genetic Transformation of N. benthamiana
4.3. RNA Extraction and Quantitative Reverse Transcription-PCR (qRT-PCR) Assays
4.4. Western Blot Assay
4.5. Yeast Two-Hybrid Assay
4.6. Bioinformatics Analysis of Differentially Expressed Genes (DEGs)
4.7. Virus-Induced Gene Silencing (VIGS)
4.8. Identification of Acetylation Site in NbHAG1 Histone H3 through LC-MS/MS
4.9. Isolation of Nuclei and Cut&Tag
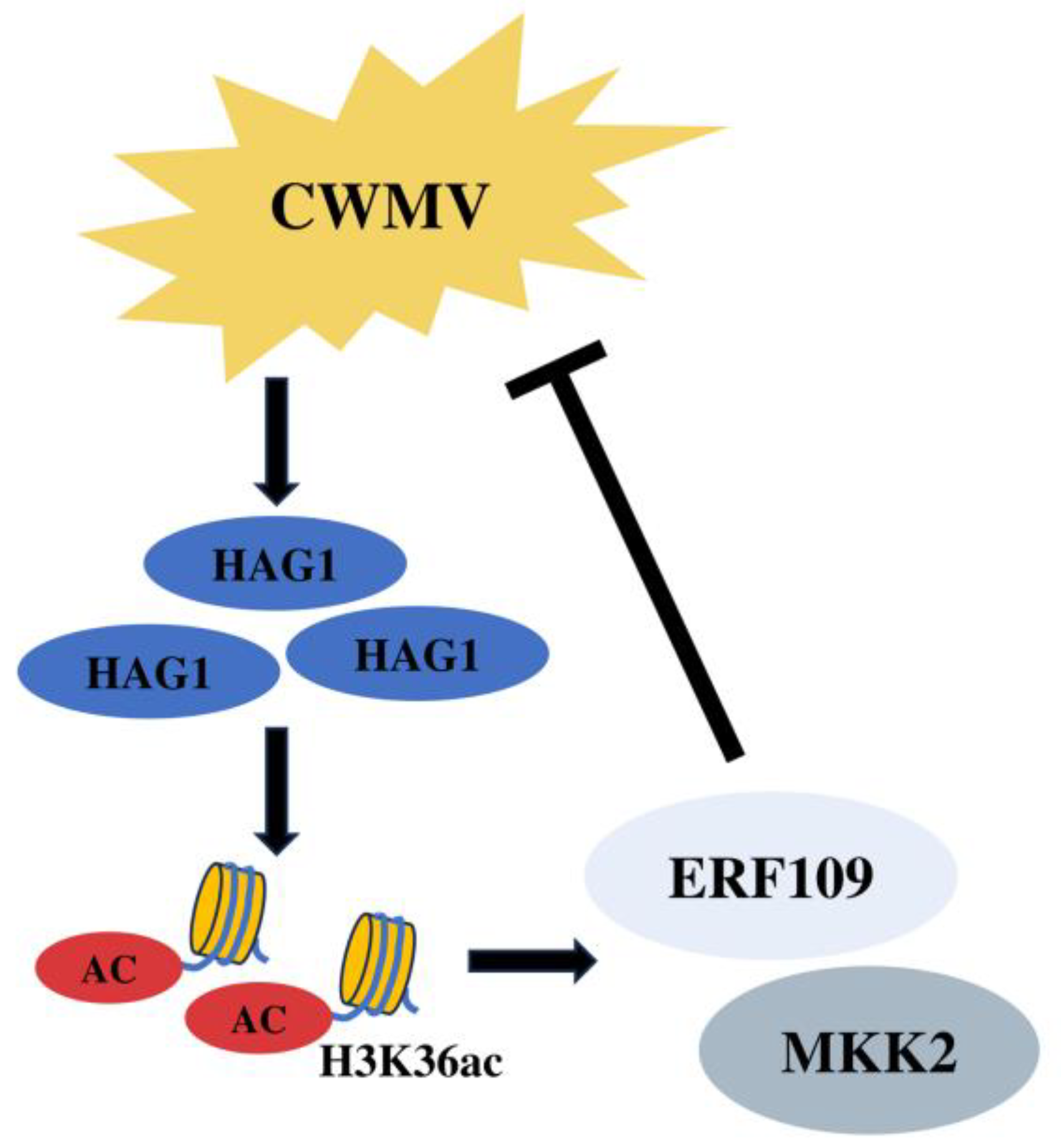
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Millar, A.H.; Heazlewood, J.L.; Giglione, C.; Holdsworth, M.J.; Bachmair, A.; Schulze, W.X. The Scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 2019, 70, 119–151. [Google Scholar] [CrossRef] [PubMed]
- Tahir, J.; Rashid, M.; Afzal, A.J. Post-translational modifications in effectors and plant proteins involved in host–pathogen conflicts. Plant Pathol. 2019, 68, 628–644. [Google Scholar] [CrossRef]
- Sun, W.; Poschmann, J.; Cruz-Herrera del Rosario, R.; Parikshak, N.N.; Hajan, H.S.; Kumar, V.; Ramasamy, R.; Belgard, T.G.; Elanggovan, B.; Wong, C.C.Y.; et al. Histone acetylome-wide association study of autism spectrum disorder. Cell 2016, 167, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Tao, Y.; Li, M.; Che, T.; Qu, J. Protein acetylation and deacetylation: An important regulatory modification in gene transcription (Review). Exp. Ther. Med. 2020, 20, 2923–2940. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, J.K.; Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 2021, 78, 4467–4486. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2014, 16, 258–264. [Google Scholar] [CrossRef]
- Carré, C.; Szymczak, D.; Pidoux, J.; Antoniewski, C. The histone H3 acetylase dGcn5 is a key player in Drosophila melanogaster metamorphosis. Mol. Cell Biol. 2023, 25, 8228–8238. [Google Scholar] [CrossRef]
- Grant, P.A.; Duggan, L.; Côté, J.; Roberts, S.M.; Brownell, J.E.; Candau, R.; Ohba, R.; Owen-Hughes, T.; Allis, C.D.; Winston, F.; et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones characterization of an Ada complex and the SAGA (SptAda) complex. Genes Dev. 1997, 11, 1640–1650. [Google Scholar] [CrossRef]
- Yamauchi, T.; Yamauchi, J.; Kuwata, T.; Tamura, T.; Yamashita, T.; Bae, N.; Westphal, H.; Ozato, K.; Nakatani, Y. Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-BGCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 2000, 97, 11303–11306. [Google Scholar] [CrossRef]
- Helmlinger, D.; Tora, L. Sharing the SAGA. Trends Biochem. Sci. 2017, 42, 850–861. [Google Scholar] [CrossRef]
- Wu, C.J.; Liu, Z.Z.; Wei, L.; Zhou, J.X.; Cai, X.W.; Su, Y.N.; Li, L.; Chen, S.; He, X.J. Three functionally redundant plant-specific paralogs are core subunits of the SAGA histone acetyltransferase complex in Arabidopsis. Mol. Plant 2021, 14, 1071–1087. [Google Scholar] [CrossRef]
- Pandey, R.; MuÈller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef]
- Kornet, N.; Scheres, B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef]
- Servet, C.; Conde e Silva, N.; Zhou, D.-X. Histone Acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Benhamed, M.; Martin-Magniette, M.L.; Taconnat, L.; Bitton, F.; Servet, C.; De Clercq, R.; De Meyer, B.; Buysschaert, C.; Rombauts, S.; Villarroel, R.; et al. Genome-scale Arabidopsis promoter array identifies targets of the histone acetyltransferase GCN5. Plant J. 2008, 56, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Jiang, W.; Long, F.; Cheng, S.; Yang, W.; Zhao, Y.; Zhou, D.-X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 2017, 29, 1088–1104. [Google Scholar] [CrossRef]
- Grasser, K.D.; Rubio, V.; Barneche, F. Multifaceted activities of the plant SAGA complex. Biochim. Biophys. Acta Gene Regul. Mech. 2021, 1864, 194613. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. Occurrence of fungally transmitted wheat mosaic viruses in China. Ann. Appl. Biol. 1993, 123, 55–61. [Google Scholar]
- Adams, M.J.; Antoniw, J.F.; Kreuze, J. Virgaviridae: A new family of rod-shaped plant viruses. Arch. Virol. 2009, 154, 1967–1972. [Google Scholar] [CrossRef]
- Yang, Y.-H.; Chen, H.; Jiang, M.; Zhao, S.; Adams. Sequence of a second isolate of Chinese wheat mosaic Furovirus. J. Phytopathol. 2001, 149, 135–140. [Google Scholar] [CrossRef]
- Diao, A.; Chen, J.; Ye, R.; Zheng, T.; Yu, S.; Antoniw, J.F.; Adams, M.J. Complete sequence and genome properties of Chinese wheat mosaic virus, a new furovirus from China. J. Gen. Virol. 1999, 80, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, F.; Xie, L.; Song, X.-J.; Li, J.; Chen, J.-P.; Zhang, H.-M. Functional identification of two minor capsid proteins from Chinese wheat mosaic virus using its infectious full-length cDNA clones. J. Gen. Virol. 2016, 97, 2441–2450. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Liu, T.; Cheng, Y.; Gao, S.; Li, L.; Cai, L.; Yang, J.; Chen, J.; Zhong, K. Comprehensive proteomic analysis of lysine acetylation in Nicotiana benthamiana after sensing CWMV infection. Front. Microbiol. 2021, 12, 672559. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Li, L.; Han, X.; Liu, T.; Jin, P.; Cai, L.; Xu, M.; Zhang, T.; Zhang, F.; Chen, J.; et al. Genome-wide identification of the histone acetyltransferase gene family in Triticum aestivum. BMC Genom. 2021, 22, 49. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, F.; Chen, D.; Chu, W.; Liu, H.; Xiang, Y. Genome-wide identification and analysis of the Populus trichocarpa TIFY gene family. Plant Physiol. Biochem. 2017, 115, 360–371. [Google Scholar] [CrossRef]
- Sterner, D.; Berger, S. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.J.E.; Brind’Amour, J.; Kuzmin, A.; Jensen, K.N.; Liu, Z.C.; Lorincz, M.; Howe, L.J. Transcription shapes genome-wide histone acetylation patterns. Nat. Commun. 2021, 12, 210. [Google Scholar] [CrossRef]
- Mahrez, W.; Arellano, M.S.T.; Moreno-Romero, J.; Nakamura, M.; Shu, H.; Nanni, P.; Köhler, C.; Gruissem, W.; Hennig, L. H3K36ac is an evolutionary conserved plant histone modification that marks active genes. Plant Physiol. 2016, 170, 1566–1577. [Google Scholar] [CrossRef]
- Boycheva, I.; Vassileva, V.; Iantcheva, A. Histone acetyltransferases in plant development and plasticity. Curr. Genom. 2014, 15, 28–37. [Google Scholar] [CrossRef]
- Wang, Z.; Cao, H.; Chen, F.; Liu, Y. The roles of histone acetylation in seed performance and plant development. Plant Physiol. Biochem. 2014, 84, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, M.; Zhang, W.; Zhao, J.; Zhang, J.; Wu, K.; Tian, L.; Duan, J. Histone acetyltransferases in rice (Oryza sativa L.): Phylogenetic analysis, subcellular localization and expression. BMC Plant Biol. 2012, 12, 145. [Google Scholar] [CrossRef]
- Papaefthimiou, D.; Likotrafiti, E.; Kapazoglou, A.; Bladenopoulos, K.; Tsaftaris, A. Epigenetic chromatin modifiers in barley: Isolation and characterization of the barley GNAT-MYST family of histone acetyltransferases and responses to exogenous ABA. Plant Physiol. Biochem. 2010, 48, 98–107. [Google Scholar] [CrossRef]
- Aiese Cigliano, R.; Sanseverino, W.; Cremona, G.; Ercolano, M.R.; Conicella, C.; Consiglio, F.M. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles. BMC Genom. 2013, 14, 57. [Google Scholar] [CrossRef]
- Aksnes, H.; Drazic, A.; Marie, M.; Arnesen, T. First things first: Vital protein marks by N-terminal acetyltransferases. Trends Biochem. Sci. 2016, 41, 746–760. [Google Scholar] [CrossRef]
- Linster, E.; Forero Ruiz, F.L.; Miklankova, P.; Ruppert, T.; Mueller, J.; Armbruster, L.; Gong, X.; Serino, G.; Mann, M.; Hell, R.; et al. Cotranslational N-degron masking by acetylation promotes proteome stability in plants. Nat. Commun. 2022, 13, 810. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, T.; He, Y.; Su, C.; Wang, Z.; Zhou, X. N-terminal acetylation of the βC1 protein encoded by the betasatellite of tomato yellow leaf curl China virus is critical for its viral pathogenicity. Virology 2023, 586, 1–11. [Google Scholar] [CrossRef]
- Yang, L.; Li, B.; Zheng, X.Y.; Li, J.; Yang, M.; Dong, X.; He, G.; An, C.; Deng, X.W. Salicylic acid biosynthesis is enhanced and contributes to increased biotrophic pathogen resistance in Arabidopsis hybrids. Nat. Commun. 2015, 6, 7309. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Lin, J.; Liu, X.; Liu, Z.; Liu, D.; Chu, W.; Li, J.; Chen, Y.; Chang, S.; Yang, Q.; et al. Histone acetyltransferase TaHAG1 interacts with TaPLATZ5 to activate TaPAD4 expression and positively contributes to powdery mildew resistance in wheat. New Phytol. 2022, 236, 590–607. [Google Scholar] [CrossRef]
- Jin, J.H.; Wang, M.; Zhang, H.X.; Khan, A.; Wei, A.M.; Luo, D.X.; Gong, Z.H. Genome-wide identification of the AP2/ERF transcription factor family in pepper (Capsicum annuum L.). Genome 2018, 61, 663–674. [Google Scholar] [CrossRef]
- Hoehenwarter, W.; Thomas, M.; Nukarinen, E.; Egelhofer, V.; Röhrig, H.; Weckwerth, W.; Conrath, U.; Beckers, G.J. Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell Proteom. 2013, 12, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, T.; Li, J.; Wu, N.; Wu, G.; Yang, J.; Chen, X.; He, L.; Chen, J. Chinese wheat mosaic virus-derived vsiRNA-20 can regulate virus infection in wheat through inhibition of vacuolar-(H+)-PPase induced cell death. New Phytol. 2020, 226, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Kuang, J.; Zhai, Z.; Li, P.; Shi, R.; Guo, W.; Yao, Y.; Guo, J.; Zhao, G.; He, J.; Xu, S.; et al. SS18 regulates pluripotent-somatic transition through phase separation. Nat. Commun. 2021, 12, 4090. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, J.; Wang, C.; Dai, H.; Du, X.; Li, Q.; Pan, Z. The super-enhancer repertoire in porcine liver. J. Anim. Sci. 2023, 101, skad056. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tu, A.; Wu, M.; Jiang, Y.; Guo, L.; Guo, Y.; Wang, J.; Xu, G.; Shi, J.; Chen, J.; Yang, J.; et al. Regulation of Disease-Resistance Genes against CWMV Infection by NbHAG1-Mediated H3K36ac. Int. J. Mol. Sci. 2024, 25, 2800. https://doi.org/10.3390/ijms25052800
Tu A, Wu M, Jiang Y, Guo L, Guo Y, Wang J, Xu G, Shi J, Chen J, Yang J, et al. Regulation of Disease-Resistance Genes against CWMV Infection by NbHAG1-Mediated H3K36ac. International Journal of Molecular Sciences. 2024; 25(5):2800. https://doi.org/10.3390/ijms25052800
Chicago/Turabian StyleTu, Aizhu, Mila Wu, Yaoyao Jiang, Lidan Guo, Yunfei Guo, Jinnan Wang, Gecheng Xu, Jingjing Shi, Jianping Chen, Jian Yang, and et al. 2024. "Regulation of Disease-Resistance Genes against CWMV Infection by NbHAG1-Mediated H3K36ac" International Journal of Molecular Sciences 25, no. 5: 2800. https://doi.org/10.3390/ijms25052800
APA StyleTu, A., Wu, M., Jiang, Y., Guo, L., Guo, Y., Wang, J., Xu, G., Shi, J., Chen, J., Yang, J., & Zhong, K. (2024). Regulation of Disease-Resistance Genes against CWMV Infection by NbHAG1-Mediated H3K36ac. International Journal of Molecular Sciences, 25(5), 2800. https://doi.org/10.3390/ijms25052800






