Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza
Abstract
1. Introduction
2. Results
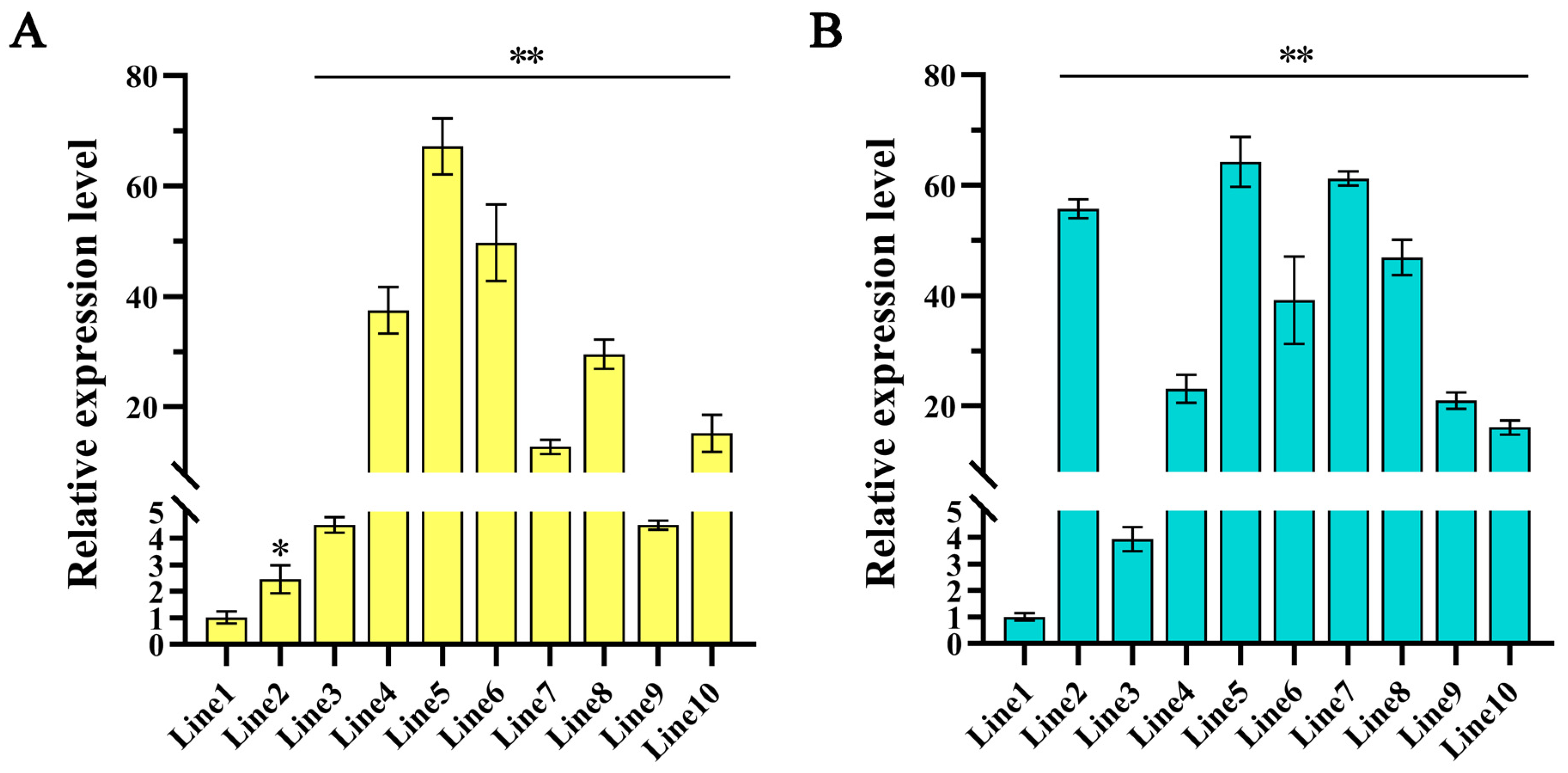
2.1. Generation and Molecular Characterization of Transgenic Hairy Roots and Plants
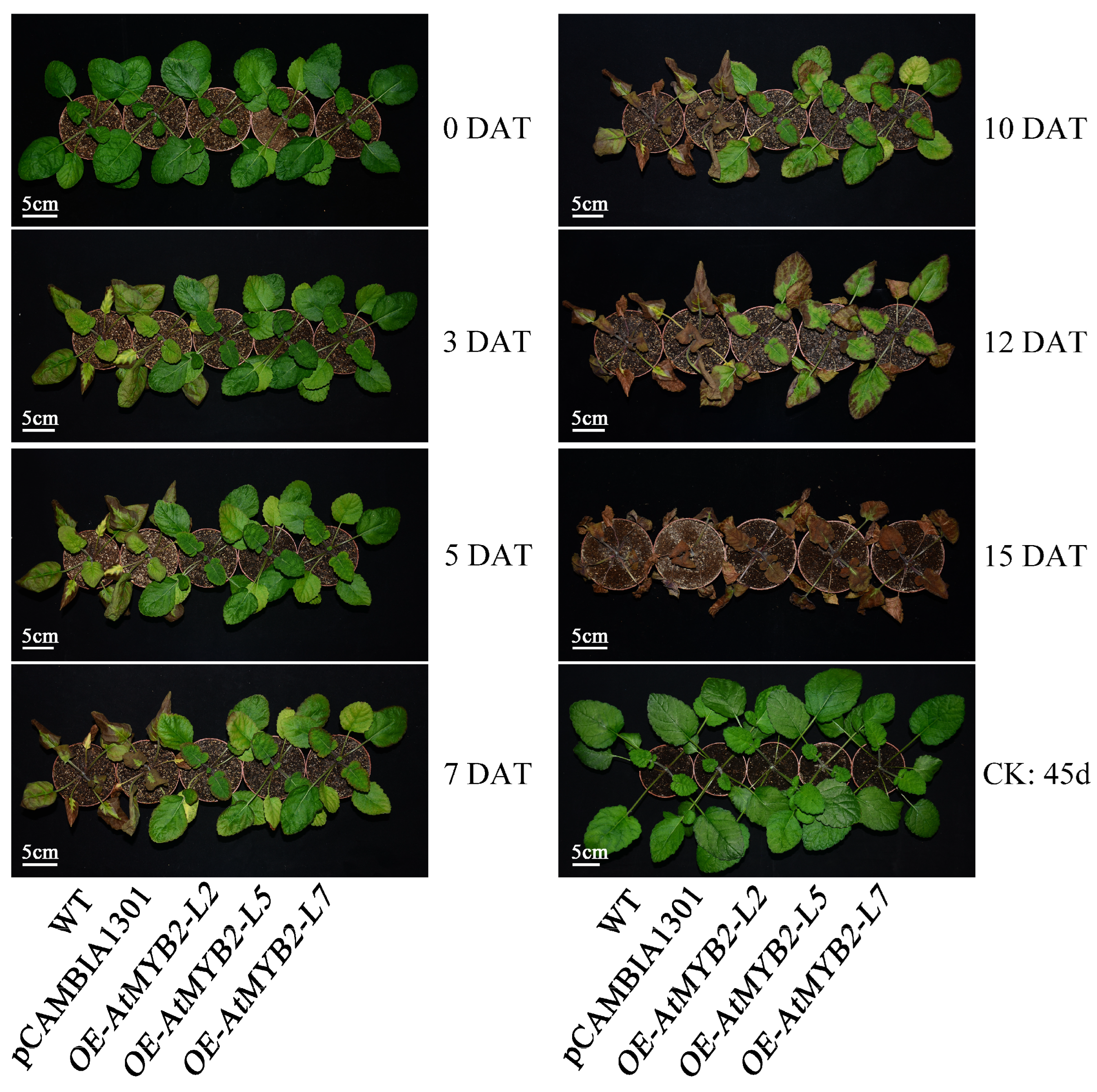
2.2. AtMYB2 Promotes Tolerance to Salt Stress in S. miltiorrhiza
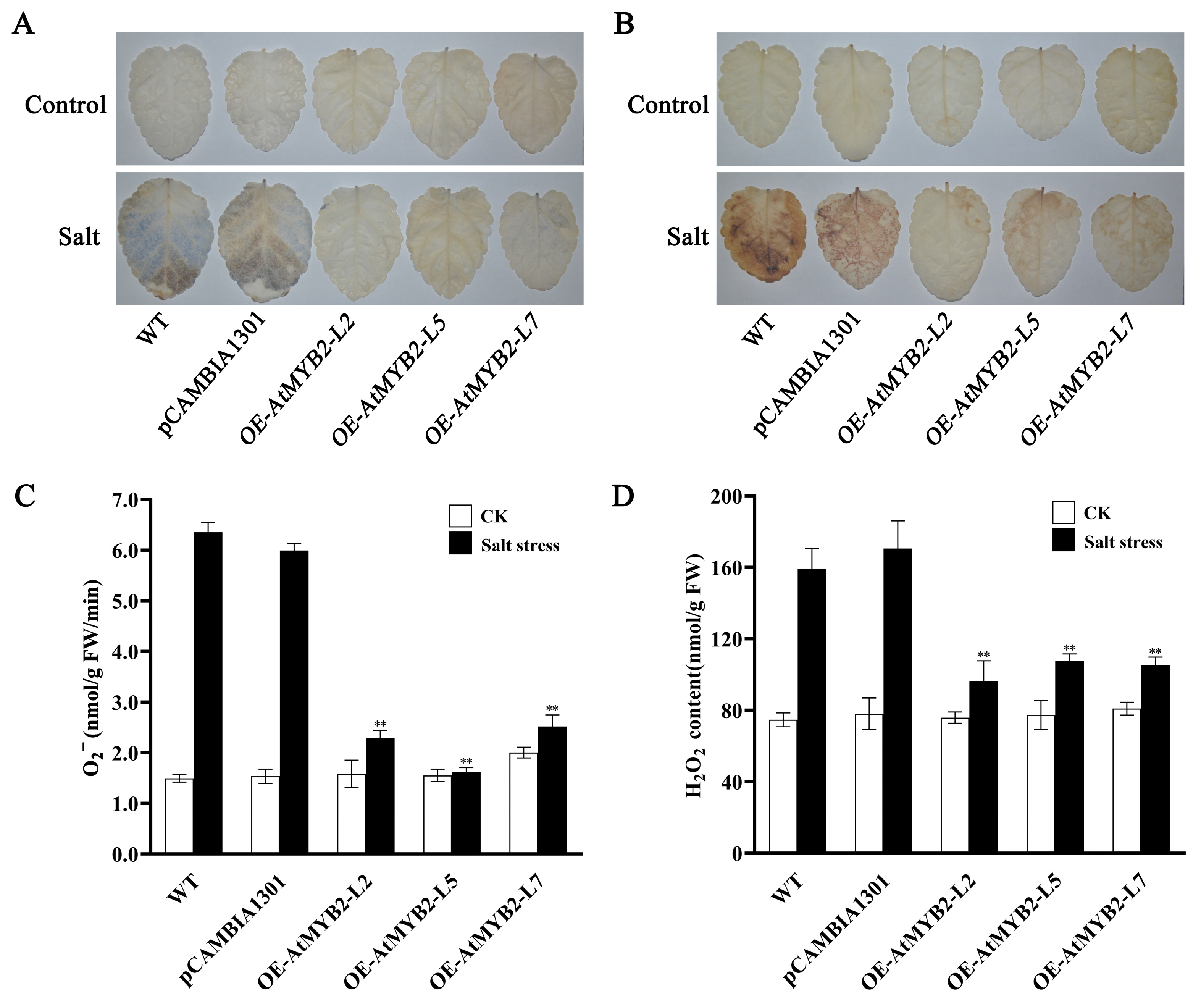
2.3. AtMYB2 Reduces ROS Accumulation under Salt Stress
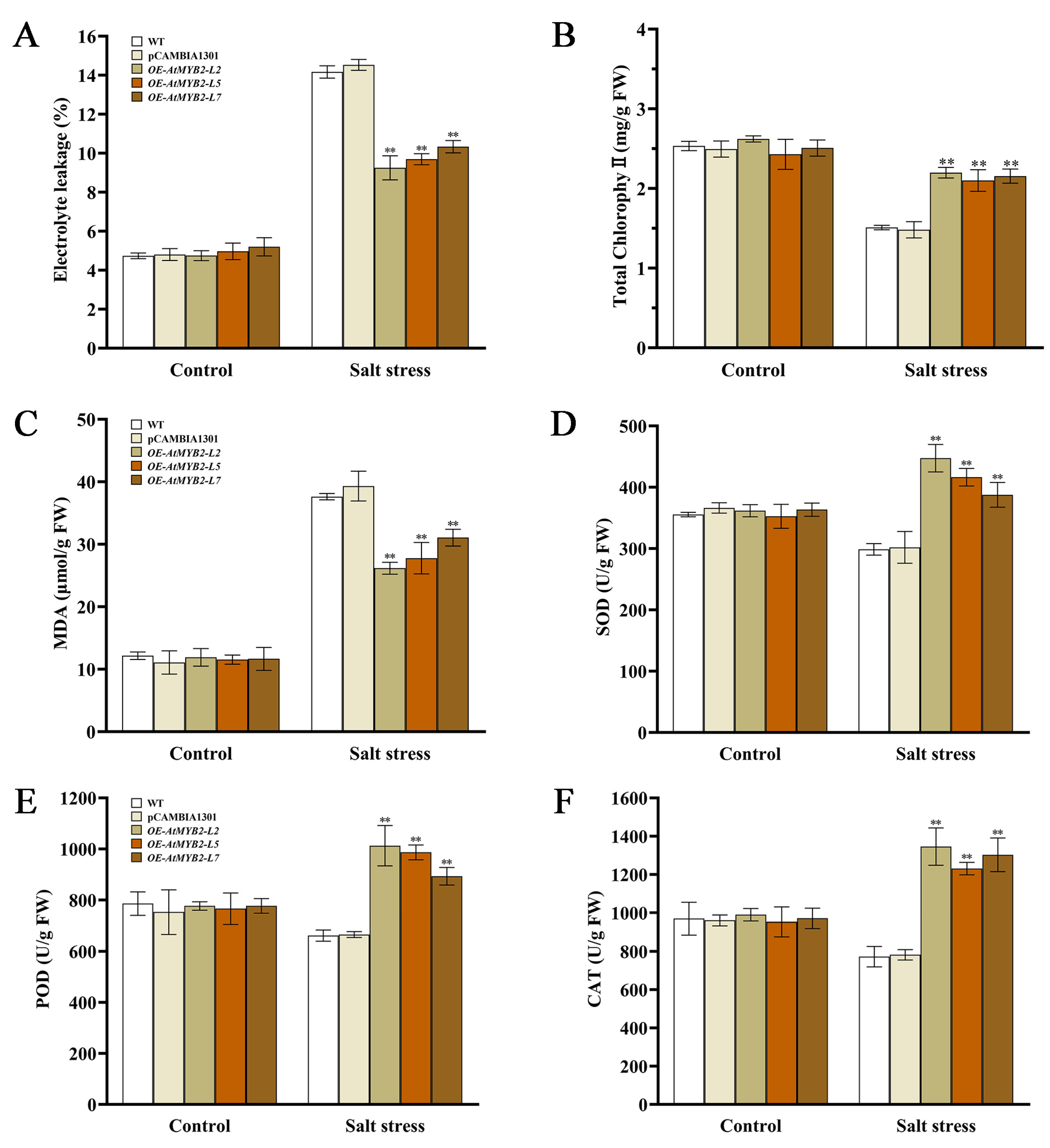
2.4. AtMYB2 Improves Antioxidant Capacity under Salt Stress
2.5. AtMYB2 Promotes the Accumulation of Tanshinones and Salvianolic Acid
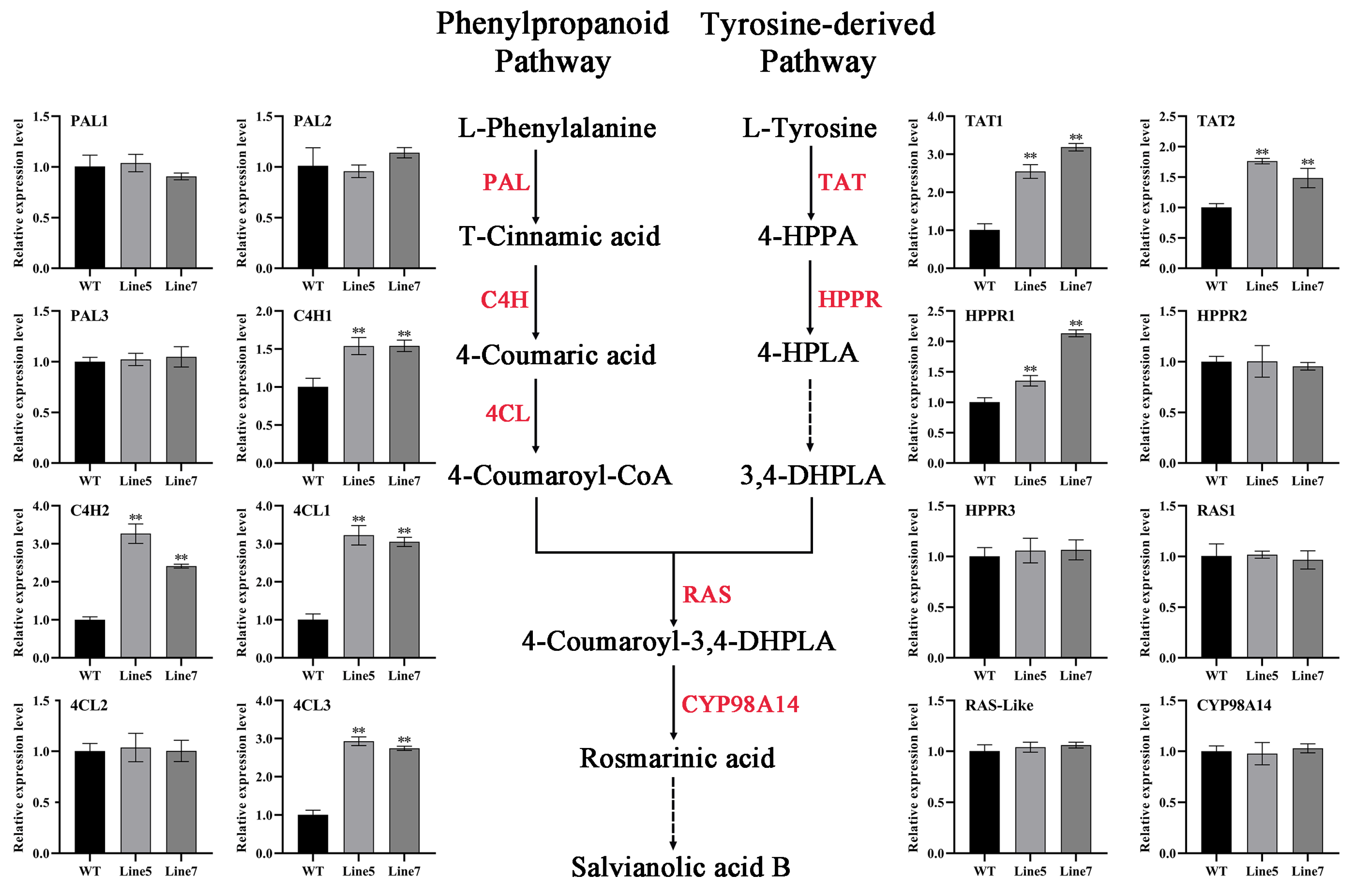
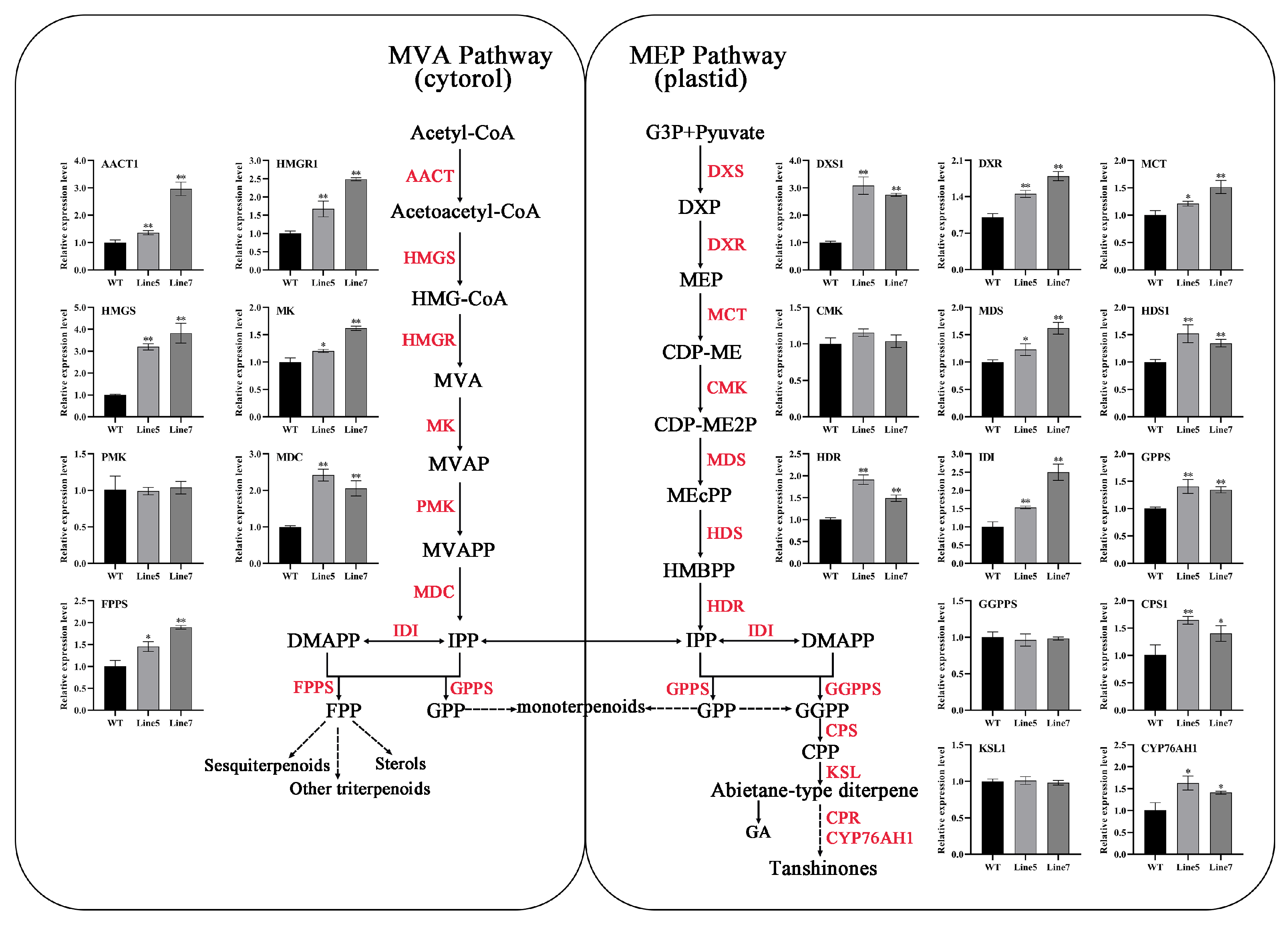
2.6. AtMYB2 Promotes Tanshinones and Salvianolic Acid Content by Upregulating the Expression of Biosynthesis Genes
2.7. Transcriptomic Analysis of AtMYB2 Transgenic S. miltiorrhiza
3. Discussion
3.1. AtMYB2 Promotes Tolerance to Salt Stress by Improving Antioxidant System Function
3.2. AtMYB2 Promotes Tanshinones and Salvianolic Acid Content by Upregulating the Expression of Biosynthesis Genes
4. Materials and Methods
4.1. Plant Materials and Vector Construction
4.2. Genetic Transformation and Molecular Characterization
4.3. RNA Isolation and Detection of Gene Transcription Level
4.4. Stress Treatment
4.5. Determining the Physiological Indices of Leaves and the Active Ingredients in Roots
4.6. Transcriptomic Analysis of AtMYB2 Transgenic S. miltiorrhiza
4.7. Statistical AnalysisOK
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Zhou, H.; Shi, H.; Yang, Y.; Feng, X.; Chen, X.; Xiao, F.; Lin, H.; Guo, Y. Insights into plant salt stress signaling and tolerance. J. Genet. Genom. 2024, 51, 16–34. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, G.; Chen, S.; Jia, Z.; Zhang, S.; He, F.; Ren, M. Genome-wide analysis of the Tritipyrum NAC gene family and the response of TtNAC477 in salt tolerance. BMC Plant Biol. 2024, 24, 40. [Google Scholar] [CrossRef]
- Fu, H.; Yang, Y. How Plants Tolerate Salt Stress. Curr. Issues Mol. Biol. 2023, 45, 5914–5934. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Raihan, M.R.H.; Masud, A.A.C.; Rahman, K.; Nowroz, F.; Rahman, M.; Nahar, K.; Fujita, M. Regulation of Reactive Oxygen Species and Antioxidant Defense in Plants under Salinity. Int. J. Mol. Sci. 2021, 22, 9326. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, H.; He, Y.; Shen, X.; Lin, S.; Huang, L. MYB transcription factors and their roles in the male reproductive development of flowering plants. Plant Sci. 2023, 335, 111811. [Google Scholar] [CrossRef]
- Yu, Y.; Guo, D.D.; Min, D.H.; Cao, T.; Ning, L.; Jiang, Q.Y.; Sun, X.J.; Zhang, H.; Tang, W.S.; Gao, S.Q.; et al. Foxtail millet MYB-like transcription factor SiMYB16 confers salt tolerance in transgenic rice by regulating phenylpropane pathway. Plant Physiol. Biochem. 2023, 195, 310–321. [Google Scholar] [CrossRef]
- Bian, S.; Jin, D.; Sun, G.; Shan, B.; Zhou, H.; Wang, J.; Zhai, L.; Li, X. Characterization of the soybean R2R3-MYB transcription factor GmMYB81 and its functional roles under abiotic stresses. Gene 2020, 753, 144803. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef]
- XD, M.E.; Cao, Y.F.; Che, Y.Y.; Li, J.; Shang, Z.P.; Zhao, W.J.; Qiao, Y.J.; Zhang, J.Y. Danshen: A phytochemical and pharmacological overview. Chin. J. Nat. Med. 2019, 17, 59–80. [Google Scholar] [CrossRef]
- Ren, J.; Fu, L.; Nile, S.H.; Zhang, J.; Kai, G. Salvia miltiorrhiza in Treating Cardiovascular Diseases: A Review on Its Pharmacological and Clinical Applications. Front. Pharmacol. 2019, 10, 753. [Google Scholar] [CrossRef]
- Wei, B.; Sun, C.; Wan, H.; Shou, Q.; Han, B.; Sheng, M.; Li, L.; Kai, G. Bioactive components and molecular mechanisms of Salvia miltiorrhiza Bunge in promoting blood circulation to remove blood stasis. J. Ethnopharmacol. 2023, 317, 116697. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, L.; Zheng, X.; Zhang, J.; Yang, L.; Tan, R.; Zhao, S. Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep. 2017, 36, 1297–1309. [Google Scholar] [CrossRef]
- Liu, L.; Yang, D.; Xing, B.; Zhang, C.; Liang, Z. SmMYB98b positive regulation to tanshinones in Salvia miltiorrhiza Bunge hairy roots. Plant Cell Tissue Organ Cult. 2019, 140, 459–467. [Google Scholar] [CrossRef]
- Li, S.; Wu, Y.; Kuang, J.; Wang, H.; Du, T.; Huang, Y.; Zhang, Y.; Cao, X.; Wang, Z. SmMYB111 Is a Key Factor to Phenolic Acid Biosynthesis and Interacts with Both SmTTG1 and SmbHLH51 in Salvia miltiorrhiza. J. Agric. Food Chem. 2018, 66, 8069–8078. [Google Scholar] [CrossRef]
- Deng, C.; Wang, Y.; Huang, F.; Lu, S.; Zhao, L.; Ma, X.; Kai, G. SmMYB2 promotes salvianolic acid biosynthesis in the medicinal herb Salvia miltiorrhiza. J. Integr. Plant Biol. 2020, 62, 1688–1702. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, Y.; Li, L.; Guo, X.; Wang, B.; Cao, X.; Wang, Z. AtPAP1 Interacts with and Activates SmbHLH51, a Positive Regulator to Phenolic Acids Biosynthesis in Salvia miltiorrhiza. Front. Plant Sci. 2018, 9, 1687. [Google Scholar] [CrossRef]
- Li, T.; Zhang, S.; Li, Y.; Zhang, L.; Song, W.; Chen, C.; Ruan, W. Simultaneous Promotion of Salt Tolerance and Phenolic Acid Biosynthesis in Salvia miltiorrhiza via Overexpression of Arabidopsis MYB12. Int. J. Mol. Sci. 2023, 24, 15506. [Google Scholar] [CrossRef]
- Urao, T.; Yamaguchi-Shinozaki, K.; Urao, S.; Shinozaki, K. An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 1993, 5, 1529–1539. [Google Scholar] [CrossRef]
- Baek, D.; Kim, M.C.; Chun, H.J.; Kang, S.; Park, H.C.; Shin, G.; Park, J.; Shen, M.; Hong, H.; Kim, W.-Y.; et al. Regulation of miR399f Transcription by AtMYB2 Affects Phosphate Starvation Responses in Arabidopsis. Plant Physiol. 2013, 161, 362–373. [Google Scholar] [CrossRef]
- Hoeren, F.U.; Dolferus, R.; Wu, Y.; Peacock, W.J.; Dennis, E.S. Evidence for a role for AtMYB2 in the induction of the Arabidopsis alcohol dehydrogenase gene (ADH1) by low oxygen. Genetics 1998, 149, 479–490. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, M.; Yang, Q.; Liu, K.; Cui, J.; Chen, J.; Ren, Y.; Shao, Y.; Wang, R.; Li, G. The S40 family members delay leaf senescence by promoting cytokinin synthesis. Plant Physiol. Biochem. 2022, 191, 99–109. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtMYB2 Regulates Whole Plant Senescence by Inhibiting Cytokinin-Mediated Branching at Late Stages of Development in Arabidopsis. Plant Physiol. 2011, 156, 1612–1619. [Google Scholar] [CrossRef]
- Lim, G.H.; Kim, S.W.; Ryu, J.; Kang, S.Y.; Kim, J.B.; Kim, S.H. Upregulation of the MYB2 Transcription Factor Is Associated with Increased Accumulation of Anthocyanin in the Leaves of Dendrobium bigibbum. Int. J. Mol. Sci. 2020, 21, 5653. [Google Scholar] [CrossRef]
- Jun, J.H.; Liu, C.; Xiao, X.; Dixon, R.A. The Transcriptional Repressor MYB2 Regulates Both Spatial and Temporal Patterns of Proanthocyandin and Anthocyanin Pigmentation in Medicago truncatula. Plant Cell 2015, 27, 2860–2879. [Google Scholar] [CrossRef]
- Jia, T.; Zhang, K.; Li, F.; Huang, Y.; Fan, M.; Huang, T. The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Rep. 2020, 39, 1755–1765. [Google Scholar] [CrossRef]
- Guo, X.; Li, L.; Liu, X.; Zhang, C.; Yao, X.; Xun, Z.; Zhao, Z.; Yan, W.; Zou, Y.; Liu, D.; et al. MYB2 Is Important for Tapetal PCD and Pollen Development by Directly Activating Protease Expression in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3563. [Google Scholar] [CrossRef]
- Kawase, Y.; Imamura, S.; Tanaka, K. A MYB-type transcription factor, MYB2, represses light-harvesting protein genes in Cyanidioschyzon merolae. FEBS Lett. 2017, 591, 2439–2448. [Google Scholar] [CrossRef]
- Urao, T.; Noji, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A transcriptional activation domain of ATMYB2, a drought-inducible Arabidopsis Myb-related protein. Plant J. Cell Mol. Biol. 1996, 10, 1145–1148. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Urao, T.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef]
- Yoo, J.H.; Park, C.Y.; Kim, J.C.; Do Heo, W.; Cheong, M.S.; Park, H.C.; Kim, M.C.; Moon, B.C.; Choi, M.S.; Kang, Y.H.; et al. Direct Interaction of a Divergent CaM Isoform and the Transcription Factor, MYB2, Enhances Salt Tolerance in Arabidopsis. J. Biol. Chem. 2005, 280, 3697–3706. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Guan, Q.; Ma, X.; Zheng, X.; Zou, C.; Li, Q. AtMYB2 transcription factor can interact with the CMO promoter and regulate its downstream gene expression. Biotechnol. Lett. 2012, 34, 1749–1755. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 2018, 60, 796–804. [Google Scholar] [CrossRef]
- Abogadallah, G.M. Antioxidative defense under salt stress. Plant Signal. Behav. 2010, 5, 369–374. [Google Scholar] [CrossRef]
- Yang, R.; Yang, Z.; Xing, M.; Jing, Y.; Zhang, Y.; Zhang, K.; Zhou, Y.; Zhao, H.; Qiao, W.; Sun, J. TaBZR1 enhances wheat salt tolerance via promoting ABA biosynthesis and ROS scavenging. J. Genet. Genom. 2023, 50, 861–871. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, R.; Yang, X.; Ju, Q.; Li, W.; Lü, S.; Tran, L.P.; Xu, J. The R2R3-MYB transcription factor AtMYB49 modulates salt tolerance in Arabidopsis by modulating the cuticle formation and antioxidant defence. Plant Cell Environ. 2020, 43, 1925–1943. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lan, X.; Zhou, J.; Gao, K.; Zhong, C.; Xie, J. Dioscorea composita WRKY3 positively regulates salt-stress tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 2022, 269, 153592. [Google Scholar] [CrossRef]
- Olsson, M.E.; Olofsson, L.M.; Lindahl, A.-L.; Lundgren, A.; Brodelius, M.; Brodelius, P.E. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 2009, 70, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-H.; Wang, J.-W.; Wang, S.; Wang, J.-Y.; Chen, X.-Y. Characterization of GaWRKY1, a Cotton Transcription Factor That Regulates the Sesquiterpene Synthase Gene (+)-δ-Cadinene Synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Wang, Y.; Wang, X.; Deng, C.; Cao, W.; Hua, Q.; Kai, G. Simultaneous promotion of tanshinone and phenolic acid biosynthesis in Salvia miltiorrhiza hairy roots by overexpressing Arabidopsis MYC2. Ind. Crops Prod. 2020, 155, 112826. [Google Scholar] [CrossRef]
- Alfieri, M.; Vaccaro, M.C.; Cappetta, E.; Ambrosone, A.; De Tommasi, N.; Leone, A. Coactivation of MEP-biosynthetic genes and accumulation of abietane diterpenes in Salvia sclarea by heterologous expression of WRKY and MYC2 transcription factors. Sci. Rep. 2018, 8, 11009. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Dong, Q.; Duan, D.; Zhao, S.; Li, M.; van Nocker, S.; Ma, F.; Mao, K. Comprehensive genomic analysis of the TYROSINE AMINOTRANSFERASE (TAT) genes in apple (Malus domestica) allows the identification of MdTAT2 conferring tolerance to drought and osmotic stresses in plants. Plant Physiol. Biochem. 2018, 133, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Liu, J.; Zhang, C.; Liang, Z. Regulation of Water-Soluble Phenolic Acid Biosynthesis in Salvia miltiorrhiza Bunge. Appl. Biochem. Biotechnol. 2013, 170, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Sun, M.; Yuan, T.; Wang, Y.; Shi, M.; Lu, S.; Tang, B.; Pan, J.; Wang, Y.; Kai, G. The AP2/ERF transcription factor SmERF1L1 regulates the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Food Chem. 2019, 274, 368–375. [Google Scholar] [CrossRef]
- Zheng, H.; Jing, L.; Jiang, X.; Pu, C.; Zhao, S.; Yang, J.; Guo, J.; Cui, G.; Tang, J.; Ma, Y.; et al. The ERF-VII transcription factor SmERF73 coordinately regulates tanshinone biosynthesis in response to stress elicitors in Salvia miltiorrhiza. New Phytol. 2021, 231, 1940–1955. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Deng, K.; Zhang, Q.; Gao, Y.; Liu, Y.; Yang, M.; Zhang, L.; Zheng, X.; Wang, C.; Liu, Z.; et al. Modulating AtDREB1C Expression Improves Drought Tolerance in Salvia miltiorrhiza. Front. Plant Sci. 2017, 8, 52. [Google Scholar] [CrossRef]
- Wei, T.; Deng, K.; Gao, Y.; Chen, L.; Song, W.; Zhang, Y.; Wang, C.; Chen, C. SmKSL overexpression combined with elicitor treatment enhances tanshinone production from Salvia miltiorrhiza hairy roots. Biochem. Eng. J. 2020, 158, 107562. [Google Scholar] [CrossRef]
- Zhang, X.; Tan, B.; Zhu, D.; Dufresne, D.; Jiang, T.; Chen, S. Proteomics of Homeobox7 Enhanced Salt Tolerance in Mesembryanthemum crystallinum. Int. J. Mol. Sci. 2021, 22, 6390. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Deng, K.; Gao, Y.; Liu, Y.; Yang, M.; Zhang, L.; Zheng, X.; Wang, C.; Song, W.; Chen, C.; et al. Arabidopsis DREB1B in transgenic Salvia miltiorrhiza increased tolerance to drought stress without stunting growth. Plant Physiol. Biochem. 2016, 104, 17–28. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Zhang, S.; Li, Y.; Zhang, L.; Song, W.; Chen, C. Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza. Int. J. Mol. Sci. 2024, 25, 4111. https://doi.org/10.3390/ijms25074111
Li T, Zhang S, Li Y, Zhang L, Song W, Chen C. Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza. International Journal of Molecular Sciences. 2024; 25(7):4111. https://doi.org/10.3390/ijms25074111
Chicago/Turabian StyleLi, Tianyu, Shuangshuang Zhang, Yidan Li, Lipeng Zhang, Wenqin Song, and Chengbin Chen. 2024. "Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza" International Journal of Molecular Sciences 25, no. 7: 4111. https://doi.org/10.3390/ijms25074111
APA StyleLi, T., Zhang, S., Li, Y., Zhang, L., Song, W., & Chen, C. (2024). Overexpression of AtMYB2 Promotes Tolerance to Salt Stress and Accumulations of Tanshinones and Phenolic Acid in Salvia miltiorrhiza. International Journal of Molecular Sciences, 25(7), 4111. https://doi.org/10.3390/ijms25074111





