Small Molecules Promote the Rapid Generation of Dental Epithelial Cells from Human-Induced Pluripotent Stem Cells
Abstract
:1. Introduction
2. Results
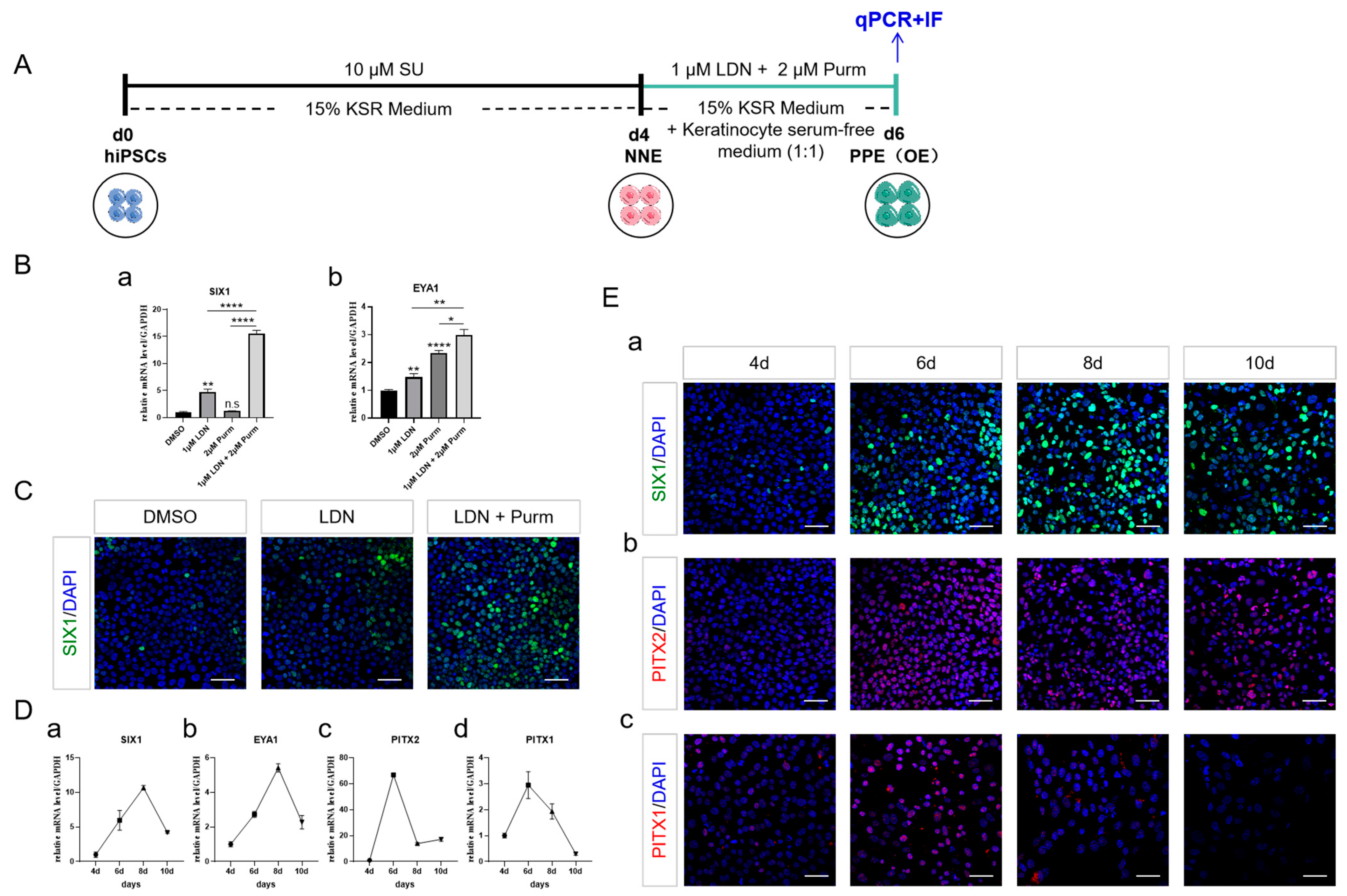
2.1. NNE Cell Fate Determination through FGF Inhibition
2.2. Cell Density and the Duration of Differentiation Are Important for NNE Differentiation
2.3. PPE Specification and the Emergence of OE through BMP Inhibition and SHH Activation
2.4. DE Formation through SHH and BMP Activation, Alongside with WNT Inhibition
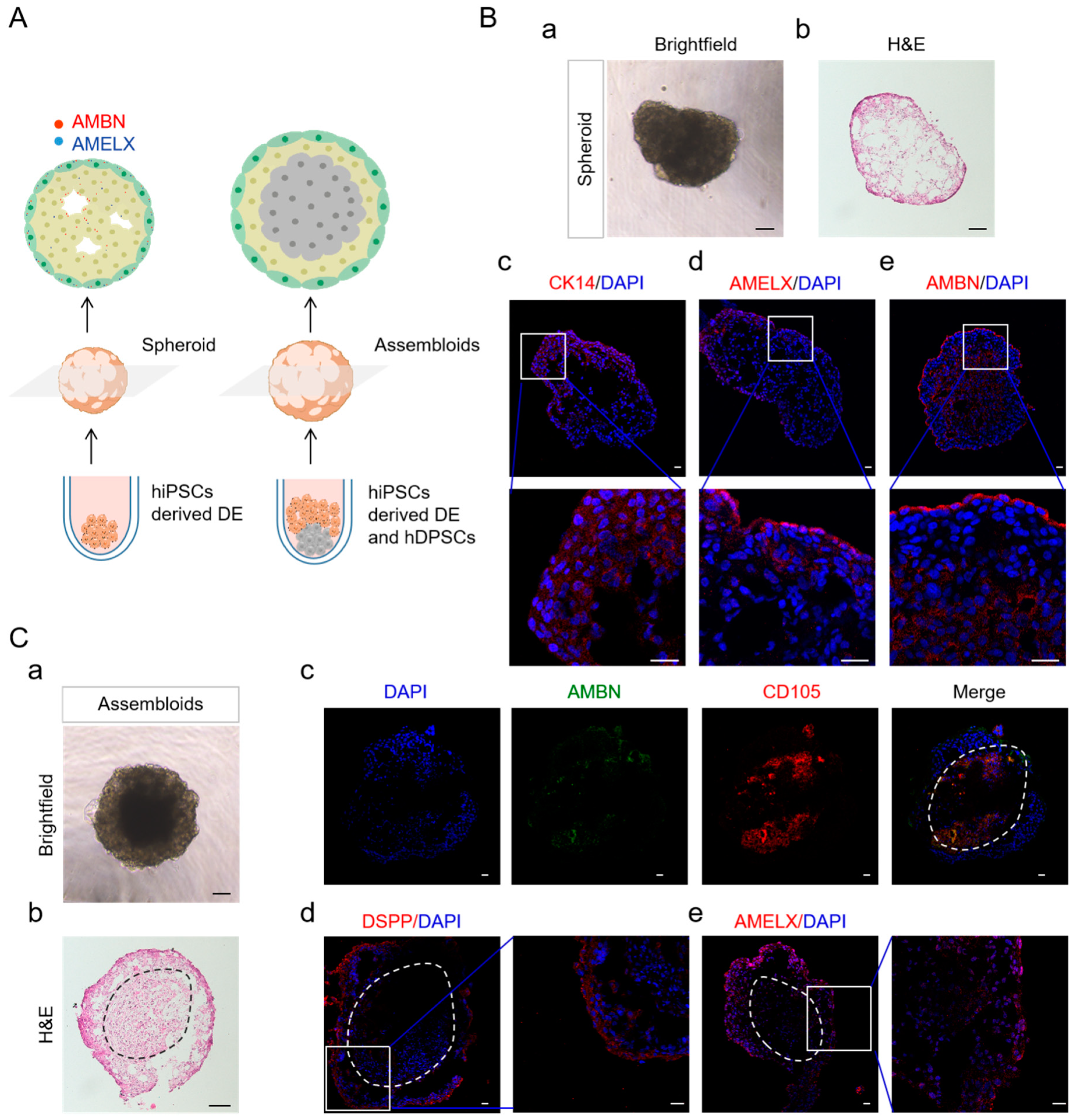
2.5. DE in Spheroid Culture and Assembled with hDPSCs Allows Further Differentiation
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Culture Conditions
4.2. Dental Epithelial (DE) Cell Differentiation in Monolayer
4.3. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR) Analysis
4.4. Immunostaining
4.5. Development of hiPSCs Derived DE Cells and Co-Culturing hiPSCs Derived DE with hDPSCs in Sphere Culture
4.6. Cryosectioning and Immunostaining for the Sphere
4.7. Alizarin Red Staining
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houari, S.; DeRocher, K.; Thuy, T.T.; Coradin, T.; Srot, V.; van Aken, P.A.; Lecoq, H.; Sauvage, T.; Balan, E.; Aufort, J.; et al. Multi-scale characterization of Developmental Defects of Enamel and their clinical significance for diagnosis and treatment. Acta Biomater. 2023, 169, 155–167. [Google Scholar] [CrossRef] [PubMed]
- Jokisaari, J.R.; Wang, C.; Qiao, Q.; Hu, X.; Reed, D.A.; Bleher, R.; Luan, X.; Klie, R.F.; Diekwisch, T.G. Particle-Attachment-Mediated and Matrix/Lattice-Guided Enamel Apatite Crystal Growth. ACS Nano 2019, 13, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.L.; Tavares, A.L.P.; Peterson, M.; Sullivan, C.H.; Moody, S.A. Repressive Interactions Between Transcription Factors Separate Different Embryonic Ectodermal Domains. Front. Cell Dev. Biol. 2022, 10, 786052. [Google Scholar] [CrossRef] [PubMed]
- Heasman, J. Patterning the early Xenopus embryo. Development 2006, 133, 1205–1217. [Google Scholar] [CrossRef]
- Britton, G.; Heemskerk, I.; Hodge, R.; Qutub, A.A.; Warmflash, A. A novel self-organizing embryonic stem cell system reveals signaling logic underlying the patterning of human ectoderm. Development 2019, 146, dev179093. [Google Scholar] [CrossRef]
- Ong, A.L.C.; Kokaji, T.; Kishi, A.; Takihara, Y.; Shinozuka, T.; Shimamoto, R.; Isotani, A.; Shirai, M.; Sasai, N. Acquisition of neural fate by combination of BMP blockade and chromatin modification. iScience 2023, 26, 107887. [Google Scholar] [CrossRef]
- Wei, S.; Wang, Q. Molecular regulation of Nodal signaling during mesendoderm formation. Acta Biochim. Biophys. Sin. 2017, 50, 74–81. [Google Scholar] [CrossRef]
- Zhong, H.; Ren, Z.; Wang, X.; Miao, K.; Ni, W.; Meng, Y.; Lu, L.; Wang, C.; Liu, W.; Deng, C.-X.; et al. Stagewise keratinocyte differentiation from human embryonic stem cells by defined signal transduction modulators. Int. J. Biol. Sci. 2020, 16, 1450–1462. [Google Scholar] [CrossRef]
- Mammadova, A.; Zhou, H.; Carels, C.E.; Hoff, J.W.V.D. Retinoic acid signalling in the development of the epidermis, the limbs and the secondary palate. Differentiation 2016, 92, 326–335. [Google Scholar] [CrossRef]
- Metallo, C.M.; Ji, L.; de Pablo, J.J.; Palecek, S.P. Retinoic acid and bone morphogenetic protein signaling synergize to efficiently direct epithelial differentiation of human embryonic stem cells. Stem Cells 2008, 26, 372–380. [Google Scholar] [CrossRef]
- Guzzetta, A.; Koska, M.; Rowton, M.; Sullivan, K.R.; Jacobs-Li, J.; Kweon, J.; Hidalgo, H.; Eckart, H.; Hoffmann, A.D.; Back, R.; et al. Hedgehog–FGF signaling axis patterns anterior mesoderm during gastrulation. Proc. Natl. Acad. Sci. USA 2020, 117, 15712–15723. [Google Scholar] [CrossRef] [PubMed]
- Hongo, I.; Okamoto, H. FGF/MAPK/Ets signaling in Xenopus ectoderm contributes to neural induction and patterning in an autonomous and paracrine manner, respectively. Cells Dev. 2022, 170, 203769. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.H.; Amirruddin, N.S.; Loo, L.S.W.; Chan, J.W.; Iich, E.; Krishnan, V.G.; Hoon, S.; Teo, A.K.K. FGFR-mediated ERK1/2 signaling contributes to mesendoderm and definitive endoderm formation in vitro. iScience 2023, 26, 107265. [Google Scholar] [CrossRef] [PubMed]
- Tchieu, J.; Zimmer, B.; Fattahi, F.; Amin, S.; Zeltner, N.; Chen, S.; Studer, L. A Modular Platform for Differentiation of Human PSCs into All Major Ectodermal Lineages. Cell Stem Cell 2017, 21, 399–410.e7. [Google Scholar] [CrossRef] [PubMed]
- Griffin, C.; Saint-Jeannet, J.-P. In vitro modeling of cranial placode differentiation: Recent advances, challenges, and perspectives. Dev. Biol. 2024, 506, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Jacox, L.; Chen, J.; Rothman, A.; Lathrop-Marshall, H.; Sive, H. Formation of a “Pre-mouth Array” from the Extreme Anterior Domain Is Directed by Neural Crest and Wnt/PCP Signaling. Cell Rep. 2016, 16, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Michiue, T.; Tsukano, K. Feedback Regulation of Signaling Pathways for Precise Pre-Placodal Ectoderm Formation in Vertebrate Embryos. J. Dev. Biol. 2022, 10, 35. [Google Scholar] [CrossRef]
- Seo, H.; Amano, T.; Seki, R.; Sagai, T.; Kim, J.; Cho, S.; Shiroishi, T. Upstream Enhancer Elements of Shh Regulate Oral and Dental Patterning. J. Dent. Res. 2018, 97, 1055–1063. [Google Scholar] [CrossRef]
- Tabler, J.M.; Bolger, T.G.; Wallingford, J.; Liu, K.J. Hedgehog activity controls opening of the primary mouth. Dev. Biol. 2014, 396, 1–7. [Google Scholar] [CrossRef]
- Koehler, K.R.; Mikosz, A.M.; Molosh, A.I.; Patel, D.; Hashino, E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 2013, 500, 217–221. [Google Scholar] [CrossRef]
- Kim, E.-J.; Mai, H.N.; Lee, D.-J.; Kim, K.-H.; Lee, S.-J.; Jung, H.-S. Strategies for differentiation of hiPSCs into dental epithelial cell lineage. Cell Tissue Res. 2021, 386, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Alghadeer, A.; Hanson-Drury, S.; Patni, A.P.; Ehnes, D.D.; Zhao, Y.T.; Li, Z.; Phal, A.; Vincent, T.; Lim, Y.C.; O’day, D.; et al. Single-cell census of human tooth development enables generation of human enamel. Dev. Cell 2023, 58, 2163–2180.e9. [Google Scholar] [CrossRef]
- Leung, A.W.; Morest, D.K.; Li, J.Y. Differential BMP signaling controls formation and differentiation of multipotent preplacodal ectoderm progenitors from human embryonic stem cells. Dev. Biol. 2013, 379, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Soukup, V.; Horácek, I.; Cerny, R. Development and evolution of the vertebrate primary mouth. J. Anat. 2012, 222, 79–99. [Google Scholar] [CrossRef]
- Jussila, M.; Thesleff, I. Signaling Networks Regulating Tooth Organogenesis and Regeneration, and the Specification of Dental Mesenchymal and Epithelial Cell Lineages. Cold Spring Harb. Perspect. Biol. 2012, 4, a008425. [Google Scholar] [CrossRef]
- Hosoya, A.; Shalehin, N.; Takebe, H.; Shimo, T.; Irie, K. Sonic Hedgehog Signaling and Tooth Development. Int. J. Mol. Sci. 2020, 21, 1587. [Google Scholar] [CrossRef]
- Sarkar, L.; Cobourne, M.; Naylor, S.; Smalley, M.; Dale, T.; Sharpe, P.T. Wnt/Shh interactions regulate ectodermal boundary formation during mammalian tooth development. Proc. Natl. Acad. Sci. USA 2000, 97, 4520–4524. [Google Scholar] [CrossRef] [PubMed]
- Binder, M.; Biggs, L.C.; Kronenberg, M.S.; Schneider, P.; Thesleff, I.; Balic, A. Novel strategies for expansion of tooth epithelial stem cells and ameloblast generation. Sci. Rep. 2020, 10, 4963. [Google Scholar] [CrossRef]
- Li, J.; Feng, J.; Liu, Y.; Ho, T.-V.; Grimes, W.; Ho, H.A.; Park, S.; Wang, S.; Chai, Y. BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev. Cell 2015, 33, 125–135. [Google Scholar] [CrossRef]
- Bloomquist, R.F.; Parnell, N.F.; Phillips, K.A.; Fowler, T.E.; Yu, T.Y.; Sharpe, P.T.; Streelman, J.T. Coevolutionary patterning of teeth and taste buds. Proc. Natl. Acad. Sci. USA 2015, 112, E5954–E5962. [Google Scholar] [CrossRef]
- Sunil, P.M. Induced pluripotent stem cells in dentistry. J. Pharm. Bioallied Sci. 2016, 8 (Suppl. 1), S23–S27. [Google Scholar] [CrossRef] [PubMed]
- Radwan, I.A.; Rady, D.; Abbass, M.M.S.; El Moshy, S.; AbuBakr, N.; Dörfer, C.E.; El-Sayed, K.M.F. Induced Pluripotent Stem Cells in Dental and Nondental Tissue Regeneration: A Review of an Unexploited Potential. Stem Cells Int. 2020, 2020, 1941629. [Google Scholar] [CrossRef]
- Beck, H.; Härter, M.; Haß, B.; Schmeck, C.; Baerfacker, L. Small molecules and their impact in drug discovery: A perspective on the occasion of the 125th anniversary of the Bayer Chemical Research Laboratory. Drug Discov. Today 2022, 27, 1560–1574. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-J.; Bhat, N.; Sweet, E.M.; Cornell, R.A.; Riley, B.B. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010, 6, e1001133. [Google Scholar] [CrossRef] [PubMed]
- Dincer, Z.; Piao, J.; Niu, L.; Ganat, Y.; Kriks, S.; Zimmer, B.; Shi, S.-H.; Tabar, V.; Studer, L. Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Rep. 2013, 5, 1387–1402. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Klein, O.D. Molecular and cellular mechanisms of tooth development, homeostasis and repair. Development 2020, 147, dev184754. [Google Scholar] [CrossRef] [PubMed]
- Dassule, H.R.; McMahon, A.P. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 1998, 202, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, L.; Sharpe, P.T. Expression of Wnt signalling pathway genes during tooth development. Mech. Dev. 1999, 85, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Suomalainen, M.; Thesleff, I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev. Dyn. 2010, 239, 364–372. [Google Scholar] [CrossRef]
- Park, S.-J.; Lee, H.-K.; Seo, Y.-M.; Son, C.; Bae, H.S.; Park, J.-C. Dentin sialophosphoprotein expression in enamel is regulated by Copine-7, a preameloblast-derived factor. Arch. Oral Biol. 2018, 86, 131–137. [Google Scholar] [CrossRef]
- Groves, A.K.; LaBonne, C. Setting appropriate boundaries: Fate, patterning and competence at the neural plate border. Dev. Biol. 2014, 389, 2–12. [Google Scholar] [CrossRef]
- Onishi, A.; Abdullah, A.N.; Tanimoto, K.; Kato, K. Optimization of culture conditions for the efficient differentiation of mouse-induced pluripotent stem cells into dental epithelial-like cells. Vitr. Cell Dev. Biol. Anim. 2020, 56, 816–824. [Google Scholar] [CrossRef]
- Hermans, F.; Hemeryck, L.; Lambrichts, I.; Bronckaers, A.; Vankelecom, H. Intertwined Signaling Pathways Governing Tooth Development: A Give-and-Take Between Canonical Wnt and Shh. Front. Cell. Dev. Biol. 2021, 9, 758203. [Google Scholar] [CrossRef]
- Chen, X.; Liu, J.; Li, N.; Wang, Y.; Zhou, N.; Zhu, L.; Shi, Y.; Wu, Y.; Xiao, J.; Liu, C. Mesenchymal Wnt/β-catenin signaling induces Wnt and BMP antagonists in dental epithelium. Organogenesis 2019, 15, 55–67. [Google Scholar] [CrossRef]
- Kim, K.-H.; Kim, E.-J.; Kim, H.-Y.; Li, S.; Jung, H.-S. Fabrication of functional ameloblasts from hiPSCs for dental application. Front. Cell Dev. Biol. 2023, 11, 1164811. [Google Scholar] [CrossRef]
- Hemeryck, L.; Hermans, F.; Chappell, J.; Kobayashi, H.; Lambrechts, D.; Lambrichts, I.; Bronckaers, A.; Vankelecom, H. Organoids from human tooth showing epithelial stemness phenotype and differentiation potential. Cell. Mol. Life Sci. 2022, 79, 153. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-Y.; Cooley, V.; Kim, E.-J.; Li, S.; Lee, J.-M.; Sheyfer, D.; Liu, W.; Klein, O.D.; Joester, D.; Jung, H.-S. Adult dental epithelial stem cell-derived organoids deposit hydroxylapatite biomineral. Int. J. Oral Sci. 2023, 15, 55. [Google Scholar] [CrossRef]
- Yen, B.L.; Hsieh, C.-C.; Hsu, P.-J.; Chang, C.-C.; Wang, L.-T.; Yen, M.-L. Three-Dimensional Spheroid Culture of Human Mesenchymal Stem Cells: Offering Therapeutic Advantages and In Vitro Glimpses of the In Vivo State. Stem Cells Transl. Med. 2023, 12, 235–244. [Google Scholar] [CrossRef]
- Zeichner-David, M.; Diekwisch, T.; Fincham, A.; Lau, E.; MacDougall, M.; Moradian-Oldak, J.; Simmer, J.; Snead, M.; Slavkin, H.C. Control of ameloblast differentiation. Int. J. Dev. Biol. 1995, 39, 69–92. [Google Scholar]
- Liu, X.; Xu, C.; Tian, Y.; Sun, Y.; Zhang, J.; Bai, J.; Pan, Z.; Feng, W.; Xu, M.; Li, C.; et al. RUNX2 contributes to TGF-β1-induced expression of Wdr72 in ameloblasts during enamel mineralization. Biomed. Pharmacother. 2019, 118, 109235. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, X.; Li, Y.; Dong, Q.; Tian, C.; Gong, J.; Bai, X.; Ruan, J.; Gao, J. Small Molecules Promote the Rapid Generation of Dental Epithelial Cells from Human-Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2024, 25, 4138. https://doi.org/10.3390/ijms25084138
Zhu X, Li Y, Dong Q, Tian C, Gong J, Bai X, Ruan J, Gao J. Small Molecules Promote the Rapid Generation of Dental Epithelial Cells from Human-Induced Pluripotent Stem Cells. International Journal of Molecular Sciences. 2024; 25(8):4138. https://doi.org/10.3390/ijms25084138
Chicago/Turabian StyleZhu, Ximei, Yue Li, Qiannan Dong, Chunli Tian, Jing Gong, Xiaofan Bai, Jianping Ruan, and Jianghong Gao. 2024. "Small Molecules Promote the Rapid Generation of Dental Epithelial Cells from Human-Induced Pluripotent Stem Cells" International Journal of Molecular Sciences 25, no. 8: 4138. https://doi.org/10.3390/ijms25084138




