Effects of Slow Freezing and Vitrification of Human Semen on Post-Thaw Semen Quality and miRNA Expression
Abstract
1. Introduction
2. Results
2.1. Patient Characteristics
2.2. Changes in the Sperm Quality Parameters of Normozoospermic Samples after Semen Cryopreservation
2.3. Changes in the Sperm Quality Parameters of Oligozoospermic Samples after Semen Cryopreservation
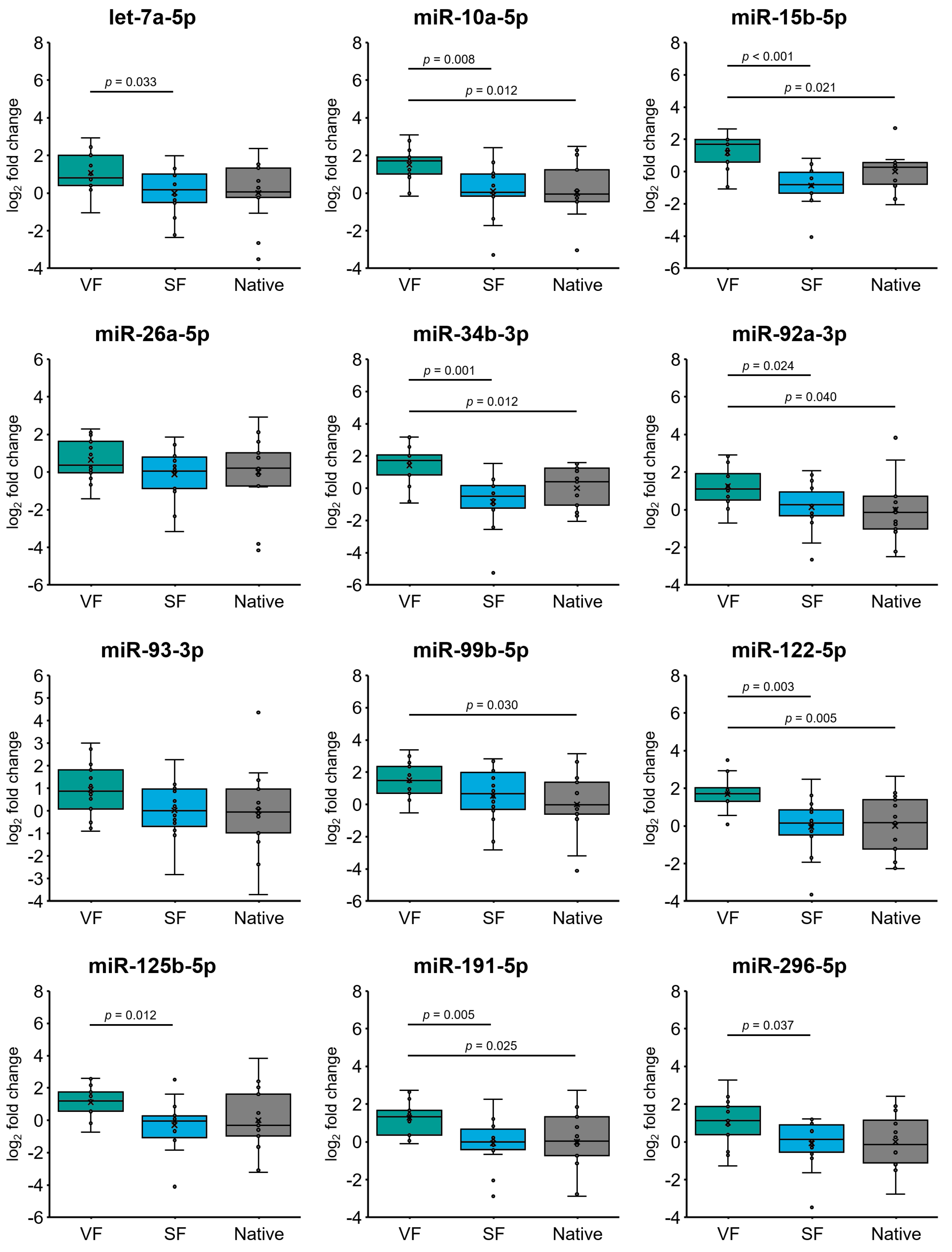
2.4. Cryopreservation Alters miRNA Expression in Spermatozoa from Normozoospermic Patients
2.5. Effect of Cryopreservation on miRNA Expression in Oligozoospermic Sperm Samples
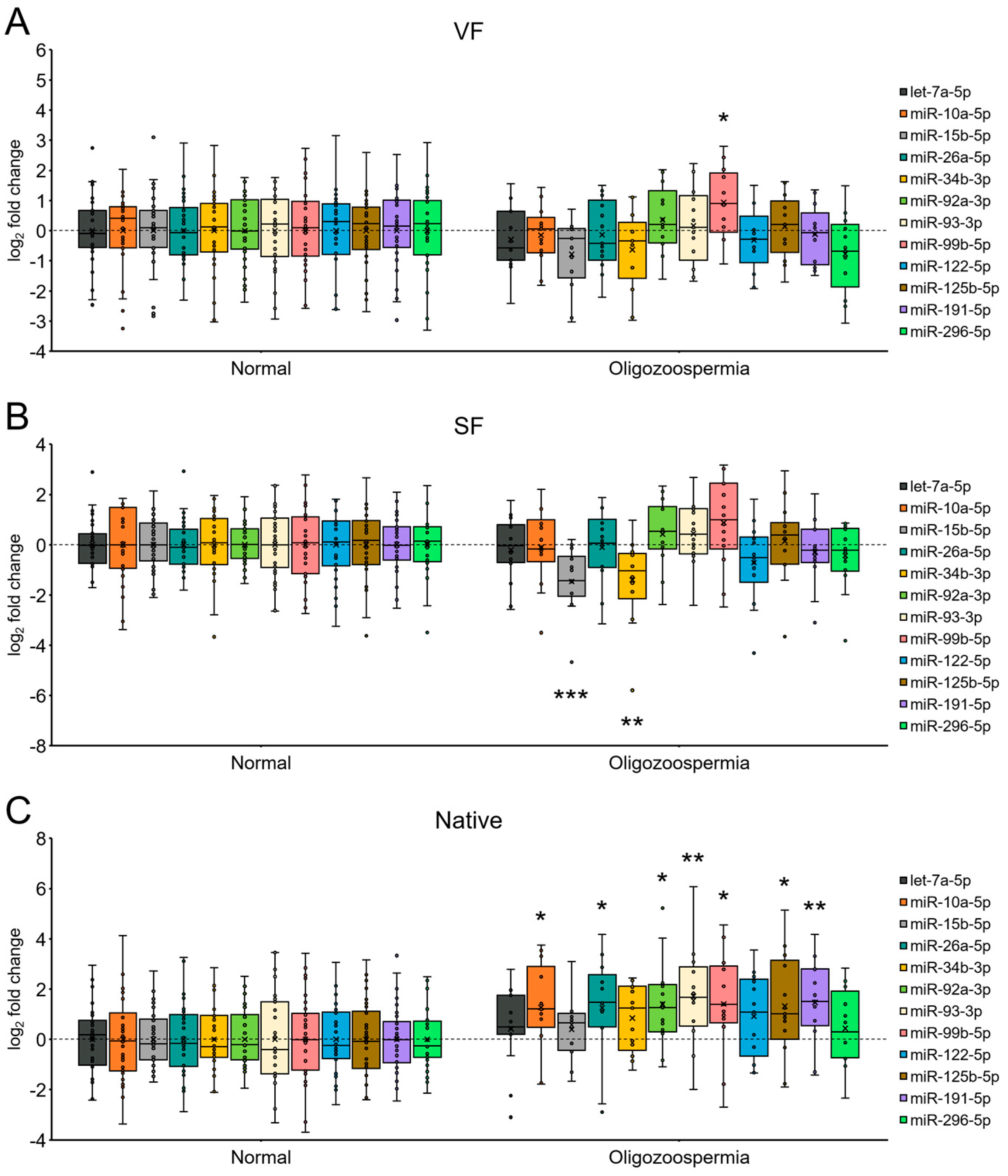
2.6. miRNA Expression in Spermatozoa from Patients with Oligozoospermia under Each Sperm Cryopreservation Condition
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Study Participants and Semen Collection
4.3. Semen Cryopreservation
4.3.1. Slow Freezing
4.3.2. Vitrification
4.4. Semen Thawing/Warming
4.4.1. Slow Freezing
4.4.2. Vitrification
4.5. Determination of Spermatozoa Viability
4.6. Sperm Hyaluronan Binding Assay
4.7. Aniline Blue Staining
4.8. Sperm DNA Fragmentation Analysis (TUNEL Analysis)
4.9. miRNA Analysis
4.9.1. Spermatozoa Preparation
4.9.2. RNA Isolation
4.9.3. Reverse Transcription
4.9.4. Quantitative Real-Time PCR
4.10. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borate, G.M.; Meshram, A. Cryopreservation of Sperm: A Review. Cureus 2022, 14, e31402. [Google Scholar] [CrossRef] [PubMed]
- Bunge, R.G.; Keettel, W.C.; Sherman, J.K. Clinical use of frozen semen: Report of four cases. Fertil. Steril. 1954, 5, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Smeenk, J.; Wyns, C.; De Geyter, C.; Bergh, C.; Cuevas, I.; de Neubourg, D.; Kupka, M.S.; Rezabek, K.; Rugescu, I.; Tandler-Schneider, A.; et al. Assisted Reproductive Technology (ART) in Europe 2020 and development of a strategy of vigilance: Preliminary results generated from European registers by the ESHRE EIM Consortium. Hum. Reprod. 2023, 38 (Suppl. S1), dead093.014. [Google Scholar] [CrossRef]
- Huang, C.; Tang, Y.L.; Hu, J.L.; Zhou, W.J.; Huang, Z.H.; Luo, X.F.; Li, Z.; Zhu, W.B. Update on techniques for cryopreservation of human spermatozoa. Asian J. Androl. 2022, 24, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Sanger, E.; Saewu, A.; Leveille, M.C. Human sperm vitrification: The state of the art. Reprod. Biol. Endocrinol. 2020, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Ali Mohamed, M.S. Slow cryopreservation is not superior to vitrification in human spermatozoa; an experimental controlled study. Iran. J. Reprod. Med. 2015, 13, 633–644. [Google Scholar] [PubMed]
- Ozimic, S.; Ban-Frangez, H.; Stimpfel, M. Sperm Cryopreservation Today: Approaches, Efficiency, and Pitfalls. Curr. Issues Mol. Biol. 2023, 45, 4716–4734. [Google Scholar] [CrossRef]
- Tamburrino, L.; Traini, G.; Marcellini, A.; Vignozzi, L.; Baldi, E.; Marchiani, S. Cryopreservation of Human Spermatozoa: Functional, Molecular and Clinical Aspects. Int. J. Mol. Sci. 2023, 24, 4656. [Google Scholar] [CrossRef] [PubMed]
- Raad, G.; Lteif, L.; Lahoud, R.; Azoury, J.; Azoury, J.; Tanios, J.; Hazzouri, M.; Azoury, J. Cryopreservation media differentially affect sperm motility, morphology and DNA integrity. Andrology 2018, 6, 836–845. [Google Scholar] [CrossRef]
- O’Connell, M.; McClure, N.; Lewis, S.E. The effects of cryopreservation on sperm morphology, motility and mitochondrial function. Hum. Reprod. 2002, 17, 704–709. [Google Scholar] [CrossRef]
- Ozkavukcu, S.; Erdemli, E.; Isik, A.; Oztuna, D.; Karahuseyinoglu, S. Effects of cryopreservation on sperm parameters and ultrastructural morphology of human spermatozoa. J. Assist. Reprod. Genet. 2008, 25, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Pabón, D.; Meseguer, M.; Sevillano, G.; Cobo, A.; Romero, J.L.; Remohí, J.; de Los Santos, M.J. A new system of sperm cryopreservation: Evaluation of survival, motility, DNA oxidation, and mitochondrial activity. Andrology 2019, 7, 293–301. [Google Scholar] [CrossRef]
- Gómez-Torres, M.J.; Medrano, L.; Romero, A.; Fernández-Colom, P.J.; Aizpurúa, J. Effectiveness of human spermatozoa biomarkers as indicators of structural damage during cryopreservation. Cryobiology 2017, 78, 90–94. [Google Scholar] [CrossRef] [PubMed]
- de Paula, T.S.; Bertolla, R.P.; Spaine, D.M.; Cunha, M.A.; Schor, N.; Cedenho, A.P. Effect of cryopreservation on sperm apoptotic deoxyribonucleic acid fragmentation in patients with oligozoospermia. Fertil. Steril. 2006, 86, 597–600. [Google Scholar] [CrossRef]
- Zribi, N.; Feki Chakroun, N.; El Euch, H.; Gargouri, J.; Bahloul, A.; Ammar Keskes, L. Effects of cryopreservation on human sperm deoxyribonucleic acid integrity. Fertil. Steril. 2010, 93, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Tongdee, P.; Sukprasert, M.; Satirapod, C.; Wongkularb, A.; Choktanasiri, W. Comparison of Cryopreserved Human Sperm between Ultra Rapid Freezing and Slow Programmable Freezing: Effect on Motility, Morphology and DNA Integrity. J. Med. Assoc. Thai. 2015, 98 (Suppl. S4), S33–S42. [Google Scholar]
- Cankut, S.; Dinc, T.; Cincik, M.; Ozturk, G.; Selam, B. Evaluation of Sperm DNA Fragmentation via Halosperm Technique and TUNEL Assay Before and After Cryopreservation. Reprod. Sci. 2019, 26, 1575–1581. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, L.; Jalali, S.; Shami, S.A.; Akram, Z.; Batool, S.; Kalsoom, O. Effects of cryopreservation on sperm DNA integrity in normospermic and four categories of infertile males. Syst. Biol. Reprod. Med. 2010, 56, 74–83. [Google Scholar] [CrossRef][Green Version]
- Rahiminia, T.; Hosseini, A.; Anvari, M.; Ghasemi-Esmailabad, S.; Talebi, A.R. Modern human sperm freezing: Effect on DNA, chromatin and acrosome integrity. Taiwan J. Obstet. Gynecol. 2017, 56, 472–476. [Google Scholar] [CrossRef]
- O’Neill, H.C.; Nikoloska, M.; Ho, H.; Doshi, A.; Maalouf, W. Improved cryopreservation of spermatozoa using vitrification: Comparison of cryoprotectants and a novel device for long-term storage. J. Assist. Reprod. Genet. 2019, 36, 1713–1720. [Google Scholar] [CrossRef]
- Tvrdá, E.; Gosálvez, J.; Arroyo, F.; Sánchez, P.; de Jesús Risco Delgado, R.; Sánchez, R. Dynamic assessment of human sperm DNA damage III: The effect of sperm freezing techniques. Cell Tissue Bank. 2021, 22, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wang, X.M.; Li, R.X.; Wang, Y.Z.; Chao, Y.C.; Liu, Z.Z.; Huang, Z.H.; Nie, H.C.; Zhu, W.B.; Tan, Y.Q.; et al. Improving native human sperm freezing protection by using a modified vitrification method. Asian J. Androl. 2021, 23, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Riva, N.S.; Ruhlmann, C.; Iaizzo, R.S.; Marcial López, C.A.; Martínez, A.G. Comparative analysis between slow freezing and ultra-rapid freezing for human sperm cryopreservation. JBRA Assist. Reprod. 2018, 22, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, F.; Isachenko, V.; Dessole, S.; Rahimi, G.; Farina, M.; Vargiu, N.; Mallmann, P.; Dattena, M.; Capobianco, G.; Peters, D.; et al. Vitrification of human spermatozoa without cryoprotectants. Cryo Lett. 2002, 23, 93–102. [Google Scholar]
- Vutyavanich, T.; Piromlertamorn, W.; Nunta, S. Rapid freezing versus slow programmable freezing of human spermatozoa. Fertil. Steril. 2010, 93, 1921–1928. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Jin, R.T.; Wu, L.M.; Johansson, L.; Guo, T.H.; Liu, Y.S.; Tong, X.H. Cryoprotectant-free ultra-rapid freezing of human spermatozoa in cryogenic vials. Andrologia 2014, 46, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Aizpurua, J.; Medrano, L.; Enciso, M.; Sarasa, J.; Romero, A.; Fernández, M.A.; Gómez-Torres, M.J. New permeable cryoprotectant-free vitrification method for native human sperm. Hum. Reprod. 2017, 32, 2007–2015. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, M.; Arakkal, D.; Mangalaraj, A.M.; Kamath, M.S. Comparison of Conventional Slow Freeze versus Permeable Cryoprotectant-Free Vitrification of Abnormal Semen Sample: A Randomized Controlled Trial. J. Hum. Reprod. Sci. 2019, 12, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Spis, E.; Bushkovskaia, A.; Isachenko, E.; Todorov, P.; Sanchez, R.; Skopets, V.; Isachenko, V. Conventional freezing vs. cryoprotectant-free vitrification of epididymal (MESA) and testicular (TESE) spermatozoa: Three live births. Cryobiology 2019, 90, 100–102. [Google Scholar] [CrossRef]
- Isachenko, E.; Isachenko, V.; Katkov, I.I.; Rahimi, G.; Schöndorf, T.; Mallmann, P.; Dessole, S.; Nawroth, F. DNA integrity and motility of human spermatozoa after standard slow freezing versus cryoprotectant-free vitrification. Hum. Reprod. 2004, 19, 932–939. [Google Scholar] [CrossRef]
- Isachenko, V.; Isachenko, E.; Katkov, I.I.; Montag, M.; Dessole, S.; Nawroth, F.; Van Der Ven, H. Cryoprotectant-free cryopreservation of human spermatozoa by vitrification and freezing in vapor: Effect on motility, DNA integrity, and fertilization ability. Biol. Reprod. 2004, 71, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Saritha, K.R.; Bongso, A. Comparative evaluation of fresh and washed human sperm cryopreserved in vapor and liquid phases of liquid nitrogen. J. Androl. 2001, 22, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.J.; Lee, J.R.; Chae, S.J.; Jee, B.C.; Suh, C.S.; Kim, S.H. Comparative study of two cryopreservation methods of human spermatozoa: Vitrification versus slow freezing. Fertil. Steril. 2008, 90, S280. [Google Scholar] [CrossRef]
- Hosseini, A.; Khalili, M.A.; Talebi, A.R.; Agha-Rahimi, A.; Ghasemi-Esmailabad, S.; Woodward, B.; Yari, N. Cryopreservation of Low Number of Human Spermatozoa; Which is Better: Vapor Phase or Direct Submerging in Liquid Nitrogen? Hum. Fertil. 2019, 22, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Agha-Rahimi, A.; Khalili, M.A.; Nabi, A.; Ashourzadeh, S. Vitrification is not superior to rapid freezing of normozoospermic spermatozoa: Effects on sperm parameters, DNA fragmentation and hyaluronan binding. Reprod. Biomed. Online 2014, 28, 352–358. [Google Scholar] [CrossRef][Green Version]
- Le, M.T.; Nguyen, T.T.T.; Nguyen, T.T.; Nguyen, V.T.; Nguyen, T.T.A.; Nguyen, V.Q.H.; Cao, N.T. Cryopreservation of human spermatozoa by vitrification versus conventional rapid freezing: Effects on motility, viability, morphology and cellular defects. Eur. J. Obstet. Gynecol. Reprod. Biol. 2019, 234, 14–20. [Google Scholar] [CrossRef]
- Rayea, M.; Amer, M.; Mousa, A.; Farghaly, M. Comparative Study between Slow Conventional Freezing and Cryoprotectant-Free Vitrification of Human Spermatozoa in Large Volume. Egypt. J. Hosp. Med. 2019, 77, 5983–5988. [Google Scholar] [CrossRef]
- Chatterjee, A.; Saha, D.; Niemann, H.; Gryshkov, O.; Glasmacher, B.; Hofmann, N. Effects of cryopreservation on the epigenetic profile of cells. Cryobiology 2017, 74, 1–7. [Google Scholar] [CrossRef]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef]
- Walker, W.H. Regulation of mammalian spermatogenesis by miRNAs. Semin. Cell Dev. Biol. 2022, 121, 24–31. [Google Scholar] [CrossRef]
- McIver, S.C.; Roman, S.D.; Nixon, B.; McLaughlin, E.A. miRNA and mammalian male germ cells. Hum. Reprod. Update 2012, 18, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Khawar, M.B.; Mehmood, R.; Roohi, N. MicroRNAs: Recent insights towards their role in male infertility and reproductive cancers. Bosn. J. Basic Med. Sci. 2019, 19, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wen, H.; Cao, C.; Dong, D.; Yang, C.; Xie, S.; Zhang, J.; Huang, X.; Huang, X.; Yuan, S.; et al. Overexpression of MicroRNA-10a in Germ Cells Causes Male Infertility by Targeting Rad51 in Mouse and Human. Front. Physiol. 2019, 10, 765. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Meng, H.; Wang, X.G.; Jin, H.X.; Yao, G.D.; Shi, S.L.; Wu, L.; Zhang, X.Y.; Sun, Y.P. Reduced microRNA-188-3p expression contributes to apoptosis of spermatogenic cells in patients with azoospermia. Cell Prolif. 2017, 50, e12297. [Google Scholar] [CrossRef] [PubMed]
- Norioun, H.; Motovali-Bashi, M.; Javadirad, S.M. Hsa-miR-27a-3p overexpression in men with nonobstructive azoospermia: A case-control study. Int. J. Reprod. Biomed. 2020, 18, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, W.; Zhang, L.; Ji, Y.; Qin, J.; Wang, L.; Wang, M.; Qi, L.; Xue, J.; Lv, B.; et al. Effect of Sperm Cryopreservation on miRNA Expression and Early Embryonic Development. Front. Cell Dev. Biol. 2021, 9, 749486. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zeng, C.J.; He, L.; Ding, L.; Tang, K.Y.; Peng, W.P. Selection of endogenous reference microRNA genes for quantitative reverse transcription polymerase chain reaction studies of boar spermatozoa cryopreservation. Theriogenology 2015, 83, 634–641. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, D.; Chang, Y.; Li, Y.; Zhang, M.; Zhou, G.; Peng, Z.; Zeng, C. Cryopreservation of boar sperm induces differential microRNAs expression. Cryobiology 2017, 76, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.H.; Qazi, I.H.; Ran, M.X.; Liang, K.; Zhang, Y.; Zhang, M.; Zhou, G.B.; Angel, C.; Zeng, C.J. Exploration of miRNA and mRNA Profiles in Fresh and Frozen-Thawed Boar Sperm by Transcriptome and Small RNA Sequencing. Int. J. Mol. Sci. 2019, 20, 802. [Google Scholar] [CrossRef]
- Pedrosa, A.C.; Andrade Torres, M.; Vilela Alkmin, D.; Pinzon, J.E.P.; Kitamura Martins, S.M.M.; Coelho da Silveira, J.; Furugen Cesar de Andrade, A. Spermatozoa and seminal plasma small extracellular vesicles miRNAs as biomarkers of boar semen cryotolerance. Theriogenology 2021, 174, 60–72. [Google Scholar] [CrossRef]
- Ran, M.X.; Zhou, Y.M.; Liang, K.; Wang, W.C.; Zhang, Y.; Zhang, M.; Yang, J.D.; Zhou, G.B.; Wu, K.; Wang, C.D.; et al. Comparative Analysis of MicroRNA and mRNA Profiles of Sperm with Different Freeze Tolerance Capacities in Boar (Susscrofa) and Giant Panda (Ailuropodamelanoleuca). Biomolecules 2019, 9, 432. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, A.; Zhou, H.; Sun, W.; Ding, R.; Li, X.; Liu, J.; Zhou, Y.; Chen, X.; Ding, F.; Yang, L.; et al. Cryopreservation Induces Alterations of miRNA and mRNA Fragment Profiles of Bull Sperm. Front. Genet. 2020, 11, 419. [Google Scholar] [CrossRef] [PubMed]
- Capra, E.; Turri, F.; Lazzari, B.; Cremonesi, P.; Gliozzi, T.M.; Fojadelli, I.; Stella, A.; Pizzi, F. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genom. 2017, 18, 14. [Google Scholar] [CrossRef] [PubMed]
- Güngör, B.H.; Tektemur, A.; Arkali, G.; Dayan Cinkara, S.; Acisu, T.C.; Koca, R.H.; Etem Önalan, E.; Özer Kaya, S.; Kizil, M.; Sönmez, M.; et al. Effect of freeze-thawing process on lipid peroxidation, miRNAs, ion channels, apoptosis and global DNA methylation in ram spermatozoa. Reprod. Fertil. Dev. 2021, 33, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Ezzati, M.; Shanehbandi, D.; Bahramzadeh, B.; Hamdi, K.; Pashaiasl, M. Investigation of molecular cryopreservation, fertility potential and microRNA-mediated apoptosis in Oligoasthenoteratozoospermia men. Cell Tissue Bank. 2021, 22, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Ji, X.R.; Huang, Z.H.; Liu, Q.; Wang, R.J.; Fan, L.Q.; Wu, H.L.; Bo, H.; Zhu, W.B. Long-term storage modifies the microRNA expression profile of cryopreserved human semen. Biomol. Biomed. 2024, 24, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Pinho, P.; Arantes-Rodrigues, R.; Gaivão, I.; Peixoto, F.; Gomes, Z.; Brito, M.; Moutinho, O.; Colaço, B.; Pinto-Leite, R. Mitochondrial Effects, DNA Damage, and Antioxidant Enzyme Activity in Cryopreserved Human Sperm Samples: A Pilot Study. Physiologia 2022, 2, 80–93. [Google Scholar] [CrossRef]
- Gonzalez, M.; Prashar, T.; Connaughton, H.; Barry, M.; Robker, R.; Rose, R. Restoring Sperm Quality Post-Cryopreservation Using Mitochondrial-Targeted Compounds. Antioxidants 2022, 11, 1808. [Google Scholar] [CrossRef]
- Shi, H.; Li, Q.Y.; Li, H.; Wang, H.Y.; Fan, C.X.; Dong, Q.Y.; Pan, B.C.; Ji, Z.L.; Li, J.Y. ROS-induced oxidative stress is a major contributor to sperm cryoinjury. Hum. Reprod. 2024, 39, 310–325. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, Y.; Wu, C.; Qiu, S.; Chen, X.; Cai, B.; Xie, H. Freeze-thawing impairs the motility, plasma membrane integrity and mitochondria function of boar spermatozoa through generating excessive ROS. BMC Vet. Res. 2021, 17, 127. [Google Scholar] [CrossRef]
- Sawyer, D.E.; Mercer, B.G.; Wiklendt, A.M.; Aitken, R.J. Quantitative analysis of gene-specific DNA damage in human spermatozoa. Mutat. Res. 2003, 529, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Liu, J.; Zheng, Q.Y.; Liu, N.; Huang, X.L.; Wu, Y.Y.; Yao, X.F.; Tan, Q.Y.; Huang, Y.; Hu, C.H.; et al. The effect of the mitochondria-targeted antioxidant Mito-tempo during sperm ultra-rapid freezing. Cryobiology 2024, 114, 104860. [Google Scholar] [CrossRef]
- Corral-Vazquez, C.; Salas-Huetos, A.; Blanco, J.; Vidal, F.; Sarrate, Z.; Anton, E. Sperm microRNA pairs: New perspectives in the search for male fertility biomarkers. Fertil. Steril. 2019, 112, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Godo, A.; Grossmann, M.; Pons, M.C.; F-Fernández, S.; Garrido, N.; Anton, E. Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil. Steril. 2015, 104, 591–601. [Google Scholar] [CrossRef]
- Mokánszki, A.; Molnár, Z.; Varga Tóthné, E.; Bodnár, B.; Jakab, A.; Bálint, B.L.; Balogh, I. Altered microRNAs expression levels of sperm and seminal plasma in patients with infertile ejaculates compared with normozoospermic males. Hum. Fertil. 2020, 23, 246–255. [Google Scholar] [CrossRef]
- Yeh, L.Y.; Lee, R.K.; Lin, M.H.; Huang, C.H.; Li, S.H. Correlation between Sperm Micro Ribonucleic Acid-34b and -34c Levels and Clinical Outcomes of Intracytoplasmic Sperm Injection in Men with Male Factor Infertility. Int. J. Mol. Sci. 2022, 23, 12381. [Google Scholar] [CrossRef] [PubMed]
- Corney, D.C.; Flesken-Nikitin, A.; Godwin, A.K.; Wang, W.; Nikitin, A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007, 67, 8433–8438. [Google Scholar] [CrossRef]
- Kumamoto, K.; Spillare, E.A.; Fujita, K.; Horikawa, I.; Yamashita, T.; Appella, E.; Nagashima, M.; Takenoshita, S.; Yokota, J.; Harris, C.C. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008, 68, 3193–3203. [Google Scholar] [CrossRef]
- Vogt, M.; Munding, J.; Grüner, M.; Liffers, S.T.; Verdoodt, B.; Hauk, J.; Steinstraesser, L.; Tannapfel, A.; Hermeking, H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011, 458, 313–322. [Google Scholar] [CrossRef]
- Córdova-Rivas, S.; Fraire-Soto, I.; Mercado-Casas Torres, A.; Servín-González, L.S.; Granados-López, A.J.; López-Hernández, Y.; Reyes-Estrada, C.A.; Gutiérrez-Hernández, R.; Castañeda-Delgado, J.E.; Ramírez-Hernández, L.; et al. 5p and 3p Strands of miR-34 Family Members Have Differential Effects in Cell Proliferation, Migration, and Invasion in Cervical Cancer Cells. Int. J. Mol. Sci. 2019, 20, 545. [Google Scholar] [CrossRef]
- Yan, N.; Lu, Y.; Sun, H.; Qiu, W.; Tao, D.; Liu, Y.; Chen, H.; Yang, Y.; Zhang, S.; Li, X.; et al. Microarray profiling of microRNAs expressed in testis tissues of developing primates. J. Assist. Reprod. Genet. 2009, 26, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Batista, M.R.; Diniz, P.; Torres, A.; Murta, D.; Lopes-da-Costa, L.; Silva, E. Notch signaling in mouse blastocyst development and hatching. BMC Dev. Biol. 2020, 20, 9. [Google Scholar] [CrossRef] [PubMed]
- Swiatek, P.J.; Lindsell, C.E.; del Amo, F.F.; Weinmaster, G.; Gridley, T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994, 8, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Conlon, R.A.; Reaume, A.G.; Rossant, J. Notch1 is required for the coordinate segmentation of somites. Development 1995, 121, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Ye, L.; Liu, G.; Shao, G.; Zheng, R.; Ren, Z.; Zuo, B.; Xu, D.; Lei, M.; Jiang, S.; et al. Microarray-based approach identifies differentially expressed microRNAs in porcine sexually immature and mature testes. PLoS ONE 2010, 5, e11744. [Google Scholar] [CrossRef] [PubMed]
- Tscherner, A.; Gilchrist, G.; Smith, N.; Blondin, P.; Gillis, D.; LaMarre, J. MicroRNA-34 family expression in bovine gametes and preimplantation embryos. Reprod. Biol. Endocrinol. 2014, 12, 85. [Google Scholar] [CrossRef] [PubMed]
- Bidarimath, M.; Edwards, A.K.; Wessels, J.M.; Khalaj, K.; Kridli, R.T.; Tayade, C. Distinct microRNA expression in endometrial lymphocytes, endometrium, and trophoblast during spontaneous porcine fetal loss. J. Reprod. Immunol. 2015, 107, 64–79. [Google Scholar] [CrossRef] [PubMed]
- Mayor-Lynn, K.; Toloubeydokhti, T.; Cruz, A.C.; Chegini, N. Expression profile of microRNAs and mRNAs in human placentas from pregnancies complicated by preeclampsia and preterm labor. Reprod. Sci. 2011, 18, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, W.; Xu, J.; Guo, Y.; Yan, J.; Meng, L.; Jiang, C.; Lu, S. Interpreting the MicroRNA-15/107 family: Interaction identification by combining network based and experiment supported approach. BMC Med. Genet. 2019, 20, 96. [Google Scholar] [CrossRef]
- Salas-Huetos, A.; Blanco, J.; Vidal, F.; Grossmann, M.; Pons, M.C.; Garrido, N.; Anton, E. Spermatozoa from normozoospermic fertile and infertile individuals convey a distinct miRNA cargo. Andrology 2016, 4, 1028–1036. [Google Scholar] [CrossRef]
- Tomic, M.; Bolha, L.; Pizem, J.; Ban-Frangez, H.; Vrtacnik-Bokal, E.; Stimpfel, M. Association between Sperm Morphology and Altered Sperm microRNA Expression. Biology 2022, 11, 1671. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, Z.; Jin, Y.; Dragas, D.; Zhang, L.; Adjei, B.S.; Wang, A.; Dai, Y.; Zhou, X. MicroRNA-99 family members suppress Homeobox A1 expression in epithelial cells. PLoS ONE 2013, 8, e80625. [Google Scholar] [CrossRef] [PubMed]
- Makki, N.; Capecchi, M.R. Hoxa1 lineage tracing indicates a direct role for Hoxa1 in the development of the inner ear, the heart, and the third rhombomere. Dev. Biol. 2010, 341, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Draime, A.; Bridoux, L.; Belpaire, M.; Pringels, T.; Tys, J.; Rezsohazy, R. PRDM14, a putative histone methyl-transferase, interacts with and decreases the stability and activity of the HOXA1 transcription factor. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 534–542. [Google Scholar] [CrossRef] [PubMed]
- Correia, B.; Sousa, M.I.; Ramalho-Santos, J. The mTOR pathway in reproduction: From gonadal function to developmental coordination. Reproduction 2020, 159, R173–R188. [Google Scholar] [CrossRef]
- Stopka, T.; Skoultchi, A.I. The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. USA 2003, 100, 14097–14102. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Chen, Z.; Yin, Q.; Fu, X.; Li, Y.; Stopka, T.; Skoultchi, A.I.; Zhang, Y. The chromatin remodeler Snf2h is essential for oocyte meiotic cell cycle progression. Genes Dev. 2020, 34, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Hu, Z.; Qin, Y.; Dong, J.; Dai, J.; Lu, C.; Zhang, W.; Shen, H.; Xia, Y.; Wang, X. Seminal plasma microRNAs: Potential biomarkers for spermatogenesis status. Mol. Hum. Reprod. 2012, 18, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Joshi, M.; Andrabi, S.W.; Yadav, R.K.; Sankhwar, S.N.; Gupta, G.; Rajender, S. Qualitative and quantitative assessment of sperm miRNAs identifies hsa-miR-9-3p, hsa-miR-30b-5p and hsa-miR-122-5p as potential biomarkers of male infertility and sperm quality. Reprod. Biol. Endocrinol. 2022, 20, 122. [Google Scholar] [CrossRef]
- Dorostghoal, M.; Galehdari, H.; Moramezi, F.; Danyari, R. Sperm miR-26a-5p and its target PTEN transcripts content in men with unexplained infertility. Andrology 2020, 8, 1167–1173. [Google Scholar] [CrossRef]
- Palak, E.; Lebiedzińska, W.; Anisimowicz, S.; Sztachelska, M.; Pierzyński, P.; Wiczkowski, W.; Żelazowska-Rutkowska, B.; Niklińska, G.N.; Ponikwicka-Tyszko, D.; Wołczyński, S. The Association between Bisphenol, A, Steroid Hormones, and Selected MicroRNAs Levels in Seminal Plasma of Men with Infertility. J. Clin. Med. 2021, 10, 5945. [Google Scholar] [CrossRef] [PubMed]
- Larriba, S.; Sánchez-Herrero, J.F.; Pluvinet, R.; López-Rodrigo, O.; Bassas, L.; Sumoy, L. Seminal extracellular vesicle sncRNA sequencing reveals altered miRNA/isomiR profiles as sperm retrieval biomarkers for azoospermia. Andrology 2024, 12, 137–156. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Ramírez, M.; Calvo-Anguiano, G.; Lugo-Trampe, J.J.; Martínez-de-Villarreal, L.E.; Rodríguez-Torres, D.; Nistal, M.; González-Peramato, P. Expression profile of microRNAs in the testes of patients with Klinefelter syndrome. Sci. Rep. 2020, 10, 11470. [Google Scholar] [CrossRef] [PubMed]
- Abu-Halima, M.; Hammadeh, M.; Schmitt, J.; Leidinger, P.; Keller, A.; Meese, E.; Backes, C. Altered microRNA expression profiles of human spermatozoa in patients with different spermatogenic impairments. Fertil. Steril. 2013, 99, 1249–1255.e16. [Google Scholar] [CrossRef] [PubMed]
- Rahbar, S.; Pashaiasl, M.; Ezzati, M.; Ahmadi AsrBadr, Y.; Mohammadi-Dehcheshmeh, M.; Mohammadi, S.A.; Ghaffari Novin, M. MicroRNA-based regulatory circuit involved in sperm infertility. Andrologia 2020, 52, e13453. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Cheng, W.; Gao, Y.; Wang, H.; Liu, Z. Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol. Med. Rep. 2012, 6, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Finocchi, F.; Pelloni, M.; Balercia, G.; Pallotti, F.; Radicioni, A.F.; Lenzi, A.; Lombardo, F.; Paoli, D. Seminal plasma miRNAs in Klinefelter syndrome and in obstructive and non-obstructive azoospermia. Mol. Biol. Rep. 2020, 47, 4373–4382. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tang, Y.; Huang, J.; Liu, H.; Liu, X.; Zhou, Y.; Ma, C.; Wang, Q.; Yang, J.; Sun, F.; et al. Circulating microRNAs in seminal plasma as predictors of sperm retrieval in microdissection testicular sperm extraction. Ann. Transl. Med. 2022, 10, 392. [Google Scholar] [CrossRef]
- Hamilton, M.; Russell, S.; Menezes, K.; Moskovtsev, S.I.; Librach, C. Assessing spermatozoal small ribonucleic acids and their relationship to blastocyst development in idiopathic infertile males. Sci. Rep. 2022, 12, 20010. [Google Scholar] [CrossRef]
- Abu-Halima, M.; Backes, C.; Leidinger, P.; Keller, A.; Lubbad, A.M.; Hammadeh, M.; Meese, E. MicroRNA expression profiles in human testicular tissues of infertile men with different histopathologic patterns. Fertil. Steril. 2014, 101, 78–86.e2. [Google Scholar] [CrossRef]
- Zhu, M.; Fei, L.; Li, D.; Chen, D. Correlation Analysis of miR-122-5p and Occludin with Sperm Density in Oligospermia Patients’ Sperm. Clin. Lab. 2019, 65, 3. [Google Scholar] [CrossRef] [PubMed]
- Mostafa, T.; Rashed, L.A.; Nabil, N.I.; Osman, I.; Mostafa, R.; Farag, M. Seminal miRNA Relationship with Apoptotic Markers and Oxidative Stress in Infertile Men with Varicocele. Biomed. Res. Int. 2016, 2016, 4302754. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, X.; Wang, Z.; Li, J.; Xu, Z.; Miao, M.; Chen, G.; Lei, X.; Wu, J.; Shi, H.; et al. MicroRNA expression profile analysis in sperm reveals hsa-mir-191 as an auspicious omen of in vitro fertilization. BMC Genom. 2020, 21, 165. [Google Scholar] [CrossRef] [PubMed]
- Metzler-Guillemain, C.; Victorero, G.; Lepoivre, C.; Bergon, A.; Yammine, M.; Perrin, J.; Sari-Minodier, I.; Boulanger, N.; Rihet, P.; Nguyen, C. Sperm mRNAs and microRNAs as candidate markers for the impact of toxicants on human spermatogenesis: An application to tobacco smoking. Syst. Biol. Reprod. Med. 2015, 61, 139–149. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen, 6th ed.; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Sharma, R.; Iovine, C.; Agarwal, A.; Henkel, R. TUNEL assay-Standardized method for testing sperm DNA fragmentation. Andrologia 2021, 53, e13738. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


| Native Semen | Slow Freezing of Semen | Vitrification of Semen | p Value | |
|---|---|---|---|---|
| Total motility (%) | 60.0 (50.0–70.0) | 18.9 (11.9–24.3) | 13.8 (7.9–20.9) | <0.001 |
| Fast progressive motility (%) | 45.0 (33.8–50.0) | 7.5 (3.4–11.8) | 5.4 (1.7–9.3) | <0.001 |
| Slow progressive motility (%) | 10.0 (10.0–10.0) | 6.0 (3.5–8.6) | 4.3 (2.5–9.5) | <0.001 |
| Non-progressive motility (%) | 5.0 (5.0–10.0) | 3.2 (2.1–5.3) | 2.7 (1.4–4.4) | <0.001 |
| Viability (mean ± SD) | / | 19.7 ± 11.4 | 21.3 ± 11.9 | 0.346 |
| Protamine staining (mean ± SD) | 53.6 ± 15.1 | 53.9 ± 16.0 | 52.4 ± 15.2 | 0.905 |
| Sperm DNA fragmentation (mean ± SD) | 18.7 ± 9.5 | 24.7 ± 11.6 | 27.1 ± 15.7 | 0.049 |
| Mature spermatozoa according to HBA assay | 78.0 (67.5–82.4) | 79.4 (71.7–82.9) | 75.3 (67.5–81.1) | 0.607 |
| Native Semen before Slow Freezing | Semen after Slow Freezing | p Value | Native Semen before Vitrification | Semen after Vitrification | p Value | |
|---|---|---|---|---|---|---|
| Sperm concentration (×106/mL) | 11 (6.5–12.5) | / | 9.5 (7–12) | / | 0.976 | |
| Total sperm count (×106) | 45 (27–58.5) | / | 44.9 (31.7–50.5) | / | 0.741 | |
| Total motility (%) | 45 (25–55) | 9.4 (5.8–20.0) | <0.001 | 40 (30–47.5) | 7.9 (6.1–15.2) | <0.001 |
| Fast progressive motility (%) | 30 (12.5–37.5) | 3 (0.4–4.1) | <0.001 | 25 (17.5–30) | 2.4 (0–4.9) | 0.002 |
| Slow progressive motility (%) | 10 (5–12.5) | 3.6 (2.2–6.4) | 0.003 | 10 (7.5–10) | 2.5 (0.9–3.3) | <0.001 |
| Non-progressive motility (%) | 5 (5–5) | 2.7 (1.6–5.8) | 0.016 | 5 (5–5) | 3.7 (1.5–5.3) | 0.112 |
| Viability (mean ± SD) | 15.1 ± 7.7 | 12.4 ± 6.5 |
| miRNA | miRNA Expression a | Reference |
|---|---|---|
| let-7a-5p |
| [81] |
| [88] | |
| [65] | |
| [91] | |
| [89] | |
| miR-10a-5p |
| [81] |
| [92] | |
| [43] | |
| [93] | |
| miR-15b-5p |
| [65] |
| [94] | |
| [64] | |
| [80] | |
| [95] | |
| [81] | |
| [93] | |
| miR-26a-5p |
| [90] |
| [96] | |
| [81] | |
| [94] | |
| miR-34b-3p |
| [63] |
| [97] | |
| [98] | |
| [66] | |
| [65] | |
| [81] | |
| [64] | |
| [94] | |
| miR-92a-3p |
| [99] |
| [94] | |
| [93] | |
| [100] | |
| miR-93-3p |
| [63] |
| [80] | |
| miR-99b-5p |
| [81] |
| [94] | |
| [100] | |
| miR-122-5p |
| [101] |
| [97] | |
| [89] | |
| [96] | |
| [65] | |
| [66] | |
| [81] | |
| [102] | |
| miR-125b-5p |
| [81] |
| [93] | |
| [100] | |
| miR-191-5p |
| [103] |
| [81] | |
| miR-296-5p |
| [63] |
| [81] | |
| [80] | |
| [104] |
| miRNA | Catalog Number a |
|---|---|
| let-7a-5p | YP00205727 |
| miR-10a-5p | YP00204778 |
| miR-15b-5p | YP00204243 |
| miR-26a-5p | YP00206023 |
| miR-34b-3p | YP00204005 |
| miR-92a-3p | YP00204258 |
| miR-93-3p | YP00204470 |
| miR-99b-5p | YP00205983 |
| miR-122-5p | YP00205664 |
| miR-125b-5p | YP00205713 |
| miR-191-5p | YP00204306 |
| miR-296-5p | YP00204436 |
| SNORD38B b | YP00203901 |
| SNORD44 b | YP00203902 |
| SNORD49A b | YP00203904 |
| UniSp6 c | YP00203954 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Podgrajsek, R.; Bolha, L.; Pungert, T.; Pizem, J.; Jazbec, K.; Malicev, E.; Stimpfel, M. Effects of Slow Freezing and Vitrification of Human Semen on Post-Thaw Semen Quality and miRNA Expression. Int. J. Mol. Sci. 2024, 25, 4157. https://doi.org/10.3390/ijms25084157
Podgrajsek R, Bolha L, Pungert T, Pizem J, Jazbec K, Malicev E, Stimpfel M. Effects of Slow Freezing and Vitrification of Human Semen on Post-Thaw Semen Quality and miRNA Expression. International Journal of Molecular Sciences. 2024; 25(8):4157. https://doi.org/10.3390/ijms25084157
Chicago/Turabian StylePodgrajsek, Rebeka, Luka Bolha, Tjasa Pungert, Joze Pizem, Katerina Jazbec, Elvira Malicev, and Martin Stimpfel. 2024. "Effects of Slow Freezing and Vitrification of Human Semen on Post-Thaw Semen Quality and miRNA Expression" International Journal of Molecular Sciences 25, no. 8: 4157. https://doi.org/10.3390/ijms25084157
APA StylePodgrajsek, R., Bolha, L., Pungert, T., Pizem, J., Jazbec, K., Malicev, E., & Stimpfel, M. (2024). Effects of Slow Freezing and Vitrification of Human Semen on Post-Thaw Semen Quality and miRNA Expression. International Journal of Molecular Sciences, 25(8), 4157. https://doi.org/10.3390/ijms25084157








