The Experimental and In Silico-Based Evaluation of NRF2 Modulators, Sulforaphane and Brusatol, on the Transcriptome of Immortalized Bovine Mammary Alveolar Cells
Abstract
1. Introduction
2. Results
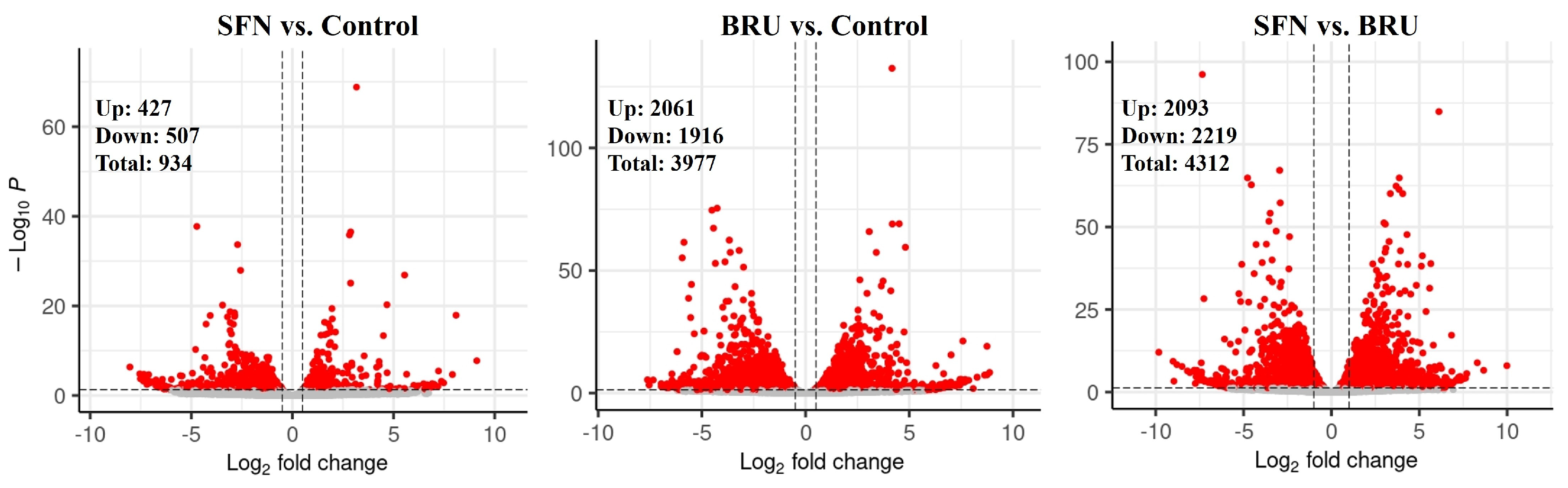
2.1. Differentially Expressed Genes Determined by RNA Sequencing
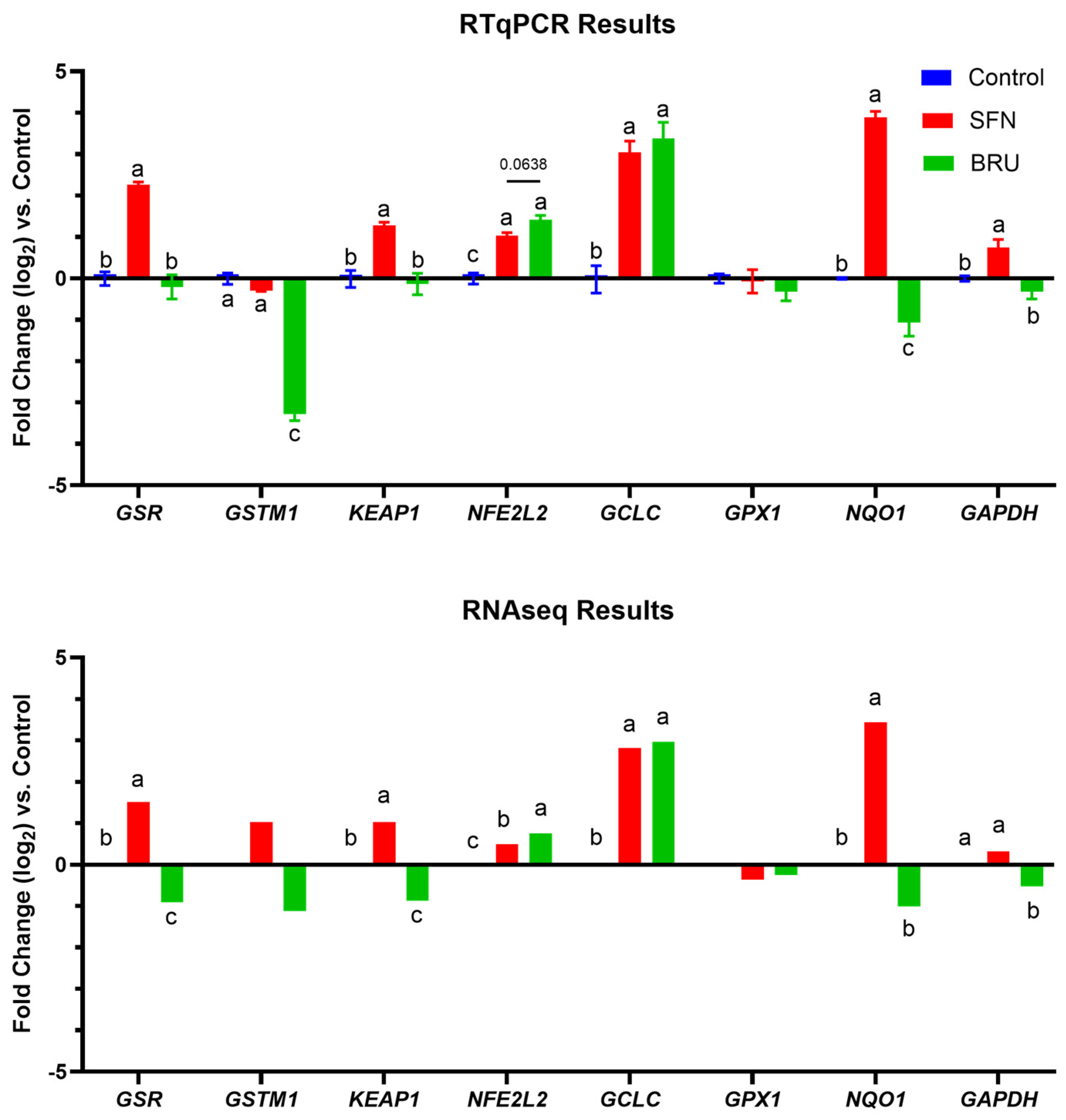
2.2. Expression of Putative NRF2 Target Genes
2.3. Functional Analysis of Differentially Expressed Genes
2.3.1. SFN vs. CTR
2.3.2. BRU vs. CTR
2.3.3. SFN vs. BRU
3. Discussion
3.1. Gene Targets of NRF2 in MACT Cells
3.2. NRF2 Modulation Has Varying Effects on Glutathione System Genes
3.3. Cellular Stress and Feedback Loops Regulate NRF2 and KEAP1 Expression
3.4. SFN Treatment Promotes NRF2-Related Pathways While Inhibiting the Cell Cycle and Affecting mRNA Splicing
3.5. The General Protein Synthesis Inhibitor Brusatol Negatively Affects Metabolism While Stimulating Inflammatory Pathways
4. Materials and Methods
4.1. Reagents and Cell Culture
4.2. RNA Isolation and RTqPCR of Putative Target Genes
4.3. RNA Sequencing and Analysis
4.4. Bioinformatics Analysis
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abuelo Sebio, Á.; Hernández Bermúdez, J.; Benedito Castellote, J.L.; Castillo Rodríguez, C. The importance of the oxidative status of dairy cattle in the periparturient period: Revisiting antioxidant supplementation. J. Anim. Physiol. Anim. Nutr. 2015, 99, 1003–1016. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Ott, M.; Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria, oxidative stress and cell death. Apoptosis 2007, 12, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Drackley, J.K. Biology of dairy cows during the transition period: The final frontier? J. Dairy Sci. 1999, 82, 2259–2273. [Google Scholar] [CrossRef] [PubMed]
- Capuco, A.; Wood, D.; Baldwin, R.; Mcleod, K.; Paape, M. Mammary cell number, proliferation, and apoptosis during a bovine lactation: Relation to milk production and effect of bST. J. Dairy Sci. 2001, 84, 2177–2187. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.M.; Aitken, S.L. Impact of oxidative stress on the health and immune function of dairy cattle. Vet. Immunol. Immunopathol. 2009, 128, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, A.K. Antioxidant response element. Biochem. Pharmacol. 1994, 48, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef] [PubMed]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Bellezza, I. Oxidative stress in age-related macular degeneration: Nrf2 as therapeutic target. Front. Pharmacol. 2018, 9, 1280. [Google Scholar] [CrossRef]
- Ford, H.R.; Busato, S.; Bionaz, M. In vitro-In vivo Hybrid Approach for Studying Modulation of NRF2 in Immortalized Bovine Mammary Cells. Front. Anim. Sci. 2021, 2, 674355. [Google Scholar] [CrossRef]
- Jin, X.; Wang, K.; Liu, L.; Liu, H.; Zhao, F.-Q.; Liu, J. Nuclear factor-like factor 2-antioxidant response element signaling activation by tert-butylhydroquinone attenuates acute heat stress in bovine mammary epithelial cells. J. Dairy Sci. 2016, 99, 9094–9103. [Google Scholar] [CrossRef] [PubMed]
- Sohel, M.M.H.; Amin, A.; Prastowo, S.; Linares-Otoya, L.; Hoelker, M.; Schellander, K.; Tesfaye, D. Sulforaphane protects granulosa cells against oxidative stress via activation of NRF2-ARE pathway. Cell Tissue Res. 2018, 374, 629–641. [Google Scholar] [CrossRef]
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 talks, who’s listening? Antioxid. Redox Signal. 2010, 13, 1649–1663. [Google Scholar] [CrossRef] [PubMed]
- Fakhri, S.; Pesce, M.; Patruno, A.; Moradi, S.Z.; Iranpanah, A.; Farzaei, M.H.; Sobarzo-Sánchez, E. Attenuation of Nrf2/Keap1/ARE in Alzheimer’s disease by plant secondary metabolites: A mechanistic review. Molecules 2020, 25, 4926. [Google Scholar] [CrossRef]
- Han, L.; Batistel, F.; Ma, Y.; Alharthi, A.; Parys, C.; Loor, J. Methionine supply alters mammary gland antioxidant gene networks via phosphorylation of nuclear factor erythroid 2-like 2 (NFE2L2) protein in dairy cows during the periparturient period. J. Dairy Sci. 2018, 101, 8505–8512. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Buttari, B.; Panieri, E.; Profumo, E.; Saso, L. An overview of Nrf2 signaling pathway and its role in inflammation. Molecules 2020, 25, 5474. [Google Scholar] [CrossRef]
- Sashida, G.; Bazzoli, E.; Menendez, S.; Stephen, D.N. The oncogenic role of the ETS transcription factors MEF and ERG. Cell Cycle 2010, 9, 3457–3459. [Google Scholar] [CrossRef]
- Townsend, B.E.; Johnson, R.W. Sulforaphane induces Nrf2 target genes and attenuates inflammatory gene expression in microglia from brain of young adult and aged mice. Exp. Gerontol. 2016, 73, 42–48. [Google Scholar] [CrossRef]
- Yang, S.-H.; Li, P.; Yu, L.-H.; Li, L.; Long, M.; Liu, M.-D.; He, J.-B. Sulforaphane protect against cadmium-induced oxidative damage in mouse Leydigs cells by activating Nrf2/ARE signaling pathway. Int. J. Mol. Sci. 2019, 20, 630. [Google Scholar] [CrossRef] [PubMed]
- Harvey, C.; Thimmulappa, R.; Singh, A.; Blake, D.; Ling, G.; Wakabayashi, N.; Fujii, J.; Myers, A.; Biswal, S. Nrf2-regulated glutathione recycling independent of biosynthesis is critical for cell survival during oxidative stress. Free Radic. Biol. Med. 2009, 46, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci. Rep. 2017, 7, 1559. [Google Scholar] [CrossRef] [PubMed]
- Harder, B.; Tian, W.; La Clair, J.J.; Tan, A.C.; Ooi, A.; Chapman, E.; Zhang, D.D. Brusatol overcomes chemoresistance through inhibition of protein translation. Mol. Carcinog. 2017, 56, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and inhibitors of NRF2: A review of their potential for clinical development. Oxidative Med. Cell. Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Plácido, A.; Pereira, C.; Duarte, A.; Candeias, E.; Correia, S.; Santos, R.; Carvalho, C.; Cardoso, S.; Oliveira, C.; Moreira, P. The role of endoplasmic reticulum in amyloid precursor protein processing and trafficking: Implications for Alzheimer’s disease. Biochim. Et Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Cominacini, L.; Mozzini, C.; Garbin, U.; Pasini, A.; Stranieri, C.; Solani, E.; Vallerio, P.; Tinelli, I.A.; Pasini, A.F. Endoplasmic reticulum stress and Nrf2 signaling in cardiovascular diseases. Free Radic. Biol. Med. 2015, 88, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.; Lu, G.; Ota, A.; Zhou, Z.H.; Vondriska, T.M.; Lane, T.F.; Wang, Y. IRE1 phosphatase PP2Ce regulates adaptive ER stress response in the postpartum mammary gland. PLoS ONE 2014, 9, e111606. [Google Scholar] [CrossRef] [PubMed]
- Invernizzi, G.; Naeem, A.; Loor, J. Endoplasmic reticulum stress gene network expression in bovine mammary tissue during the lactation cycle. J. Dairy Sci. 2012, 95, 2562–2566. [Google Scholar] [CrossRef] [PubMed]
- Kreß, J.K.C.; Jessen, C.; Hufnagel, A.; Schmitz, W.; da Silva, T.N.X.; Dos Santos, A.F.; Mosteo, L.; Goding, C.R.; Angeli, J.P.F.; Meierjohann, S. The integrated stress response effector ATF4 is an obligatory metabolic activator of NRF2. Cell Rep. 2023, 42, 112724. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Kostov, R.V.; Kensler, T.W. KEAP1 and done? Targeting the NRF2 pathway with sulforaphane. Trends Food Sci. Technol. 2017, 69, 257–269. [Google Scholar] [CrossRef]
- Capece, D.; Verzella, D.; Flati, I.; Arboretto, P.; Cornice, J.; Franzoso, G. NF-κB: Blending metabolism, immunity, and inflammation. Trends Immunol. 2022, 43, 757–775. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Guo, L.; Yang, Y.; Wang, Y.; Xia, S.; Gong, H.; Zhang, B.-K.; Yan, M. Dissecting the crosstalk between Nrf2 and NF-κB response pathways in drug-induced toxicity. Front. Cell Dev. Biol. 2022, 9, 809952. [Google Scholar] [CrossRef]
- Gado, F.; Ferrario, G.; Della Vedova, L.; Zoanni, B.; Altomare, A.; Carini, M.; Aldini, G.; D’Amato, A.; Baron, G. Targeting Nrf2 and NF-κB Signaling Pathways in Cancer Prevention: The Role of Apple Phytochemicals. Molecules 2023, 28, 1356. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhao, K.; Sun, L.; Yin, X.; Zhang, J.; Liu, C.; Li, B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021, 19, 367. [Google Scholar] [CrossRef] [PubMed]
- Fantone, S.; Piani, F.; Olivieri, F.; Rippo, M.R.; Sirico, A.; Di Simone, N.; Marzioni, D.; Tossetta, G. Role of SLC7A11/xCT in Ovarian Cancer. Int. J. Mol. Sci. 2024, 25, 587. [Google Scholar] [CrossRef]
- Huang, Y.; Dai, Z.; Barbacioru, C.; Sadée, W. Cystine-Glutamate Transporter SLC7A11 in Cancer Chemosensitivity and Chemoresistance. Cancer Res. 2005, 65, 7446–7454. [Google Scholar] [CrossRef]
- Bionaz, M.; Hurley, W.; Loor, J. Milk protein synthesis in the lactating mammary gland: Insights from transcriptomics analyses. Milk Protein 2012, 11, 285–324. [Google Scholar]
- Kurinna, S.; Muzumdar, S.; Köhler, U.A.; Kockmann, T.; Schäfer, M.; Werner, S. Autocrine and paracrine regulation of keratinocyte proliferation through a novel Nrf2–IL-36γ pathway. J. Immunol. 2016, 196, 4663–4670. [Google Scholar] [CrossRef]
- Yang, X.; Li, X.; Wu, C.; Zhang, F. Knockdown of PHLDA1 alleviates necrotizing enterocolitis by inhibiting NLRP3 inflammasome activation and pyroptosis through enhancing Nrf2 signaling. Immunol. Investig. 2023, 52, 257–269. [Google Scholar] [CrossRef]
- Chanas, S.A.; Jiang, Q.; McMahon, M.; McWalter, G.K.; McLellan, L.I.; Elcombe, C.R.; Henderson, C.J.; Wolf, C.R.; Moffat, G.J.; Itoh, K. Loss of the Nrf2 transcription factor causes a marked reduction in constitutive and inducible expression of the glutathione S-transferase Gsta1, Gsta2, Gstm1, Gstm2, Gstm3 and Gstm4 genes in the livers of male and female mice. Biochem. J. 2002, 365, 405–416. [Google Scholar] [CrossRef] [PubMed]
- McWalter, G.K.; Higgins, L.G.; McLellan, L.I.; Henderson, C.J.; Song, L.; Thornalley, P.J.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Transcription factor Nrf2 is essential for induction of NAD (P) H: Quinone oxidoreductase 1, glutathione S-transferases, and glutamate cysteine ligase by broccoli seeds and isothiocyanates. J. Nutr. 2004, 134, 3499S–3506S. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Shang, H.; Chen, Y.Q.; Pan, L.-L.; Bhatia, M.; Sun, J. Sulforaphane protects pancreatic acinar cell injury by modulating Nrf2-mediated oxidative stress and NLRP3 inflammatory pathway. Oxidative Med. Cell. Longev. 2016, 2016, 7864150. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Pu, C.; Zhou, P.; Wang, P.; Liang, D.; Wang, Q.; Hu, Y.; Li, B.; Hao, X. Cinnamaldehyde prevents endothelial dysfunction induced by high glucose by activating Nrf2. Cell. Physiol. Biochem. 2015, 36, 315–324. [Google Scholar] [CrossRef]
- Kulak, M.V.; Cyr, A.R.; Woodfield, G.W.; Bogachek, M.; Spanheimer, P.M.; Li, T.; Price, D.H.; Domann, F.E.; Weigel, R.J. Transcriptional regulation of the GPX1 gene by TFAP2C and aberrant CpG methylation in human breast cancer. Oncogene 2013, 32, 4043–4051. [Google Scholar] [CrossRef]
- Han, Y.-H.; Kim, H.-J.; Kim, E.-J.; Kim, K.-S.; Hong, S.; Park, H.-G.; Lee, M.-O. RORα decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid. Redox Signal. 2014, 21, 2083–2094. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Li, X.; Meng, W.B.; Bai, Z.T.; Rui, S.Z.; Wang, Z.F.; Zhou, W.C.; Jin, X.D. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. J. Cell. Physiol. 2019, 234, 619–631. [Google Scholar] [CrossRef]
- Faraonio, R.; Vergara, P.; Di Marzo, D.; Pierantoni, M.G.; Napolitano, M.; Russo, T.; Cimino, F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 2006, 281, 39776–39784. [Google Scholar] [CrossRef]
- Kwak, M.-K.; Itoh, K.; Yamamoto, M.; Kensler, T.W. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: Role of antioxidant response element-like sequences in the nrf2 promoter. Mol. Cell. Biol. 2002, 22, 2883–2892. [Google Scholar] [CrossRef]
- Rushworth, S.A.; Zaitseva, L.; Murray, M.Y.; Shah, N.M.; Bowles, K.M.; MacEwan, D.J. The high Nrf2 expression in human acute myeloid leukemia is driven by NF-κB and underlies its chemo-resistance. Blood J. Am. Soc. Hematol. 2012, 120, 5188–5198. [Google Scholar] [CrossRef] [PubMed]
- Lee, O.-H.; Jain, A.K.; Papusha, V.; Jaiswal, A.K. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 2007, 282, 36412–36420. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008, 58, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Cui, Q.; Wang, W.; Namani, A.; Wang, H.; Hammad, A.; Huang, P.; Gao, Y.; Elshaer, M.; Wu, Y.; Wang, X.J. NRF2 has a splicing regulatory function involving the survival of motor neuron (SMN) in non-small cell lung cancer. Oncogene 2023, 42, 2751–2763. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.-Y.; Zhu, Y.-R.; Dai, D.-J.; Wang, X.; Jin, H.-C. Epigenetic regulation of alternative splicing. Am. J. Cancer Res. 2018, 8, 2346. [Google Scholar] [PubMed]
- Marasco, L.E.; Kornblihtt, A.R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 2023, 24, 242–254. [Google Scholar] [CrossRef]
- Fan, X.; Zhang, Y.; Qiu, L.; Zhu, W.; Tu, X.; Miao, Y. Identification and characterization of alternative splicing variants of buffalo LXR α expressed in mammary gland. Sci. Rep. 2022, 12, 10588. [Google Scholar] [CrossRef] [PubMed]
- Kappeler, S.; Farah, Z.; Puhan, Z. Alternative splicing of lactophorin mRNA from lactating mammary gland of the camel (Camelus dromedarius). J. Dairy Sci. 1999, 82, 2084–2093. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Yang, S.; Cui, X.; Jiang, L.; Zhang, S.; Zhang, Q.; Zhang, Y.; Sun, D. Identification and expression pattern of two novel alternative splicing variants of EEF1D gene of dairy cattle. Gene 2014, 534, 189–196. [Google Scholar] [CrossRef]
- Wang, X.; Proud, C.G. The mTOR pathway in the control of protein synthesis. Physiology 2006, 21, 362–369. [Google Scholar] [CrossRef]
- Grootjans, J.; Kaser, A.; Kaufman, R.J.; Blumberg, R.S. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016, 16, 469–484. [Google Scholar] [CrossRef]
- Rosa, F.; Busato, S.; Avaroma, F.C.; Linville, K.; Trevisi, E.; Osorio, J.S.; Bionaz, M. Transcriptional changes detected in fecal RNA of neonatal dairy calves undergoing a mild diarrhea are associated with inflammatory biomarkers. PLoS ONE 2018, 13, e0191599. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Busato, S.; Ford, H.R.; Abdelatty, A.M.; Estill, C.T.; Bionaz, M. Peroxisome proliferator-activated receptor activation in precision-cut bovine liver slices reveals novel putative PPAR targets in periparturient dairy cows. Front. Vet. Sci. 2022, 9, 931264. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Huber, W.; Anders, S. Moderated estimation of fold changes and dispersion for RNA-seq data with Deseq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Bionaz, M.; Periasamy, K.; Rodriguez-Zas, S.L.; Hurley, W.L.; Loor, J.J. A novel dynamic impact approach (DIA) for functional analysis of time-course omics studies: Validation using the bovine mammary transcriptome. PLoS ONE 2012, 7, e32455. [Google Scholar] [CrossRef]
- Bollmann, S.; Bu, D.; Wang, J.; Bionaz, M. Unmasking Upstream Gene Expression Regulators with miRNA-corrected mRNA Data. Bioinform. Biol. Insights 2015, 9, S29332. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ford, H.R.; Bionaz, M. The Experimental and In Silico-Based Evaluation of NRF2 Modulators, Sulforaphane and Brusatol, on the Transcriptome of Immortalized Bovine Mammary Alveolar Cells. Int. J. Mol. Sci. 2024, 25, 4264. https://doi.org/10.3390/ijms25084264
Ford HR, Bionaz M. The Experimental and In Silico-Based Evaluation of NRF2 Modulators, Sulforaphane and Brusatol, on the Transcriptome of Immortalized Bovine Mammary Alveolar Cells. International Journal of Molecular Sciences. 2024; 25(8):4264. https://doi.org/10.3390/ijms25084264
Chicago/Turabian StyleFord, Hunter R., and Massimo Bionaz. 2024. "The Experimental and In Silico-Based Evaluation of NRF2 Modulators, Sulforaphane and Brusatol, on the Transcriptome of Immortalized Bovine Mammary Alveolar Cells" International Journal of Molecular Sciences 25, no. 8: 4264. https://doi.org/10.3390/ijms25084264
APA StyleFord, H. R., & Bionaz, M. (2024). The Experimental and In Silico-Based Evaluation of NRF2 Modulators, Sulforaphane and Brusatol, on the Transcriptome of Immortalized Bovine Mammary Alveolar Cells. International Journal of Molecular Sciences, 25(8), 4264. https://doi.org/10.3390/ijms25084264






