Functional Characterization of PmDXR, a Critical Rate-Limiting Enzyme, for Turpentine Biosynthesis in Masson Pine (Pinus massoniana Lamb.)
Abstract
1. Introduction
2. Results
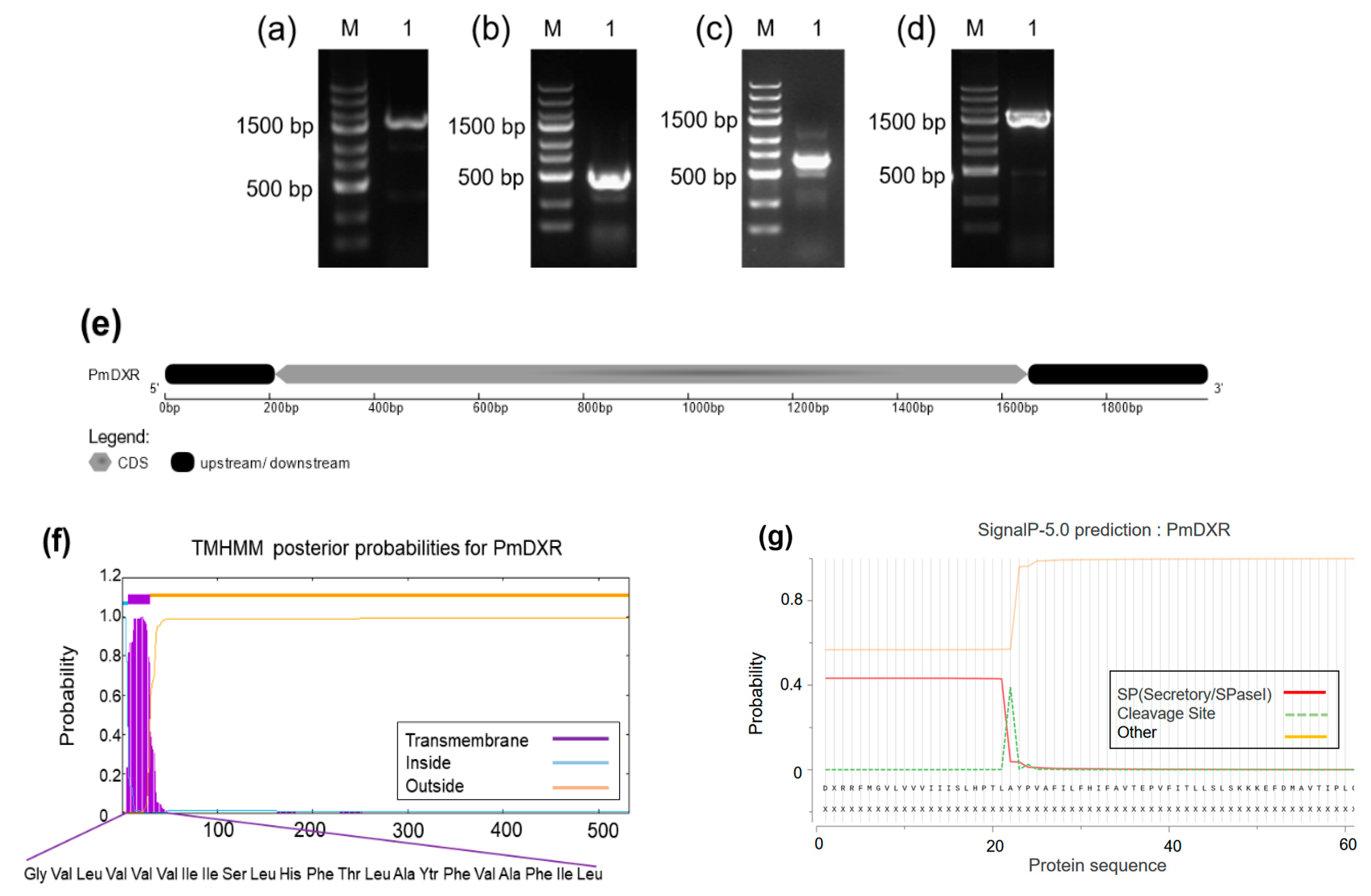
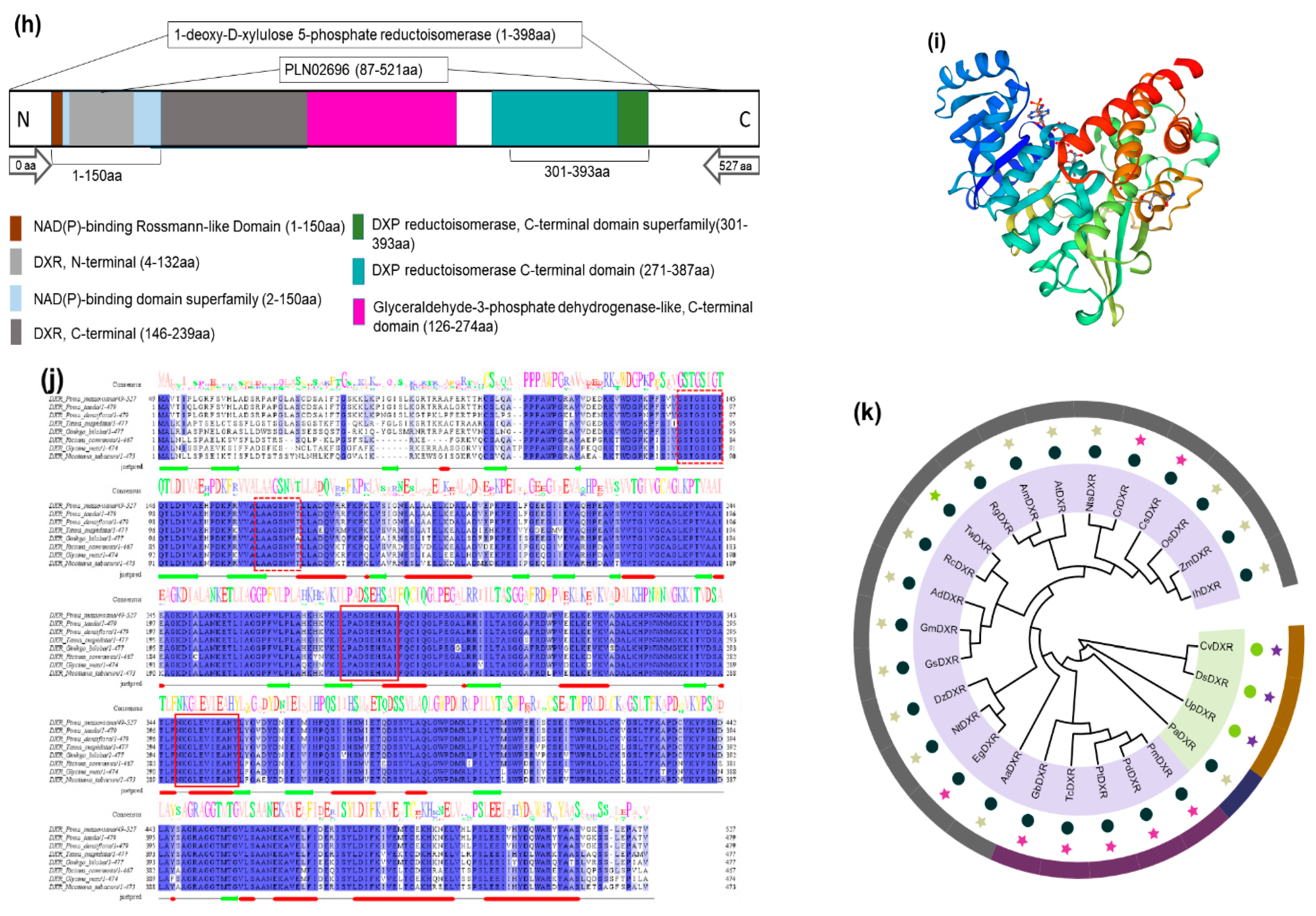
2.1. PmDXR Identification and Bioinformatics Analysis
2.2. Codon Preference Analysis of PmDXR
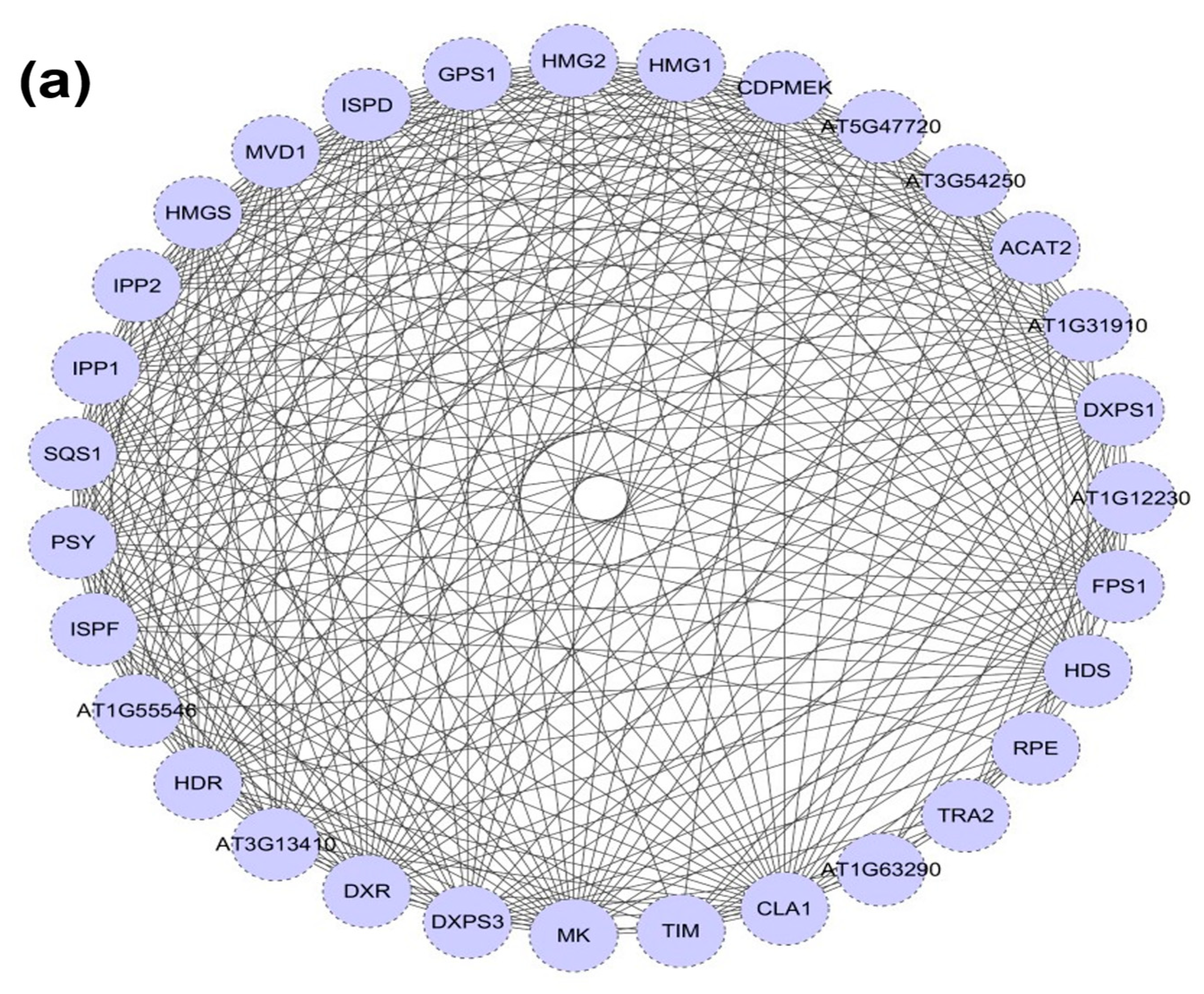
2.3. DXR Gene/Protein Interactions
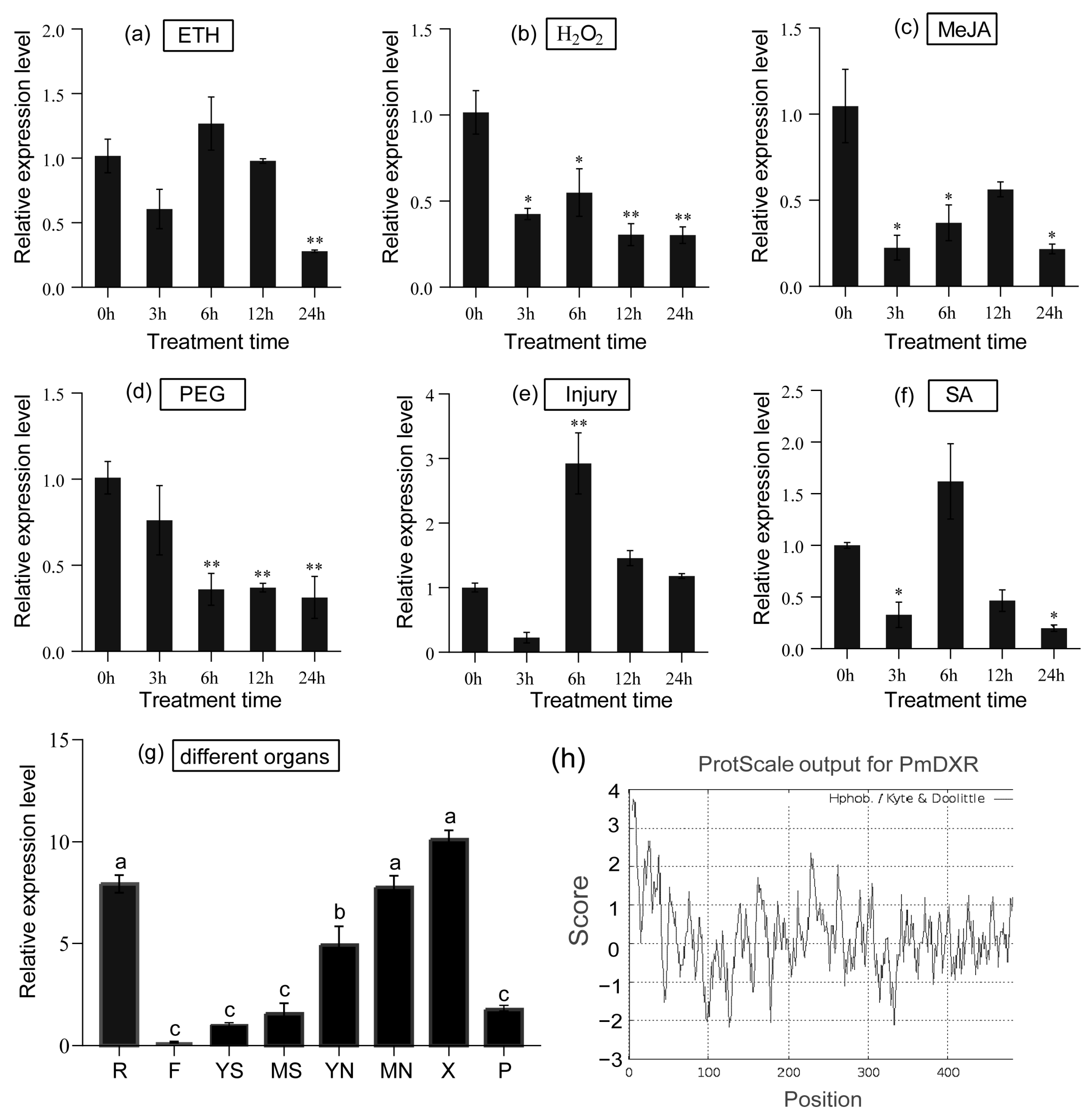
2.4. Spatial and Temporal Expression Patterns of PmDXR
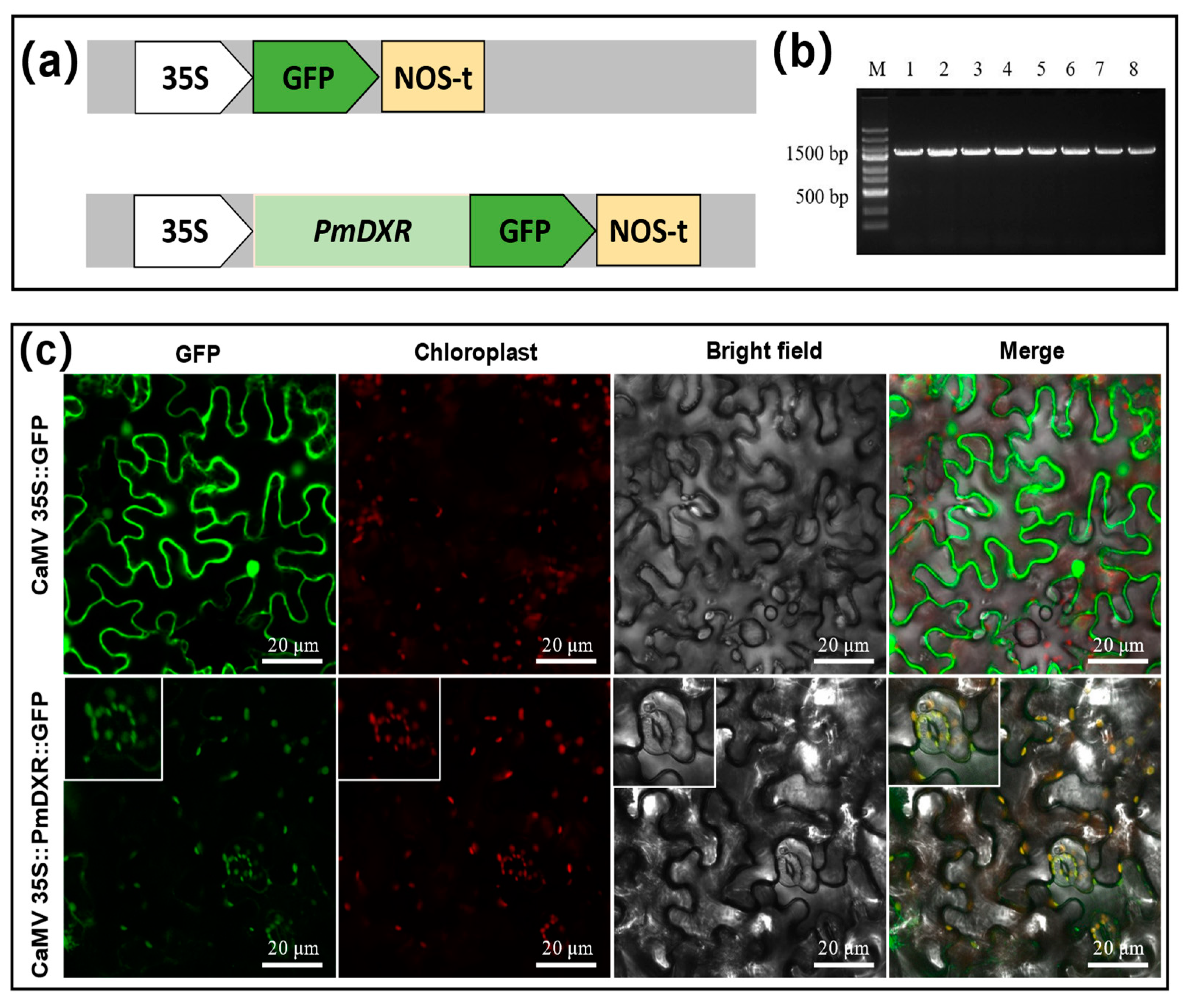
2.5. PmDXR Protein Is Localized to the Chloroplast
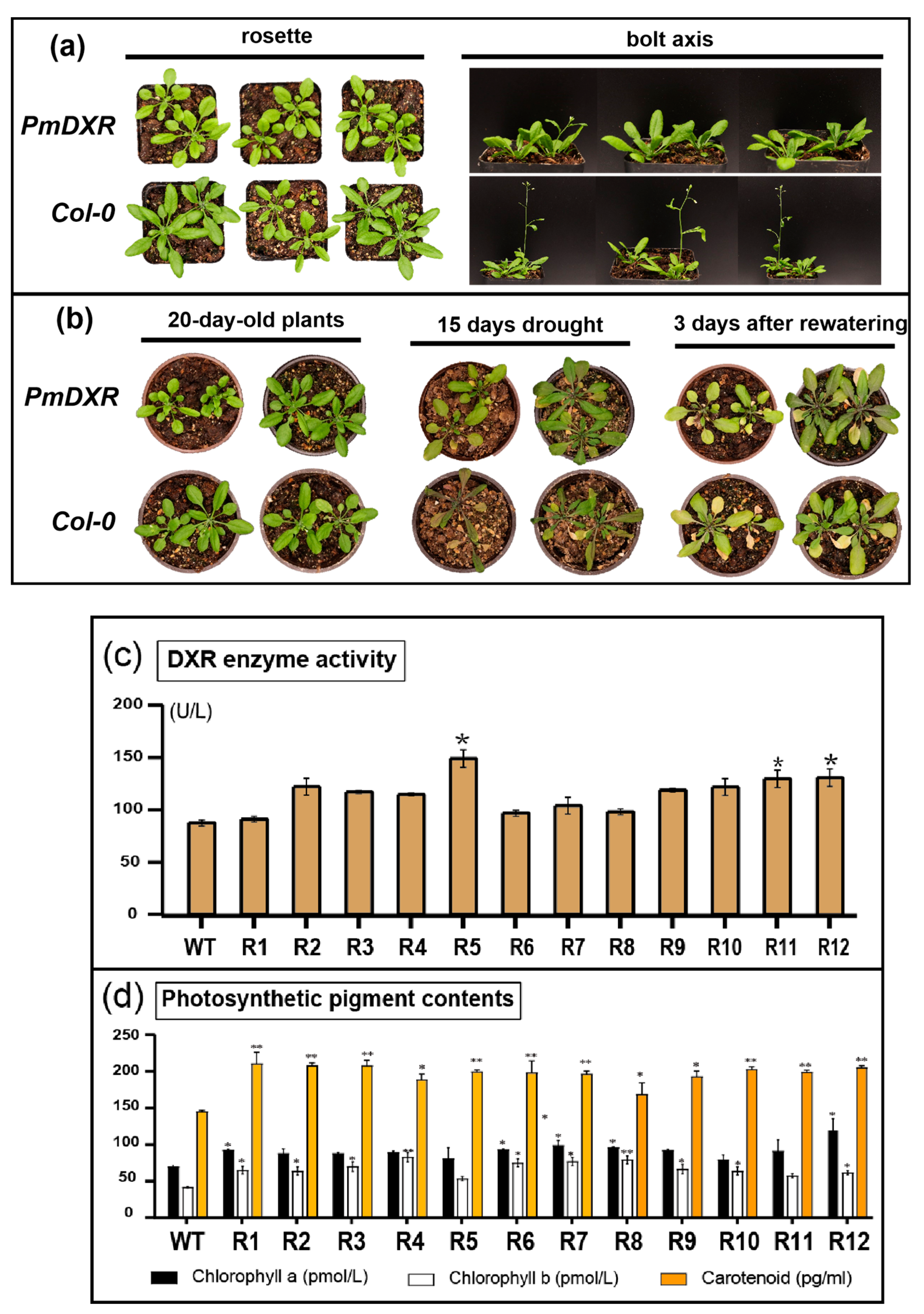
2.6. Ectopic Expression of PmDXR Promoted Arabidopsis DXR Enzyme Activity and Photosynthetic Pigment Contents
2.7. Determination of Physiological Indicators of Transgenic A. thaliana under Different Stress Treatments
2.8. GUS Staining Analysis of the ProPmDXR Promoter in N. benthamiana
3. Discussion
4. Materials
4.1. Pinus massoniana Lamb.
4.2. Arabidopsis thaliana
4.3. Nicotiana benthamiana
4.4. Reagent
5. Experimental Method
5.1. PmDXR Gene Cloning
5.2. Bioinformatics Analysis
5.3. Abiotic Stress and Hormone Treatment of Plant Materials
- (1)
- The relative expression levels of PmDXR in young stems (YS), mature stems (MS), young needles (YN), mature needles (MN), flowers (F), xylem (X), phloem (P) and roots (R) of 15-year-old P. massoniana were detected.
- (2)
- The expression patterns under abiotic stress and hormone induction of PmDXR in 2-year-old P. massoniana included the following: mechanic wound, 15% polyethylene glycol (PEG6000), 10 mM H2O2, 500 µM ethephon (ETH), 1 mM salicylic acid (SA) and 100 µM methyl jasmonate (MeJA). Mechanical damage was treated by cutting the needles at 1/2 of the upper half of the needles in the potted seedlings of P. massoniana, and collecting the cut needles at 0 h, 3 h, 6 h, 12 h, 24 h and 48 h intervals. We immediately put the sample in liquid nitrogen and put it in the refrigerator at −80 °C. In addition, the abiotic stress treatment and hormone treatment were carried out by spraying the surface of the plant. The needles of P. massoniana were collected every 0 h, 3 h, 6 h, 12 h, 24 h and 48 h, frozen in liquid nitrogen and stored at −80 °C [44,56]. The above treatments were 0 h without any treatment as a control group. Three technical and biological replicates were set up for each group of experiments, and the biological replicates for each treatment consisted of three pots (three plants per pot), with every three seedlings serving as one technical replicate.
5.4. Real-Time Quantitative PCR (qRT-PCR) Analysis
5.5. Subcellular Localization of the PmDXR
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, B.; Liu, Q.H.; Zhou, Z.C.; Yin, H.F.; Xie, Y.N. Overexpression of geranyl diphosphate synthase (PmGPPS1) boosts monoterpene and diterpene production involved in the response to pine wood nematode invasion. Tree Physiol. 2021, 2, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.R. Epidemic status of pine wilt disease in China and its prevention and control techniques and counter measures. Sci. Silvae Sin. 2019, 9, 1–10. [Google Scholar]
- Ji, K.S.; Xu, L.A.; Wang, D.B.; Ni, Z.X.; Wang, Z.R. Progress and achievement of genetic improvement on Masson Pine (Pinus massoniana Lamb.) in China. J. Nanjing For. Univ. (Nat. Sci. Ed. ) 2022, 6, 10–22. [Google Scholar]
- State Forestry and Grassland Administration. China Forestry and Grassland Statistical Yearbook; China Forestry Press: Beijing, China, 2019. [Google Scholar]
- Ye, J.R.; Wu, X.Q. Research progress of pine wilt disease. For. Pest Dis. 2022, 3, 1–10. [Google Scholar]
- Yan, X.F.; Wang, Y.; Li, Y.M. Plant secondary metabolism and its response to environment. Acta Ecol. Sin. 2007, 6, 2554–2562. [Google Scholar]
- Franceschi, V.R.; Krokene, P.; Christiansen, E.; Krekling, T. Anatomical and chemical defenses of conifer bark against bark beetles and other pests. New Phytol. 2005, 2, 353–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liu, Q.H.; Zhou, Z.C.; Xu, L.Y.; Chen, X.L.; Luo, N. Identification of candidate constitutive expressed resistant genes of pine wilt disease in Pinus massoniana based on high-throughput transcriptome sequencing. For. Res. 2019, 5, 1–10. [Google Scholar]
- Franceschi, V.R.; Krekling, T.; Christiansen, E. Application of methyl jasmonate on Picea abies (Pinaceae) stems induces defense-related responses in phloem and xylem. Am. J. Bot. 2002, 4, 578–586. [Google Scholar] [CrossRef]
- Pu, X.; Dong, X.; Li, Q.; Chen, Z.; Li, L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant Biol. 2021, 7, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, S. Effect of light intensity on the development of pine wilt disease. Can. J. Bot.-Rev. Can. De Bot. 1989, 6, 1861–1864. [Google Scholar] [CrossRef]
- Picman, A.K. Biological activities of sesquiterpene lactones. Biochem. Syst. Ecol. 1986, 3, 255–281. [Google Scholar] [CrossRef]
- Ley, S.V. Synthesis of antifeedants for insects: Novel behaviour-modifying chemicals from plants. Ciba Found. Symp. 1990, 154, 80–98. [Google Scholar] [PubMed]
- Mordue, A.J.; Blackwell, A. Azadirachtin: An update. J. Insect Physiol. 1993, 11, 903–924. [Google Scholar] [CrossRef]
- Meinwald, J.; Eisner, T. The chemistry of phyletic dominance. Proc. Natl. Acad. Sci. USA 1995, 1, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.Y.; Xu, K.; Xu, K.Q.; Ge, M.H. Study on resistance of different age classes of Pinus massoniana to pine wilt disease. J. Nanjing For. Univ. 1994, 3, 27–33. [Google Scholar]
- Tan, J.J.; Hao, D.J.; Pan, Y.W.; Qu, H.Y.; Fan, B.Q.; Ye, J.R.; Du, Y.B. Effects of several pine volatiles on the behavior of pine wood nematode. J. Northeast. For. Univ. 2009, 12, 58–59. [Google Scholar]
- Li, H.; Tian, J.; Wang, H.; Yang, S.Q.; Gao, W.Y. An improved preparation of D-Glyceraldehyde 3-Phosphate and its use in the synthesis of 1-Deoxy-D-xylulose 5-Phosphate. Helv. Chim. Acta 2010, 9, 1745–1750. [Google Scholar] [CrossRef]
- Kim, Y.B.; Kim, S.M.; Kang, M.K.; Kuzuyama, T.; Lee, J.K.; Park, S.C.; Shin, S.C.; Kim, S.U. Regulation of resin acid synthesis in Pinus densiflora by differential transcription of genes encoding multiple 1-deoxy-d-xylulose 5-phosphate synthase and 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase genes. Tree Physiol. 2009, 5, 737–749. [Google Scholar] [CrossRef]
- Gong, Y.F.; Liao, Z.H.; Chen, M.; Zuo, K.J.; Guo, L.; Tan, Q.M.; Huang, Z.S.; Kai, G.Y.; Sun, X.F.; Tan, F.; et al. Molecular cloning and characterization of a 1-deoxy-d-xylulose 5-phosphate reductoisomerase gene from Ginkgo biloba. DNA Seq. 2005, 2, 111–120. [Google Scholar] [CrossRef]
- Zheng, Q.P.; Yu, L.J.; Liu, Z.; Li, M.Y.; Xiang, F.; Yang, Q. Cloning and analysis of cDNA encoding key enzyme gene (dxr) of the non-MVA Pathway in Taxus chinensis Cell. Chin. J. Biotechnol. 2004, 4, 548–553. [Google Scholar]
- Yu, X.L. Molecular Cloning of Genes Coding Enzymes in the Upstream Biosynthesis Pathway of Taxol. Bachelor’s Thesis, Tianjin University, Tianjin, China, 2004. [Google Scholar]
- Mahmoud, S.S.; Croteau, R.B. Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. Proc. Natl. Acad. Sci. USA 2001, 15, 8915–8920. [Google Scholar] [CrossRef]
- Veau, B.; Courtois, M.; Oudin, A.; Chénieux, J.C.; Rideau, M.; Clastre, M. Cloning and expression of cDNAs encoding two enzymes of the MEP pathway in Catharanthus roseus. Biochim. Biophys. Acta 2000, 1, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Takeno, S.; Hayashi, S.; Sendai, S.; Bamba, T.; Yoshimura, S.; Tomizawa, K.-I.; Fukusaki, E.; Miyake, C. Overexpression of 1-deoxy-d-xylulose-5-phosphate reductoisomerase gene in chloroplast contributes to increment of isoprenoid production. J. Biosci. Bioeng. 2008, 5, 518–526. [Google Scholar] [CrossRef]
- Qin, G.J.; Gu, H.Y.; Ma, L.G.; Peng, Y.B.; Deng, X.W.; Chen, Z.L.; Qu, L.J. Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 2007, 5, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.F.; Miao, J.; Li, S.; Qin, G.J.; Tang, S.; Li, H.N.; Gu, H.Y.; Qu, L.J. Disruption of the 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) gene results in albino, dwarf and defects in trichome initiation and stomata closure in Arabidopsis. Cell Res. 2010, 6, 688–700. [Google Scholar] [CrossRef]
- Yang, Z.Q. Research progress of high rosin genetic improvement and breeding strategy of Pinus massoniana. Guangxi For. Sci. 2015, 4, 317–324. [Google Scholar]
- Li, L.; Dong, C.M.; Zhang, M.J.; Zhu, Y.H. Cloning and expression analysis of DXR gene in Prunella vulgaris. J. Chin. Med. Mater. 2019, 6, 1249–1254. [Google Scholar]
- Fan, W.J.; Du, C.J.; Huang, S.W.; SiTu, S.H.; Ou, H.L.; Mao, Z.P. Phylogenetic and molecular evolutionary analysis of DXR gene in plants. Mol. Plant Breed. 2020, 19, 1–16. [Google Scholar]
- Yin, S.S.; Su, X.J.; Gong, L.T.; Liao, Y.; ADiLai, A.B.D.R.Y.M.; Geng, H.W. Cloning and expression analysis of 1-deoxy-D-xylulose-5-phosphate reductoisomerase gene LaDXR in Lavandula angustifolia. Mol. Plant Breed. 2021, 7, 2193–2199. [Google Scholar]
- Yang, J.F.; Adhikari, M.N.; Liu, H.; He, G.Z.; Zhan, R.T.; Wei, J.S.; Chen, W.W. Characterization and functional analysis of the genes encoding 1-deoxy-d-xylulose-5-phosphate reductoisomerase and 1-deoxy-d-xylulose-5-phosphate synthase, the two enzymes in the MEP pathway from Amomum villosum Lour. Mol. Biol. Rep. 2012, 8, 8287–8296. [Google Scholar] [CrossRef]
- Xu, C.; Wei, H.; Movahedi, A.; Sun, W.B.; Ma, X.X.; Li, D.W. Evaluation, characterization, expression profiling, and functional analysis of DXS and DXR genes of Populus trichocarpa. Plant Physiol. Biochem. 2019, 142, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Wang, R.; Shen, J.; Yang, Z.; Chen, Y. Cloning, characterization, and expression of 1-deoxy-d-xylulose-5-phosphate reductoisomerase gene from Dioscorea zingiberensis. Biol. Plant. 2019, 1, 262–267. [Google Scholar] [CrossRef]
- Gao, W.; Wang, X.; Purente, N.; Muhammad, L.Q.; Zhou, Y.W.; He, M.l. A 1-deoxy-d-xylulose 5-phosphate reductoisomerase gene probably involved in the synthesis of terpenoids in Chrysanthemum indicum var. aromaticum. Can. J. Plant Sci. 2018, 6, 1254–1264. [Google Scholar] [CrossRef]
- Zhang, X.D.; Zhao, J.; Li, C.X.; Li, T.; Wang, Y.Z. Cloning and expression analysis of 5-phosphate deoxyxylulose reductase gene from Gentiana macrophμlla (GmDXR). Chin. Tradit. Herb. Drugs 2014, 12, 1758–1763. [Google Scholar]
- Zhang, C.R.; Yang, Q.; Chen, H.B.; Pang, Y.X.; Tang, X.M.; Cheng, X.X.; Wu, W.Y.; Chen, S.M. Cloning and expression regulation of cDNA for 1-deoxy-D-xylulose 5-phosphate reductoisomerase cDNA from Alpinia officinarum. China J. Chin. Mater. Medica 2012, 21, 3208–3214. [Google Scholar]
- Yan, X.M.; Zhang, L.; Wang, J.; Liao, P.; Zhang, Y.; Zhang, R.; Kai, G.Y. Molecular characterization and expression of 1-deoxy-d-xylulose 5-phosphate reductoisomerase (DXR) gene from Salvia miltiorrhiza. Acta Physiol. Plant. 2009, 05, 1015–1022. [Google Scholar] [CrossRef]
- Wu, Q.J. Cloning and Functional Analysis of DXS, DXR and HDR of Dendrobium ferruginum; Anhui Agricultural University: Hefei, China, 2015. [Google Scholar]
- Zhu, Y.H.; Dong, C.M.; Zu, M.H.; Li, P.D.; Zhao, L. Cloning and expression analysis of 1-deoxy-D-xylulose 5-phosphate reductoisomerase gene in Rehmannia glutino. Plant Physiol. J. 2017, 53, 563–571. [Google Scholar]
- Agarwal, P.K.; Jha, B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 2010, 2, 201–212. [Google Scholar] [CrossRef]
- Xue, R.H.; Jin, S.A. Methyl Jasmonate: A vital signaling molecule. Lett. Biotechnol. 2006, 6, 985–988. [Google Scholar]
- Fang, X.; Xu, J.N.; Meng, X.N. Construction of Arabidopsis SPM12-GFP vector and identification of transgenic lines. J. Anhui Agric. Sci. 2019, 47, 86–88. [Google Scholar]
- Li, R.; Chen, P.Z.; Zhu, L.Z.; Wu, F.; Chen, Y.; Zhu, P.H.; Ji, K.S. Characterization and function of the 1-Deoxy-D-xylose-5-phosphate synthase (DXS) gene related to terpenoid synthesis in Pinus massoniana. Int. J. Mol. Sci. 2021, 22, 848–866. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.F.; He, W.L.; Cai, W.J.; Ruan, Q.Q.; Pan, T.; Ji, K.S. Analysis on transcriptome sequenced for Pinus massoniana. Mol. Plant Breed. 2013, 11, 385–392. [Google Scholar]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the expasy server. In The Proteomics Protocols Handbook; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Sonnhammer. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.Z.; Li, R.; Zhu, L.Z.; Hao, Q.Q.; Yao, S.; Liu, J.H.; Ji, K.S. Characterization and interaction analysis of the secondary cell wall synthesis-related transcription factor PmMYB7 in Pinus massoniana Lamb. International Journal of Molecular Sciences. 2022, 23, 2079. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.H.; Ma, Y.Y.; Zhu, L.Z.; Chen, Y.; Li, R.; Ji, K.S. Selection of suitable reference genes in Pinus massoniana Lamb. Under different abiotic stresses for qPCR normalization. Forest. 2019, 10, 632–650. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods. 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, S.; Quintero, F.J.; Cubero, B.; Ruiz, M.T.; Bressan, R.A.; Hasegawa, P.M.; Pardo, J.M. Differential expression and function of Arabidopsis thaliana NHX Na+/H+ antiporters in the salt stress response. The Plant Journal. 2002, 30, 529–539. [Google Scholar] [CrossRef]



| Amino Acid Species | Quantity | Proportion | Amino Acid Species | Quantity | Proportion |
|---|---|---|---|---|---|
| Ala (A) | 53 | 10.1% | Leu (L) | 51 | 9.7% |
| Arg (R) | 20 | 3.8% | Lys (K) | 35 | 6.6% |
| Asn (N) | 9 | 1.7% | Met (M) | 10 | 1.9% |
| Asp (D) | 26 | 4.9% | Phe (F) | 19 | 3.6% |
| Cys (C) | 8 | 1.5% | Pro (P) | 34 | 6.5% |
| Gln (Q) | 10 | 1.9% | Ser (S) | 36 | 6.8% |
| Glu (E) | 34 | 6.5% | Thr (T) | 26 | 4.9% |
| Gly (G) | 35 | 6.6% | Trp (W) | 8 | 1.5% |
| His (H) | 16 | 3.0% | Tyr (Y) | 11 | 2.1% |
| Ile (I) | 41 | 7.8% | Val (V) | 45 | 8.5% |
| Codon | Amino Acid | Proportion | Frequency | Number | Relative Codon Usage | Codon | Amino Acid | Proportion | Frequency | Number | Relative Codon Usage |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GCA | Ala | 0.377 | 37.879 | 20 | 1.51 | CTT | 0.235 | 22.727 | 12 | 1.41 | |
| GCC | 0.189 | 18.939 | 10 | 0.75 | TTA | 0.176 | 17.045 | 9 | 1.06 | ||
| GCG | 0.038 | 3.788 | 2 | 0.15 | TTG | 0.196 | 18.939 | 10 | 1.18 | ||
| GCT | 0.396 | 39.773 | 21 | 1.58 | AAA | Lys | 0.543 | 35.985 | 19 | 1.09 | |
| AGA | Arg | 0.600 | 22.727 | 12 | 3.60 | AAG | 0.457 | 30.303 | 16 | 0.91 | |
| AGG | 0.150 | 5.682 | 3 | 0.90 | ATG | Met | 1.000 | 18.939 | 10 | 1.00 | |
| CGA | 0.150 | 5.682 | 3 | 0.90 | TTC | Phe | 0.316 | 11.364 | 6 | 0.63 | |
| CGC | 0.000 | 0.000 | 0 | 0.00 | TTT | 0.684 | 24.621 | 13 | 1.37 | ||
| CGG | 0.100 | 3.788 | 2 | 0.60 | CCA | Pro | 0.441 | 28.409 | 15 | 1.76 | |
| CGT | 0.000 | 0.000 | 0 | 0.00 | CCC | 0.088 | 5.682 | 3 | 0.35 | ||
| AAC | Asn | 0.222 | 3.788 | 2 | 0.44 | CCG | 0.059 | 3.788 | 2 | 0.24 | |
| AAT | 0.778 | 13.258 | 7 | 1.56 | CCT | 0.412 | 26.515 | 14 | 1.65 | ||
| GAC | Asp | 0.385 | 18.939 | 10 | 0.77 | AGC | Ser | 0.056 | 3.788 | 2 | 0.33 |
| GAT | 0.615 | 30.303 | 16 | 1.23 | AGT | 0.083 | 5.682 | 3 | 0.50 | ||
| TGC | Cys | 0.500 | 7.576 | 4 | 1.00 | TCA | 0.278 | 18.939 | 10 | 1.67 | |
| TGT | 0.500 | 7.576 | 4 | 1.00 | TCC | 0.167 | 11.364 | 6 | 1.00 | ||
| CAA | Gln | 0.600 | 11.364 | 6 | 1.20 | TCG | 0.056 | 3.788 | 2 | 0.33 | |
| CAG | 0.400 | 7.576 | 4 | 0.80 | TCT | 0.361 | 24.621 | 13 | 2.17 | ||
| GAA | Glu | 0.471 | 30.303 | 16 | 0.94 | ACA | Thr | 0.500 | 24.621 | 13 | 2.00 |
| GAG | 0.529 | 34.091 | 18 | 1.06 | ACC | 0.269 | 13.258 | 7 | 1.08 | ||
| GGA | Gly | 0.400 | 26.515 | 14 | 1.60 | ACG | 0.038 | 1.894 | 1 | 0.15 | |
| GGC | 0.143 | 9.470 | 5 | 0.57 | ACT | 0.192 | 9.470 | 5 | 0.77 | ||
| GGG | 0.200 | 13.258 | 7 | 0.80 | TGG | Trp | 1.000 | 15.152 | 8 | 1.00 | |
| GGT | 0.257 | 17.045 | 9 | 1.03 | TAC | Tyr | 0.091 | 1.894 | 1 | 0.18 | |
| CAC | His | 0.563 | 17.045 | 9 | 1.13 | TAT | 0.909 | 18.939 | 10 | 1.82 | |
| CAT | 0.438 | 13.258 | 7 | 0.88 | GTA | Val | 0.244 | 20.833 | 11 | 0.98 | |
| ATA | Ile | 0.390 | 30.303 | 16 | 1.17 | GTC | 0.089 | 7.576 | 4 | 0.36 | |
| ATC | 0.146 | 11.364 | 6 | 0.44 | GTG | 0.222 | 18.939 | 10 | 0.89 | ||
| ATT | 0.463 | 35.985 | 19 | 1.39 | GTT | 0.444 | 37.879 | 20 | 1.78 | ||
| CTA | Leu | 0.157 | 15.152 | 8 | 0.94 | TAA | TER | 0.000 | 0.000 | 0 | 0.00 |
| CTC | 0.098 | 9.470 | 5 | 0.59 | TAG | 0.000 | 0.000 | 0 | 0.00 | ||
| CTG | 0.137 | 13.258 | 7 | 0.82 | TGA | 1.000 | 1.894 | 1 | 3.00 |
| Cis Element | Sequence | Function | Quantity |
|---|---|---|---|
| AT-rich element | ATAGAAATCAA | ATBP-1-binding sites | 1 |
| CAAT-box | CAAT/CCAAT/CAAAT | Promoter and enhancer region regulatory elements | 30 |
| CAT-box | GCCACT | Syngeneic tissue expression cis-elements | 1 |
| CCAAT-box | CAACGG | MYBHv1-binding site | 1 |
| CGTCA-motif | CGTCA | Methyl jasmonate response element | 1 |
| STRE | AGGGG | Stress response element | 8 |
| Sp1 | GGGCGG | Light-responsive elements | 3 |
| TC-rich repeats | ATTCTCTAAC | Defense and stress response cis-acting elements | 1 |
| TCT-motif | TCTTAC | Partial photoresponse elements | 1 |
| TGACG-motif | TGACG | Methyl jasmonate response element | 1 |
| WUN-motif | AAATTTCTT | Damage response element | 1 |
| W-box | TTGACC | Salicylic acid response element | 1 |
| TATA-box | ATATAT/TATA/TATATA/ATTATA/ATATAA/TATACA/TATAA/TATATAA/TATATAAATC | Transcription initiation-30 core promoter element | 20 |
| Primer Name | Sequence of Primers (5′ → 3′) | Use |
|---|---|---|
| PmDXR-Mid-F | GGTGTCCAATTCCACTACTACATTGC | Gene cloning |
| PmDXR-Mid-R | CATGATAAAAGGCATCCCTTCATGGG | |
| PmDXR 5′RACE Outer | GCGGGAGGAGGTGCTTGTAGGGA | |
| PmDXR 5′RACE Inner | GGAAAGGGGCGGAGGATAAGACAAA | |
| PmDXR 3′RACE Outer | CTGGCCTCGGCTTGACCTTTGCG | |
| PmDXR 3′RACE Inner | GCTTGGAGCCTGCCACAGTCTTC | |
| PmDXR ORF F | ATGGGAGTATTAGTAGTAG | |
| PmDXR ORF R | GACTGTGGCAGGCTCCAAGC | |
| qPmDXR-F | GTTCCCTACAAGCACCTCCTC | qRT-PCR |
| qPmDXR-R | GTTCGGCAACAATGTCCAAT | |
| TUA-F | CAAACTTGGTCCCGTATCCTC | |
| TUA-R | CACAGAAAGCTGCTCATGGTAA | |
| pPmDXR-SP1 | GTGAAGATGGCAGAGTCGCAGGAA | Promoter cloning |
| pPmDXR-SP2 | GCGAGTGTAGGGTGGAGGCTTATT | |
| pPmDXR-SP3 | GGGCGGAGGATAAGACAAAGAAGA | |
| pPmDXR-F | TGGTAATGCAATGAAGTTGGGAGG | |
| pPmDXR-R | GGGGTGGAAAGGGGCGGAGGATAA | |
| pBI121-ProDXR-F | GACCATGATTACGCCAAGCTTTGGTAATGCAATGAAGTTGGGA | |
| pBI121-ProDXR-R | ACCACCCGGGGATCCTCTAGAGGGGTGGAAAGGGGCGGA | |
| PmDXR-GFP-F | GAGAACACGGGGGACTCTAGAATGGGAGTATTAGTAGTAGTAATAATAATAAG | Subcellular localization |
| PmDXR-GFP-R | GCCCTTGCTCACCATGGATCCGACTGTGGCAGGCTCCAAGC | |
| 1302-PmDXR-F | CGGGGGACTCTTGACCATGATGGGAGTATTAGTAGTAGTAATAATAATA | Overexpression of A. thaliana |
| 1302-PmDXR-R | ACTAGTCAGATCTACCATGGTCAGACTGTGGCAGGCTCCAAG | |
| 1302-CheckF | ACAGTCTCAGAAGACCAAAGGGCA | |
| AtActin2-F | ACTCTCCGCTATGTATGTCGCC | |
| AtActin2-R | ATTTCCCGCTCGCTGTTGTGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Zhu, L.; Chen, P.; Chen, Y.; Hao, Q.; Zhu, P.; Ji, K. Functional Characterization of PmDXR, a Critical Rate-Limiting Enzyme, for Turpentine Biosynthesis in Masson Pine (Pinus massoniana Lamb.). Int. J. Mol. Sci. 2024, 25, 4415. https://doi.org/10.3390/ijms25084415
Li R, Zhu L, Chen P, Chen Y, Hao Q, Zhu P, Ji K. Functional Characterization of PmDXR, a Critical Rate-Limiting Enzyme, for Turpentine Biosynthesis in Masson Pine (Pinus massoniana Lamb.). International Journal of Molecular Sciences. 2024; 25(8):4415. https://doi.org/10.3390/ijms25084415
Chicago/Turabian StyleLi, Rong, Lingzhi Zhu, Peizhen Chen, Yu Chen, Qingqing Hao, Peihuang Zhu, and Kongshu Ji. 2024. "Functional Characterization of PmDXR, a Critical Rate-Limiting Enzyme, for Turpentine Biosynthesis in Masson Pine (Pinus massoniana Lamb.)" International Journal of Molecular Sciences 25, no. 8: 4415. https://doi.org/10.3390/ijms25084415
APA StyleLi, R., Zhu, L., Chen, P., Chen, Y., Hao, Q., Zhu, P., & Ji, K. (2024). Functional Characterization of PmDXR, a Critical Rate-Limiting Enzyme, for Turpentine Biosynthesis in Masson Pine (Pinus massoniana Lamb.). International Journal of Molecular Sciences, 25(8), 4415. https://doi.org/10.3390/ijms25084415






