Molecular Circuits of Immune Sensing and Response to Oncolytic Virotherapy
Abstract
1. Introduction
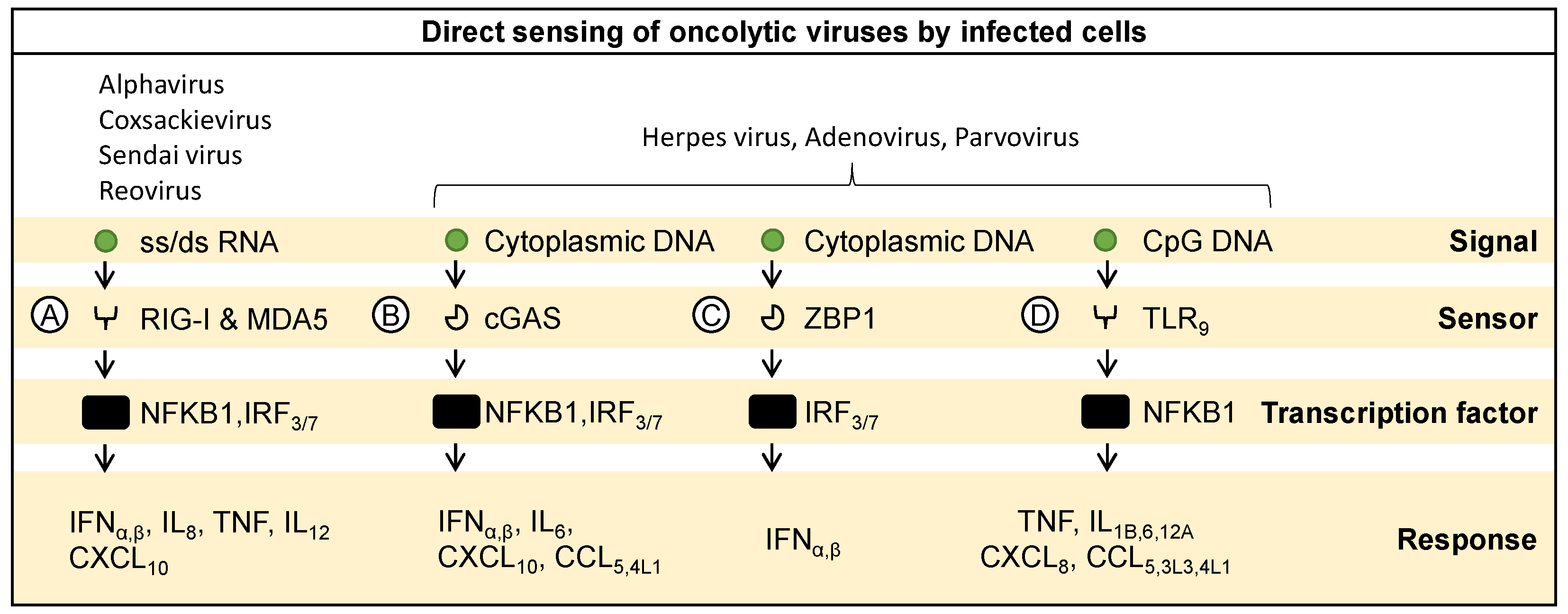
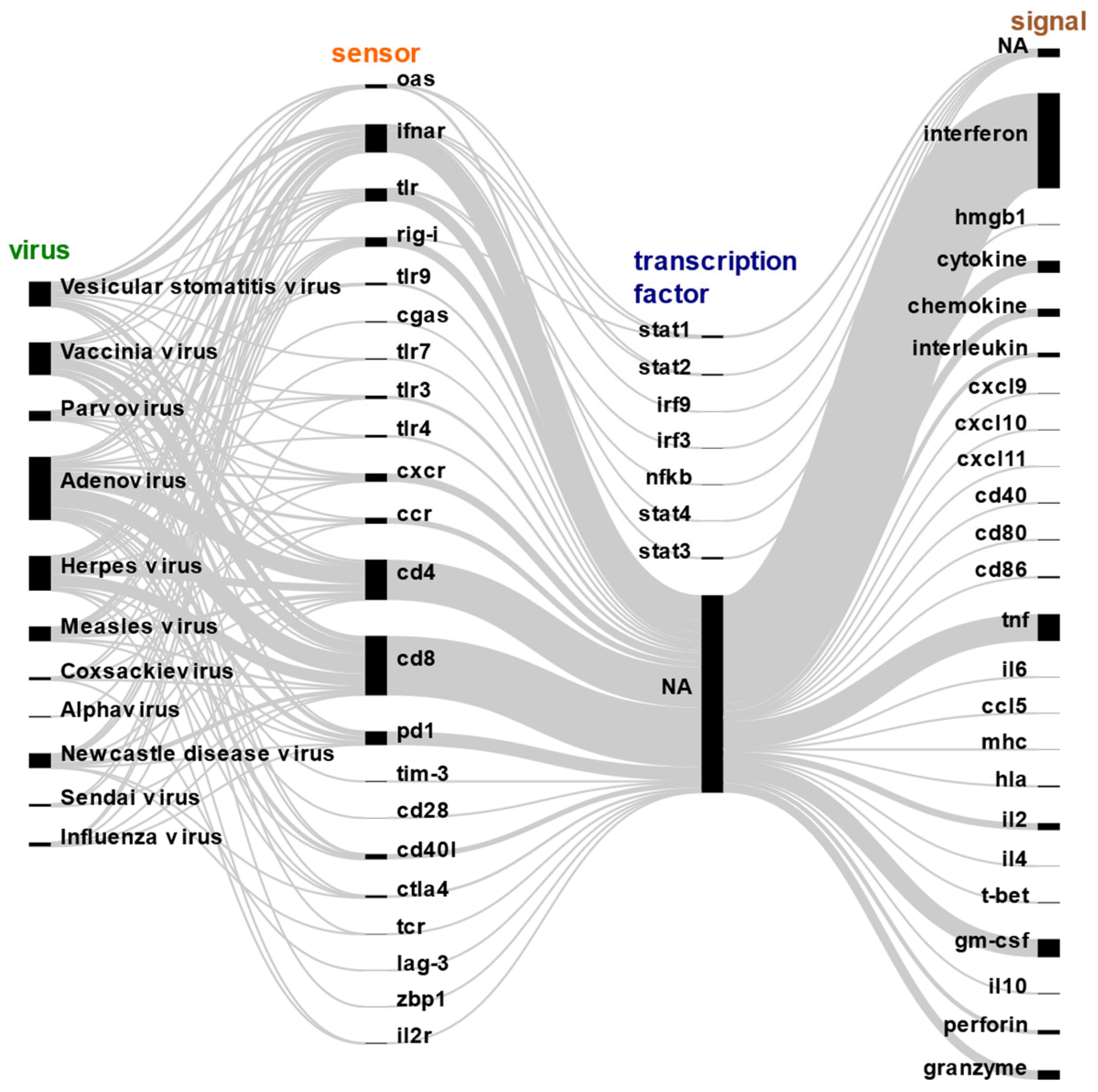
2. Molecular Sensing and Response to Oncolytic Viruses by Infected Cells
2.1. Extracellular Sensing of Oncolytic Viruses
2.2. Sensing Viral RNA
2.3. Sensing Viral DNA
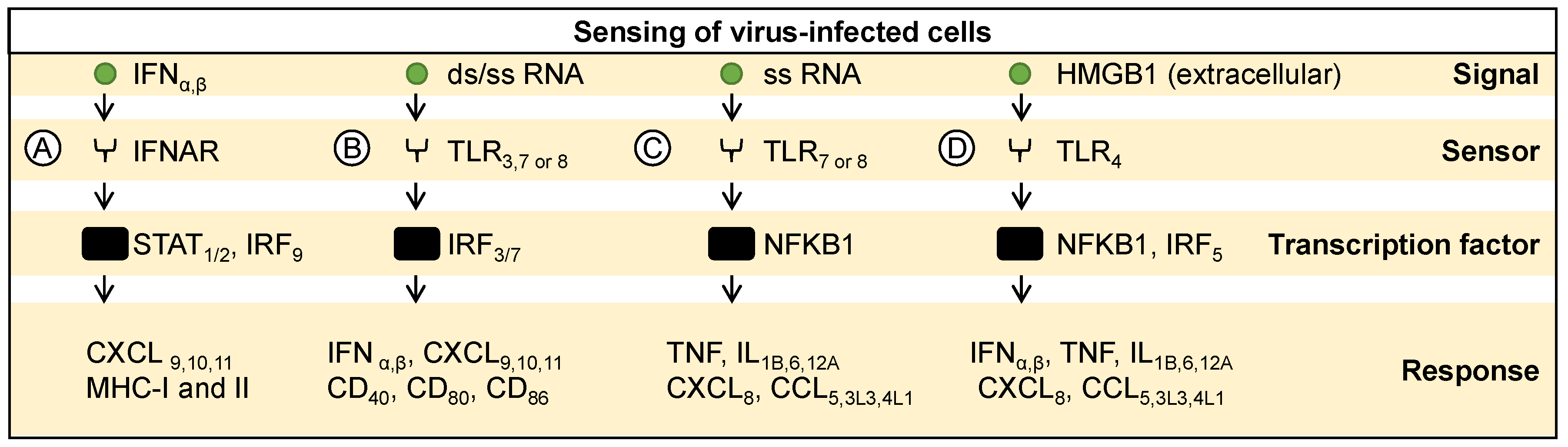
3. Molecular Sensing and Response to Infected Cells
3.1. Sensing Cytokines from Infected Cells
3.2. Sensing Pathogen-Associated Signals from Infected Cells
3.3. Sensing Danger-Associated Signals from Infected Cells
4. Molecular Signaling Response for Immune Activation
4.1. Innate Immune Signaling
4.2. Adaptive Immune Signaling
5. Strategies to Exploit Immune Signaling in Favor of Oncolytic Virotherapy
5.1. Engineering Viruses to Trigger Immune Signaling
5.2. Engineering Viruses for Tumor Targeting
5.3. Combinatorial Approaches to Boost Viral Immunogenicity
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kaufman, H.L.; Kohlhapp, F.J.; Zloza, A. Oncolytic Viruses: A New Class of Immunotherapy Drugs. Nat. Rev. Drug Discov. 2015, 14, 642–662. [Google Scholar] [CrossRef] [PubMed]
- Lemos De Matos, A.; Franco, L.S.; McFadden, G. Oncolytic Viruses and the Immune System: The Dynamic Duo. Mol. Ther. Methods Clin. Dev. 2020, 17, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Wang, L.; Song, G.; Zhou, W. Innate Immune Responses to RNA: Sensing and Signaling. Front. Immunol. 2024, 15, 1287940. [Google Scholar] [CrossRef] [PubMed]
- Mealiea, D.; McCart, J.A. Cutting Both Ways: The Innate Immune Response to Oncolytic Virotherapy. Cancer Gene Ther. 2022, 29, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Pol, J.G.; Workenhe, S.T.; Konda, P.; Gujar, S.; Kroemer, G. Cytokines in Oncolytic Virotherapy. Cytokine Growth Factor Rev. 2020, 56, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Bommareddy, P.K.; Patel, A.; Hossain, S.; Kaufman, H.L. Talimogene Laherparepvec (T-VEC) and Other Oncolytic Viruses for the Treatment of Melanoma. Am. J. Clin. Dermatol. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Andtbacka, R.H.; Collichio, F.A.; Amatruda, T.; Senzer, N.; Chesney, J.; Delman, K.; Spitler, L.; Puzanov, I.; Agarwala, S.; Milhem, M.; et al. Final Planned Overall Survival (OS) from OPTiM, a Randomized Phase III Trial of Talimogene Laherparepvec (T-VEC) versus GM-CSF for the Treatment of Unresected Stage IIIB/C/IV Melanoma (NCT00769704). J. Immunother. Cancer 2014, 2, P263. [Google Scholar] [CrossRef]
- Bhatt, D.K.; Wekema, L.; Carvalho Barros, L.R.; Chammas, R.; Daemen, T. A Systematic Analysis on the Clinical Safety and Efficacy of Onco-Virotherapy. Mol. Ther. Oncolytics 2021, 23, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Macedo, N.; Miller, D.M.; Haq, R.; Kaufman, H.L. Clinical Landscape of Oncolytic Virus Research in 2020. J. Immunother. Cancer 2020, 8, e001486. [Google Scholar] [CrossRef]
- Tian, Y.; Xie, D.; Yang, L. Engineering Strategies to Enhance Oncolytic Viruses in Cancer Immunotherapy. Sig. Transduct. Target. Ther. 2022, 7, 117. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, Z.; Zhang, Y.; Huang, X.; Liu, Q. Efficacy and Safety of Oncolytic Viruses in Randomized Controlled Trials: A Systematic Review and Meta-Analysis. Cancers 2020, 12, 1416. [Google Scholar] [CrossRef]
- Bhatt, D.K.; Chammas, R.; Daemen, T. Resistance Mechanisms Influencing Oncolytic Virotherapy, a Systematic Analysis. Vaccines 2021, 9, 1166. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Ghorbani, E.; Khazaei, M.; Avan, A.; Ryzhikov, M.; Azadmanesh, K.; Hassanian, S.M. Interferon-Mediated Tumor Resistance to Oncolytic Virotherapy. J. Cell Biochem. 2017, 118, 1994–1999. [Google Scholar] [CrossRef]
- Wongthida, P.; Diaz, R.M.; Galivo, F.; Kottke, T.; Thompson, J.; Melcher, A.; Vile, R. VSV Oncolytic Virotherapy in the B16 Model Depends upon Intact MyD88 Signaling. Mol. Ther. 2011, 19, 150–158. [Google Scholar] [CrossRef]
- Lapteva, N.; Aldrich, M.; Rollins, L.; Ren, W.; Goltsova, T.; Chen, S.-Y.; Huang, X.F. Attraction and Activation of Dendritic Cells at the Site of Tumor Elicits Potent Antitumor Immunity. Mol. Ther. 2009, 17, 1626–1636. [Google Scholar] [CrossRef]
- Ahtiainen, L.; Mirantes, C.; Jahkola, T.; Escutenaire, S.; Diaconu, I.; Osterlund, P.; Kanerva, A.; Cerullo, V.; Hemminki, A. Defects in Innate Immunity Render Breast Cancer Initiating Cells Permissive to Oncolytic Adenovirus. PLoS ONE 2010, 5, e13859. [Google Scholar] [CrossRef]
- Samudio, I.; Rezvani, K.; Shaim, H.; Hofs, E.; Ngom, M.; Bu, L.; Liu, G.; Lee, J.T.C.; Imren, S.; Lam, V.; et al. UV-Inactivated HSV-1 Potently Activates NK Cell Killing of Leukemic Cells. Blood 2016, 127, 2575–2586. [Google Scholar] [CrossRef]
- Wang, Y.; Jin, J.; Li, Y.; Zhou, Q.; Yao, R.; Wu, Z.; Hu, H.; Fang, Z.; Dong, S.; Cai, Q.; et al. NK Cell Tumor Therapy Modulated by UV-Inactivated Oncolytic Herpes Simplex Virus Type 2 and Checkpoint Inhibitors. Transl. Res. 2022, 240, 64–86. [Google Scholar] [CrossRef]
- Koellhoffer, E.C.; Mao, C.; Beiss, V.; Wang, L.; Fiering, S.N.; Boone, C.E.; Steinmetz, N.F. Inactivated Cowpea Mosaic Virus in Combination with OX40 Agonist Primes Potent Antitumor Immunity in a Bilateral Melanoma Mouse Model. Mol. Pharm. 2022, 19, 592–601. [Google Scholar] [CrossRef]
- Rojas, J.J.; Sampath, P.; Bonilla, B.; Ashley, A.; Hou, W.; Byrd, D.; Thorne, S.H. Manipulating TLR Signaling Increases the Anti-Tumor T Cell Response Induced by Viral Cancer Therapies. Cell Rep. 2016, 15, 264–273. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Toll-like Receptor and RIG-I-like Receptor Signaling. Ann. N. Y. Acad. Sci. 2008, 1143, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, D.; Wang, W.; Galls, D.; Guo, R.; Xu, L.; Pyle, A.M. The Molecular Mechanism of RIG-I Activation and Signaling. Immunol. Rev. 2021, 304, 154–168. [Google Scholar] [CrossRef]
- Cai, J.; Zhu, W.; Lin, Y.; Zhang, S.; Chen, X.; Gong, S.; He, S.; Hu, J.; Yan, G.; Liang, J. Systematic Characterization of the Biodistribution of the Oncolytic Virus M1. Hum. Gene Ther. 2020, 31, 1203–1213. [Google Scholar] [CrossRef]
- Hu, C.; Liu, Y.; Lin, Y.; Liang, J.-K.; Zhong, W.-W.; Li, K.; Huang, W.-T.; Wang, D.-J.; Yan, G.-M.; Zhu, W.-B.; et al. Intravenous Injections of the Oncolytic Virus M1 as a Novel Therapy for Muscle-Invasive Bladder Cancer. Cell Death Dis. 2018, 9, 274. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Hu, C.; Xing, F.; Gao, M.; Liang, J.; Xiao, X.; Cai, J.; Tan, Y.; Hu, J.; Zhu, W.; et al. Deficiency of the IRE1α-Autophagy Axis Enhances the Antitumor Effects of the Oncolytic Virus M1. J. Virol. 2018, 92, e01331-17. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, H.; Liang, J.; Li, K.; Zhu, W.; Fu, L.; Wang, F.; Zheng, X.; Shi, H.; Wu, S.; et al. Identification and Characterization of Alphavirus M1 as a Selective Oncolytic Virus Targeting ZAP-Defective Human Cancers. Proc. Natl. Acad. Sci. USA 2014, 111, E4504–E4512. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, K.; Zhu, W.-B.; Zhang, H.; Huang, W.-T.; Liu, X.-C.; Lin, Y.; Cai, J.; Yan, G.-M.; Qiu, J.-G.; et al. Suppression of CCDC6 Sensitizes Tumor to Oncolytic Virus M1. Neoplasia 2021, 23, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Liu, Y.; He, C.; Hu, W.; Liu, W.; Huang, X.; Wu, J.; Xie, F.; Chen, C.; Wang, J.; et al. Combining NanoKnife with M1 Oncolytic Virus Enhances Anticancer Activity in Pancreatic Cancer. Cancer Lett. 2021, 502, 9–24. [Google Scholar] [CrossRef]
- Toribio, R.; Díaz-López, I.; Berlanga, J.J.; Molina-Jiménez, F.; Majano, P.; Ventoso, I. Naturally Occurring and Engineered Alphaviruses Sensitive to Double-Stranded-RNA-Activated Protein Kinase Show Restricted Translation in Mammalian Cells, Increased Sensitivity to Interferon, and Marked Oncotropism. J. Virol. 2020, 94, e01630-19. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, H.; Zou, H.; Tian, X.; Hu, J.; Qiu, P.; Hu, H.; Yan, G. Liposome Encapsulation of Oncolytic Virus M1 To Reduce Immunogenicity and Immune Clearance In Vivo. Mol. Pharm. 2019, 16, 779–785. [Google Scholar] [CrossRef]
- Annels, N.E.; Mansfield, D.; Arif, M.; Ballesteros-Merino, C.; Simpson, G.R.; Denyer, M.; Sandhu, S.S.; Melcher, A.A.; Harrington, K.J.; Davies, B.; et al. Phase I Trial of an ICAM-1-Targeted Immunotherapeutic-Coxsackievirus A21 (CVA21) as an Oncolytic Agent Against Non Muscle-Invasive Bladder Cancer. Clin. Cancer Res. 2019, 25, 5818–5831. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Miyamoto, Y.; Inoue, T.; Kaneda, Y. Efficient Eradication of Hormone-Resistant Human Prostate Cancers by Inactivated Sendai Virus Particle. Int. J. Cancer 2009, 124, 2478–2487. [Google Scholar] [CrossRef] [PubMed]
- Oosenbrug, T.; Van Den Wollenberg, D.J.M.; Duits, E.W.; Hoeben, R.C.; Ressing, M.E. Induction of Robust Type I Interferon Levels by Oncolytic Reovirus Requires Both Viral Replication and Interferon-α/β Receptor Signaling. Hum. Gene Ther. 2021, 32, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- Matsushima-Miyagi, T.; Hatano, K.; Nomura, M.; Li-Wen, L.; Nishikawa, T.; Saga, K.; Shimbo, T.; Kaneda, Y. TRAIL and Noxa Are Selectively Upregulated in Prostate Cancer Cells Downstream of the RIG-I/MAVS Signaling Pathway by Nonreplicating Sendai Virus Particles. Clin. Cancer Res. 2012, 18, 6271–6283. [Google Scholar] [CrossRef] [PubMed]
- Froechlich, G.; Caiazza, C.; Gentile, C.; D’Alise, A.M.; De Lucia, M.; Langone, F.; Leoni, G.; Cotugno, G.; Scisciola, V.; Nicosia, A.; et al. Integrity of the Antiviral STING-Mediated DNA Sensing in Tumor Cells Is Required to Sustain the Immunotherapeutic Efficacy of Herpes Simplex Oncolytic Virus. Cancers 2020, 12, 3407. [Google Scholar] [CrossRef] [PubMed]
- Raykov, Z.; Grekova, S.; Leuchs, B.; Aprahamian, M.; Rommelaere, J. Arming Parvoviruses with CpG Motifs to Improve Their Oncosuppressive Capacity. Int. J. Cancer 2008, 122, 2880–2884. [Google Scholar] [CrossRef]
- Chen, C.; Xu, P. Cellular Functions of cGAS-STING Signaling. Trends Cell Biol. 2023, 33, 630–648. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wang, D.; Zhang, J.; Xiang, P.; Zeng, Z.; Xiong, W.; Shi, L. cGAS-STING Signaling in the Tumor Microenvironment. Cancer Lett. 2023, 577, 216409. [Google Scholar] [CrossRef] [PubMed]
- Bao, M.; Liu, Y.-J. Regulation of TLR7/9 Signaling in Plasmacytoid Dendritic Cells. Protein Cell 2013, 4, 40–52. [Google Scholar] [CrossRef]
- Efferson, C.L.; Tsuda, N.; Kawano, K.; Nistal-Villán, E.; Sellappan, S.; Yu, D.; Murray, J.L.; García-Sastre, A.; Ioannides, C.G. Prostate Tumor Cells Infected with a Recombinant Influenza Virus Expressing a Truncated NS1 Protein Activate Cytolytic CD8+ Cells to Recognize Noninfected Tumor Cells. J. Virol. 2006, 80, 383–394. [Google Scholar] [CrossRef][Green Version]
- Elankumaran, S.; Rockemann, D.; Samal, S.K. Newcastle Disease Virus Exerts Oncolysis by Both Intrinsic and Extrinsic Caspase-Dependent Pathways of Cell Death. J. Virol. 2006, 80, 7522–7534. [Google Scholar] [CrossRef] [PubMed]
- Puhlmann, J.; Puehler, F.; Mumberg, D.; Boukamp, P.; Beier, R. Rac1 Is Required for Oncolytic NDV Replication in Human Cancer Cells and Establishes a Link between Tumorigenesis and Sensitivity to Oncolytic Virus. Oncogene 2010, 29, 2205–2216. [Google Scholar] [CrossRef] [PubMed]
- Okemoto, K.; Wagner, B.; Meisen, H.; Haseley, A.; Kaur, B.; Chiocca, E.A. STAT3 Activation Promotes Oncolytic HSV1 Replication in Glioma Cells. PLoS ONE 2013, 8, e71932. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, X.; Yuan, Y.; Gong, X.; Chen, Z.; Xu, X. IPS-1 Plays a Dual Function to Directly Induce Apoptosis in Murine Melanoma Cells by Inactivated Sendai Virus. Int. J. Cancer 2014, 134, 224–234. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-W.; Nishikawa, T.; Kaneda, Y. An RNA Molecule Derived From Sendai Virus DI Particles Induces Antitumor Immunity and Cancer Cell-Selective Apoptosis. Mol. Ther. 2016, 24, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.L.; Safroneeva, E.; Mossman, K.L. The Role of ICP0-Null HSV-1 and Interferon Signaling Defects in the Effective Treatment of Breast Adenocarcinoma. Mol. Ther. 2005, 12, 1101–1110. [Google Scholar] [CrossRef]
- Nanni, P.; Gatta, V.; Menotti, L.; De Giovanni, C.; Ianzano, M.; Palladini, A.; Grosso, V.; Dall’ora, M.; Croci, S.; Nicoletti, G.; et al. Preclinical Therapy of Disseminated HER-2+ Ovarian and Breast Carcinomas with a HER-2-Retargeted Oncolytic Herpesvirus. PLoS Pathog. 2013, 9, e1003155. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of Type I Interferon Responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef]
- Di Somma, S.; Iannuzzi, C.A.; Passaro, C.; Forte, I.M.; Iannone, R.; Gigantino, V.; Indovina, P.; Botti, G.; Giordano, A.; Formisano, P.; et al. The Oncolytic Virus Dl922-947 Triggers Immunogenic Cell Death in Mesothelioma and Reduces Xenograft Growth. Front. Oncol. 2019, 9, 564. [Google Scholar] [CrossRef]
- Abraham, R.; Mudaliar, P.; Padmanabhan, A.; Sreekumar, E. Induction of Cytopathogenicity in Human Glioblastoma Cells by Chikungunya Virus. PLoS ONE 2013, 8, e75854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Tan, J.; Zhang, Y.; Wong, C.-W.; Lin, Z.; Liu, X.; Sander, M.; Yang, X.; Liang, L.; et al. Necroptotic Virotherapy of Oncolytic Alphavirus M1 Cooperated with Doxorubicin Displays Promising Therapeutic Efficacy in TNBC. Oncogene 2021, 40, 4783–4795. [Google Scholar] [CrossRef] [PubMed]
- Martikainen, M.; Ramachandran, M.; Lugano, R.; Ma, J.; Martikainen, M.-M.; Dimberg, A.; Yu, D.; Merits, A.; Essand, M. IFN-I-Tolerant Oncolytic Semliki Forest Virus in Combination with Anti-PD1 Enhances T Cell Response against Mouse Glioma. Mol. Ther. Oncolytics 2021, 21, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Hu, C.; Liu, Y.; Chen, X.; Song, D.; Shen, R.; Liu, Z.; Jia, X.; Zhang, Q.; Gao, Y.; et al. Directed Natural Evolution Generates a Next-Generation Oncolytic Virus with a High Potency and Safety Profile. Nat. Commun. 2023, 14, 3410. [Google Scholar] [CrossRef] [PubMed]
- Ghonime, M.G.; Cassady, K.A. Combination Therapy Using Ruxolitinib and Oncolytic HSV Renders Resistant MPNSTs Susceptible to Virotherapy. Cancer Immunol. Res. 2018, 6, 1499–1510. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.R.; Dash, A.; Jacobson, B.A.; Ji, Y.; Baumann, D.; Ismail, K.; Kratzke, R.A. JAK/STAT Inhibition with Ruxolitinib Enhances Oncolytic Virotherapy in Non-Small Cell Lung Cancer Models. Cancer Gene Ther. 2019, 26, 411–418. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, J.; Zhao, Y.; Ma, X.; Yi, H. Toll-like Receptor 3 (TLR3) Regulation Mechanisms and Roles in Antiviral Innate Immune Responses. J. Zhejiang Univ. Sci. B 2021, 22, 609–632. [Google Scholar] [CrossRef] [PubMed]
- Balogh, A.; Bátor, J.; Markó, L.; Németh, M.; Pap, M.; Sétáló, G.; Müller, D.N.; Csatary, L.K.; Szeberényi, J. Gene Expression Profiling in PC12 Cells Infected with an Oncolytic Newcastle Disease Virus Strain. Virus Res. 2014, 185, 10–22. [Google Scholar] [CrossRef] [PubMed]
- Sieben, M.; Schäfer, P.; Dinsart, C.; Galle, P.R.; Moehler, M. Activation of the Human Immune System via Toll-like Receptors by the Oncolytic Parvovirus H-1. Int. J. Cancer 2013, 132, 2548–2556. [Google Scholar] [CrossRef]
- Weiss, R.; Sachet, M.; Zinngrebe, J.; Aschacher, T.; Krainer, M.; Hegedus, B.; Walczak, H.; Bergmann, M. IL-24 Sensitizes Tumor Cells to TLR3-Mediated Apoptosis. Cell Death Differ. 2013, 20, 823–833. [Google Scholar] [CrossRef]
- Guillerme, J.-B.; Boisgerault, N.; Roulois, D.; Ménager, J.; Combredet, C.; Tangy, F.; Fonteneau, J.-F.; Gregoire, M. Measles Virus Vaccine–Infected Tumor Cells Induce Tumor Antigen Cross-Presentation by Human Plasmacytoid Dendritic Cells. Clin. Cancer Res. 2013, 19, 1147–1158. [Google Scholar] [CrossRef] [PubMed]
- Angelova, A.; Pierrard, K.; Detje, C.N.; Santiago, E.; Grewenig, A.; Nüesch, J.P.F.; Kalinke, U.; Ungerechts, G.; Rommelaere, J.; Daeffler, L. Oncolytic Rodent Protoparvoviruses Evade a TLR- and RLR-Independent Antiviral Response in Transformed Cells. Pathogens 2023, 12, 607. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Andersson, U. Targeting Inflammation Driven by HMGB1. Front. Immunol. 2020, 11, 484. [Google Scholar] [CrossRef] [PubMed]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in Inflammation and Cancer. Annu. Rev. Immunol. 2010, 28, 367–388. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Chen, D.S.; Powles, T.; Turley, S.J. The Cancer-Immunity Cycle: Indication, Genotype, and Immunotype. Immunity 2023, 56, 2188–2205. [Google Scholar] [CrossRef] [PubMed]
- Ross, S.H.; Cantrell, D.A. Signaling and Function of Interleukin-2 in T Lymphocytes. Annu. Rev. Immunol. 2018, 36, 411–433. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J. T Helper 2 (Th2) Cell Differentiation, Type 2 Innate Lymphoid Cell (ILC2) Development and Regulation of Interleukin-4 (IL-4) and IL-13 Production. Cytokine 2015, 75, 14–24. [Google Scholar] [CrossRef]
- Schoenborn, J.R.; Wilson, C.B. Regulation of Interferon-Gamma during Innate and Adaptive Immune Responses. Adv. Immunol. 2007, 96, 41–101. [Google Scholar] [CrossRef]
- Kato, T.; Ueda, Y.; Kinoh, H.; Yoneyama, Y.; Matsunaga, A.; Komaru, A.; Harada, Y.; Suzuki, H.; Komiya, A.; Shibata, S.; et al. RIG-I Helicase-Independent Pathway in Sendai Virus-Activated Dendritic Cells Is Critical for Preventing Lung Metastasis of AT6.3 Prostate Cancer. Neoplasia 2010, 12, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Heinzerling, L.; Künzi, V.; Oberholzer, P.A.; Kündig, T.; Naim, H.; Dummer, R. Oncolytic Measles Virus in Cutaneous T-Cell Lymphomas Mounts Antitumor Immune Responses In Vivo and Targets Interferon-Resistant Tumor Cells. Blood 2005, 106, 2287–2294. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Brzoza, K.L.; Hiltbold, E.M. Matrix Protein Mutant of Vesicular Stomatitis Virus Stimulates Maturation of Myeloid Dendritic Cells. J. Virol. 2006, 80, 2194–2205. [Google Scholar] [CrossRef] [PubMed]
- Willmon, C.L.; Saloura, V.; Fridlender, Z.G.; Wongthida, P.; Diaz, R.M.; Thompson, J.; Kottke, T.; Federspiel, M.; Barber, G.; Albelda, S.M.; et al. Expression of IFN-Beta Enhances Both Efficacy and Safety of Oncolytic Vesicular Stomatitis Virus for Therapy of Mesothelioma. Cancer Res. 2009, 69, 7713–7720. [Google Scholar] [CrossRef]
- Gaud, G.; Lesourne, R.; Love, P.E. Regulatory Mechanisms in T Cell Receptor Signalling. Nat. Rev. Immunol. 2018, 18, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Brownlie, R.J.; Zamoyska, R. T Cell Receptor Signalling Networks: Branched, Diversified and Bounded. Nat. Rev. Immunol. 2013, 13, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Prager, I.; Watzl, C. Mechanisms of Natural Killer Cell-Mediated Cellular Cytotoxicity. J. Leukoc. Biol. 2019, 105, 1319–1329. [Google Scholar] [CrossRef] [PubMed]
- Smyth, M.J.; Cretney, E.; Kelly, J.M.; Westwood, J.A.; Street, S.E.A.; Yagita, H.; Takeda, K.; van Dommelen, S.L.H.; Degli-Esposti, M.A.; Hayakawa, Y. Activation of NK Cell Cytotoxicity. Mol. Immunol. 2005, 42, 501–510. [Google Scholar] [CrossRef]
- Vivier, E.; Nunès, J.A.; Vély, F. Natural Killer Cell Signaling Pathways. Science 2004, 306, 1517–1519. [Google Scholar] [CrossRef]
- Liu, N.; Long, Y.; Liu, B.; Yang, D.; Li, C.; Chen, T.; Wang, X.; Liu, C.; Zhu, H. ISG12a Mediates Cell Response to Newcastle Disease Viral Infection. Virology 2014, 462–463, 283–294. [Google Scholar] [CrossRef]
- Koks, C.A.; Garg, A.D.; Ehrhardt, M.; Riva, M.; Vandenberk, L.; Boon, L.; De Vleeschouwer, S.; Agostinis, P.; Graf, N.; Van Gool, S.W. Newcastle Disease Virotherapy Induces Long-Term Survival and Tumor-Specific Immune Memory in Orthotopic Glioma through the Induction of Immunogenic Cell Death. Int. J. Cancer 2015, 136, E313–E325. [Google Scholar] [CrossRef] [PubMed]
- Moralès, O.; Richard, A.; Martin, N.; Mrizak, D.; Sénéchal, M.; Miroux, C.; Pancré, V.; Rommelaere, J.; Caillet-Fauquet, P.; de Launoit, Y.; et al. Activation of a Helper and Not Regulatory Human CD4+ T Cell Response by Oncolytic H-1 Parvovirus. PLoS ONE 2012, 7, e32197. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.T.; Bell, J.C. Oncolytic Virus Combination Therapy: Killing One Bird with Two Stones. Mol. Ther. 2018, 26, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- Twumasi-Boateng, K.; Pettigrew, J.L.; Kwok, Y.Y.E.; Bell, J.C.; Nelson, B.H. Oncolytic Viruses as Engineering Platforms for Combination Immunotherapy. Nat. Rev. Cancer 2018, 18, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-J.; Kim, J.-H.; Lee, Y.-S.; Kim, J.; Suh, B.-S.; Kim, H.; Cho, S.; Sohn, J.-H.; Kim, G.E.; Yun, C.-O. Concurrent Delivery of GM-CSF and B7-1 Using an Oncolytic Adenovirus Elicits Potent Antitumor Effect. Gene Ther. 2006, 13, 1010–1020. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-S.; Kim, J.-H.; Choi, K.-J.; Choi, I.-K.; Kim, H.; Cho, S.; Cho, B.C.; Yun, C.-O. Enhanced Antitumor Effect of Oncolytic Adenovirus Expressing Interleukin-12 and B7-1 in an Immunocompetent Murine Model. Clin. Cancer Res. 2006, 12, 5859–5868. [Google Scholar] [CrossRef] [PubMed]
- Lapteva, N.; Aldrich, M.; Weksberg, D.; Rollins, L.; Goltsova, T.; Chen, S.-Y.; Huang, X.F. Targeting the Intratumoral Dendritic Cells by the Oncolytic Adenoviral Vaccine Expressing RANTES Elicits Potent Antitumor Immunity. J. Immunother. 2009, 32, 145–156. [Google Scholar] [CrossRef]
- Fernandes, M.S.; Gomes, E.M.; Butcher, L.D.; Hernandez-Alcoceba, R.; Chang, D.; Kansopon, J.; Newman, J.; Stone, M.J.; Tong, A.W. Growth Inhibition of Human Multiple Myeloma Cells by an Oncolytic Adenovirus Carrying the CD40 Ligand Transgene. Clin. Cancer Res. 2009, 15, 4847–4856. [Google Scholar] [CrossRef]
- Cao, X.; Wei, R.; Liu, X.; Zeng, Y.; Huang, H.; Ding, M.; Zhang, K.; Liu, X.-Y. Cancer Targeting Gene-Viro-Therapy Specific for Liver Cancer by α-Fetoprotein-Controlled Oncolytic Adenovirus Expression of SOCS3 and IL-24. Acta Biochim. Biophys. Sin. 2011, 43, 813–821. [Google Scholar] [CrossRef]
- Choi, I.-K.; Lee, J.-S.; Zhang, S.-N.; Park, J.; Sonn, C.H.; Lee, K.-M.; Yun, C.-O. Oncolytic Adenovirus Co-Expressing IL-12 and IL-18 Improves Tumor-Specific Immunity via Differentiation of T Cells Expressing IL-12Rβ2 or IL-18Rα. Gene Ther. 2011, 18, 898–909. [Google Scholar] [CrossRef]
- Zager, J.S.; Delman, K.A.; Malhotra, S.; Ebright, M.I.; Bennett, J.J.; Kates, T.; Halterman, M.; Federoff, H.; Fong, Y. Combination Vascular Delivery of Herpes Simplex Oncolytic Viruses and Amplicon Mediated Cytokine Gene Transfer Is Effective Therapy for Experimental Liver Cancer. Mol. Med. 2001, 7, 561–568. [Google Scholar] [CrossRef]
- Wong, R.J.; Chan, M.-K.; Yu, Z.; Kim, T.H.; Bhargava, A.; Stiles, B.M.; Horsburgh, B.C.; Shah, J.P.; Ghossein, R.A.; Singh, B.; et al. Effective Intravenous Therapy of Murine Pulmonary Metastases with an Oncolytic Herpes Virus Expressing Interleukin 12. Clin. Cancer Res. 2004, 10, 251–259. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Malhotra, S.; Kim, T.; Zager, J.; Bennett, J.; Ebright, M.; D’Angelica, M.; Fong, Y. Use of an Oncolytic Virus Secreting GM-CSF as Combined Oncolytic and Immunotherapy for Treatment of Colorectal and Hepatic Adenocarcinomas. Surgery 2007, 141, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Wakimoto, H.; Tyminski, E.; Chiocca, E.A.; Saeki, Y. Development of a Rapid Method to Generate Multiple Oncolytic HSV Vectors and Their In Vivo Evaluation Using Syngeneic Mouse Tumor Models. Gene Ther. 2006, 13, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lei, G.; Deng, Z.; Sun, F.; Tian, Y.; Cheng, J.; Yu, H.; Li, C.; Bai, C.; Zhang, S.; et al. An Engineered Influenza a Virus Expressing the Co-Stimulator OX40L as an Oncolytic Agent Against Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2024, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Blechacz, B.; Splinter, P.L.; Greiner, S.; Myers, R.; Peng, K.-W.; Federspiel, M.J.; Russell, S.J.; LaRusso, N.F. Engineered Measles Virus as a Novel Oncolytic Viral Therapy System for Hepatocellular Carcinoma. Hepatology 2006, 44, 1465–1477. [Google Scholar] [CrossRef] [PubMed]
- Paraskevakou, G.; Allen, C.; Nakamura, T.; Zollman, P.; James, C.D.; Peng, K.W.; Schroeder, M.; Russell, S.J.; Galanis, E. Epidermal Growth Factor Receptor (EGFR)-Retargeted Measles Virus Strains Effectively Target EGFR- or EGFRvIII Expressing Gliomas. Mol. Ther. 2007, 15, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Allen, C.; Paraskevakou, G.; Iankov, I.; Giannini, C.; Schroeder, M.; Sarkaria, J.; Schroeder, M.; Puri, R.K.; Russell, S.J.; Galanis, E. Interleukin-13 Displaying Retargeted Oncolytic Measles Virus Strains Have Significant Activity against Gliomas with Improved Specificity. Mol. Ther. 2008, 16, 1556–1564. [Google Scholar] [CrossRef]
- Bai, F.; Niu, Z.; Tian, H.; Li, S.; Lv, Z.; Zhang, T.; Ren, G.; Li, D. Genetically Engineered Newcastle Disease Virus Expressing Interleukin 2 Is a Potential Drug Candidate for Cancer Immunotherapy. Immunol. Lett. 2014, 159, 36–46. [Google Scholar] [CrossRef]
- Niu, Z.; Bai, F.; Sun, T.; Tian, H.; Yu, D.; Yin, J.; Li, S.; Li, T.; Cao, H.; Yu, Q.; et al. Recombinant Newcastle Disease Virus Expressing IL15 Demonstrates Promising Antitumor Efficiency in Melanoma Model. Technol. Cancer Res. Treat. 2015, 14, 607–615. [Google Scholar] [CrossRef]
- Takehara, Y.; Satoh, T.; Nishizawa, A.; Saeki, K.; Nakamura, M.; Masuzawa, M.; Kaneda, Y.; Katayama, I.; Yokozeki, H. Anti-Tumor Effects of Inactivated Sendai Virus Particles with an IL-2 Gene on Angiosarcoma. Clin. Immunol. 2013, 149, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-C.S.; Lynn, R.C.; Cheng, G.; Alexander, E.; Kapoor, V.; Moon, E.K.; Sun, J.; Fridlender, Z.G.; Isaacs, S.N.; Thorne, S.H.; et al. Treating Tumors with a Vaccinia Virus Expressing IFNβ Illustrates the Complex Relationships between Oncolytic Ability and Immunogenicity. Mol. Ther. 2012, 20, 736–748. [Google Scholar] [CrossRef] [PubMed]
- Fend, L.; Yamazaki, T.; Remy, C.; Fahrner, C.; Gantzer, M.; Nourtier, V.; Préville, X.; Quéméneur, E.; Kepp, O.; Adam, J.; et al. Immune Checkpoint Blockade, Immunogenic Chemotherapy or IFN-α Blockade Boost the Local and Abscopal Effects of Oncolytic Virotherapy. Cancer Res. 2017, 77, 4146–4157. [Google Scholar] [CrossRef] [PubMed]
- John, L.B.; Howland, L.J.; Flynn, J.K.; West, A.C.; Devaud, C.; Duong, C.P.; Stewart, T.J.; Westwood, J.A.; Guo, Z.S.; Bartlett, D.L.; et al. Oncolytic Virus and Anti-4-1BB Combination Therapy Elicits Strong Antitumor Immunity against Established Cancer. Cancer Res. 2012, 72, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; O’Malley, M.; Sampath, P.; Kalinski, P.; Bartlett, D.L.; Thorne, S.H. Expression of CCL19 from Oncolytic Vaccinia Enhances Immunotherapeutic Potential While Maintaining Oncolytic Activity. Neoplasia 2012, 14, 1115–1121. [Google Scholar] [CrossRef]
- Chard, L.S.; Maniati, E.; Wang, P.; Zhang, Z.; Gao, D.; Wang, J.; Cao, F.; Ahmed, J.; El Khouri, M.; Hughes, J.; et al. A Vaccinia Virus Armed with Interleukin-10 Is a Promising Therapeutic Agent for Treatment of Murine Pancreatic Cancer. Clin. Cancer Res. 2015, 21, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Kowalsky, S.J.; Liu, Z.; Feist, M.; Berkey, S.E.; Ma, C.; Ravindranathan, R.; Dai, E.; Roy, E.J.; Guo, Z.S.; Bartlett, D.L. Superagonist IL-15-Armed Oncolytic Virus Elicits Potent Antitumor Immunity and Therapy That Are Enhanced with PD-1 Blockade. Mol. Ther. 2018, 26, 2476–2486. [Google Scholar] [CrossRef] [PubMed]
- Riederer, S.; Fux, R.; Lehmann, M.H.; Volz, A.; Sutter, G.; Rojas, J.J. Activation of Interferon Regulatory Factor 3 by Replication-Competent Vaccinia Viruses Improves Antitumor Efficacy Mediated by T Cell Responses. Mol. Ther. Oncolytics 2021, 22, 399–409. [Google Scholar] [CrossRef] [PubMed]
- DePeaux, K.; Rivadeneira, D.B.; Lontos, K.; Dean, V.G.; Gunn, W.G.; Watson, M.J.; Yao, T.; Wilfahrt, D.; Hinck, C.; Wieteska, L.; et al. An Oncolytic Virus-Delivered TGFβ Inhibitor Overcomes the Immunosuppressive Tumor Microenvironment. J. Exp. Med. 2023, 220, e20230053. [Google Scholar] [CrossRef]
- Li, F.; Sheng, Y.; Hou, W.; Sampath, P.; Byrd, D.; Thorne, S.; Zhang, Y. CCL5-Armed Oncolytic Virus Augments CCR5-Engineered NK Cell Infiltration and Antitumor Efficiency. J. Immunother. Cancer 2020, 8, e000131. [Google Scholar] [CrossRef]
- Patel, M.R.; Jacobson, B.A.; Ji, Y.; Drees, J.; Tang, S.; Xiong, K.; Wang, H.; Prigge, J.E.; Dash, A.S.; Kratzke, A.K.; et al. Vesicular Stomatitis Virus Expressing Interferon-β Is Oncolytic and Promotes Antitumor Immune Responses in a Syngeneic Murine Model of Non-Small Cell Lung Cancer. Oncotarget 2015, 6, 33165–33177. [Google Scholar] [CrossRef] [PubMed]
- Galivo, F.; Diaz, R.M.; Thanarajasingam, U.; Jevremovic, D.; Wongthida, P.; Thompson, J.; Kottke, T.; Barber, G.N.; Melcher, A.; Vile, R.G. Interference of CD40L-Mediated Tumor Immunotherapy by Oncolytic Vesicular Stomatitis Virus. Hum. Gene Ther. 2010, 21, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Stoff-Khalili, M.A.; Stoff, A.; Rivera, A.A.; Banerjee, N.S.; Everts, M.; Young, S.; Siegal, G.P.; Richter, D.F.; Wang, M.; Dall, P.; et al. Preclinical Evaluation of Transcriptional Targeting Strategies for Carcinoma of the Breast in a Tissue Slice Model System. Breast Cancer Res. 2005, 7, R1141–R1152. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Oh, J.-E.; Hong, J.; Chung, Y.; Lee, Y.; Park, K.D.; Kim, S.; Yun, C.-O. Optimized Biodegradable Polymeric Reservoir-Mediated Local and Sustained Co-Delivery of Dendritic Cells and Oncolytic Adenovirus Co-Expressing IL-12 and GM-CSF for Cancer Immunotherapy. J. Control. Release 2017, 259, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-T.; Wu, M.-H.; Chen, M.-J.; Lin, S.-P.; Yen, Y.-T.; Hung, S.-C. Combination of Mesenchymal Stem Cell-Delivered Oncolytic Virus with Prodrug Activation Increases Efficacy and Safety of Colorectal Cancer Therapy. Biomedicines 2021, 9, 548. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, S.; Coakley, B.A.; Briley-Saebo, K.; Ma, G.; Chen, H.; Meseck, M.; Ward, S.; Divino, C.; Woo, S.; Chen, S.-H.; et al. Myeloid-Derived Suppressor Cells as a Vehicle for Tumor-Specific Oncolytic Viral Therapy. Cancer Res. 2013, 73, 5003–5015. [Google Scholar] [CrossRef]
- Cervera-Carrascon, V.; Quixabeira, D.C.A.; Santos, J.M.; Havunen, R.; Milenova, I.; Verhoeff, J.; Heiniö, C.; Zafar, S.; Garcia-Vallejo, J.J.; van Beusechem, V.W.; et al. Adenovirus Armed With TNFa and IL2 Added to aPD-1 Regimen Mediates Antitumor Efficacy in Tumors Refractory to aPD-1. Front. Immunol. 2021, 12, 706517. [Google Scholar] [CrossRef]
- Esaki, S.; Goshima, F.; Kimura, H.; Murakami, S.; Nishiyama, Y. Enhanced Antitumoral Activity of Oncolytic Herpes Simplex Virus with Gemcitabine Using Colorectal Tumor Models. Int. J. Cancer 2013, 132, 1592–1601. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Ravindranathan, R.; Kalinski, P.; Guo, Z.S.; Bartlett, D.L. Rational Combination of Oncolytic Vaccinia Virus and PD-L1 Blockade Works Synergistically to Enhance Therapeutic Efficacy. Nat. Commun. 2017, 8, 14754. [Google Scholar] [CrossRef] [PubMed]
- Mistarz, A.; Komorowski, M.P.; Graczyk, M.A.; Gil, M.; Jiang, A.; Opyrchal, M.; Rokita, H.; Odunsi, K.O.; Kozbor, D. Recruitment of Intratumoral CD103+ Dendritic Cells by a CXCR4 Antagonist-Armed Virotherapy Enhances Antitumor Immunity. Mol. Ther. Oncolytics 2019, 14, 233–245. [Google Scholar] [CrossRef]
- Ishihara, M.; Seo, N.; Mitsui, J.; Muraoka, D.; Tanaka, M.; Mineno, J.; Ikeda, H.; Shiku, H. Systemic CD8+ T Cell-Mediated Tumoricidal Effects by Intratumoral Treatment of Oncolytic Herpes Simplex Virus with the Agonistic Monoclonal Antibody for Murine Glucocorticoid-Induced Tumor Necrosis Factor Receptor. PLoS ONE 2014, 9, e104669. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Meisen, W.H.; Wohleb, E.S.; Jaime-Ramirez, A.C.; Bolyard, C.; Yoo, J.Y.; Russell, L.; Hardcastle, J.; Dubin, S.; Muili, K.; Yu, J.; et al. The Impact of Macrophage- and Microglia-Secreted TNFα on Oncolytic HSV-1 Therapy in the Glioblastoma Tumor Microenvironment. Clin. Cancer Res. 2015, 21, 3274–3285. [Google Scholar] [CrossRef] [PubMed]
- Svensson-Arvelund, J.; Cuadrado-Castano, S.; Pantsulaia, G.; Kim, K.; Aleynick, M.; Hammerich, L.; Upadhyay, R.; Yellin, M.; Marsh, H.; Oreper, D.; et al. Expanding Cross-Presenting Dendritic Cells Enhances Oncolytic Virotherapy and Is Critical for Long-Term Anti-Tumor Immunity. Nat. Commun. 2022, 13, 7149. [Google Scholar] [CrossRef] [PubMed]
- Miri, S.M.; Ebrahimzadeh, M.S.; Abdolalipour, E.; Yazdi, M.; Hosseini Ravandi, H.; Ghaemi, A. Synergy between Hemagglutinin 2 (HA2) Subunit of Influenza Fusogenic Membrane Glycoprotein and Oncolytic Newcastle Disease Virus Suppressed Tumor Growth and Further Enhanced by Immune Checkpoint PD-1 Blockade. Cancer Cell Int. 2020, 20, 380. [Google Scholar] [CrossRef]
- McAusland, T.M.; van Vloten, J.P.; Santry, L.A.; Guilleman, M.M.; Rghei, A.D.; Ferreira, E.M.; Ingrao, J.C.; Arulanandam, R.; Major, P.P.; Susta, L.; et al. Combining Vanadyl Sulfate with Newcastle Disease Virus Potentiates Rapid Innate Immune-Mediated Regression with Curative Potential in Murine Cancer Models. Mol. Ther. Oncolytics 2021, 20, 306–324. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatt, D.K.; Daemen, T. Molecular Circuits of Immune Sensing and Response to Oncolytic Virotherapy. Int. J. Mol. Sci. 2024, 25, 4691. https://doi.org/10.3390/ijms25094691
Bhatt DK, Daemen T. Molecular Circuits of Immune Sensing and Response to Oncolytic Virotherapy. International Journal of Molecular Sciences. 2024; 25(9):4691. https://doi.org/10.3390/ijms25094691
Chicago/Turabian StyleBhatt, Darshak K., and Toos Daemen. 2024. "Molecular Circuits of Immune Sensing and Response to Oncolytic Virotherapy" International Journal of Molecular Sciences 25, no. 9: 4691. https://doi.org/10.3390/ijms25094691
APA StyleBhatt, D. K., & Daemen, T. (2024). Molecular Circuits of Immune Sensing and Response to Oncolytic Virotherapy. International Journal of Molecular Sciences, 25(9), 4691. https://doi.org/10.3390/ijms25094691




