Decoding Liver Fibrosis: How Omics Technologies and Innovative Modeling Can Guide Precision Medicine
Abstract
:1. Introduction: Liver Fibrosis
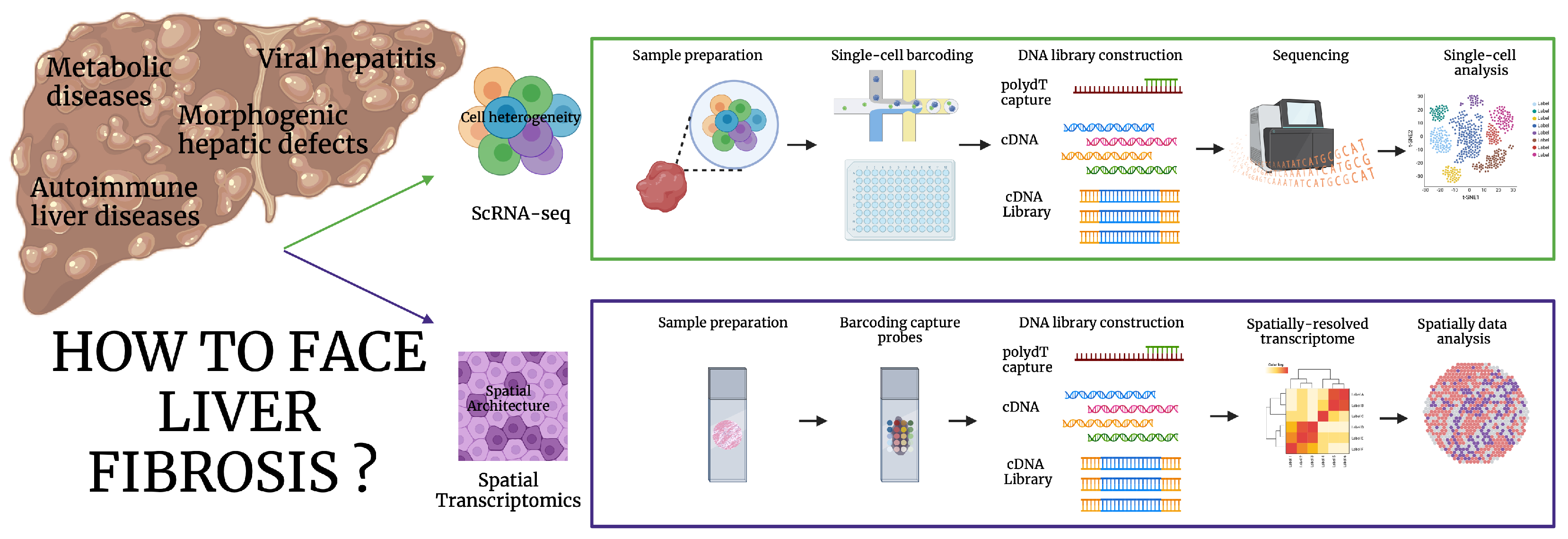
2. Unraveling the Molecular Diversity of Liver Fibrosis: Single-Cell and Spatial Transcriptomics
2.1. ScRNA-Seq
2.2. Spatial Transcriptomics
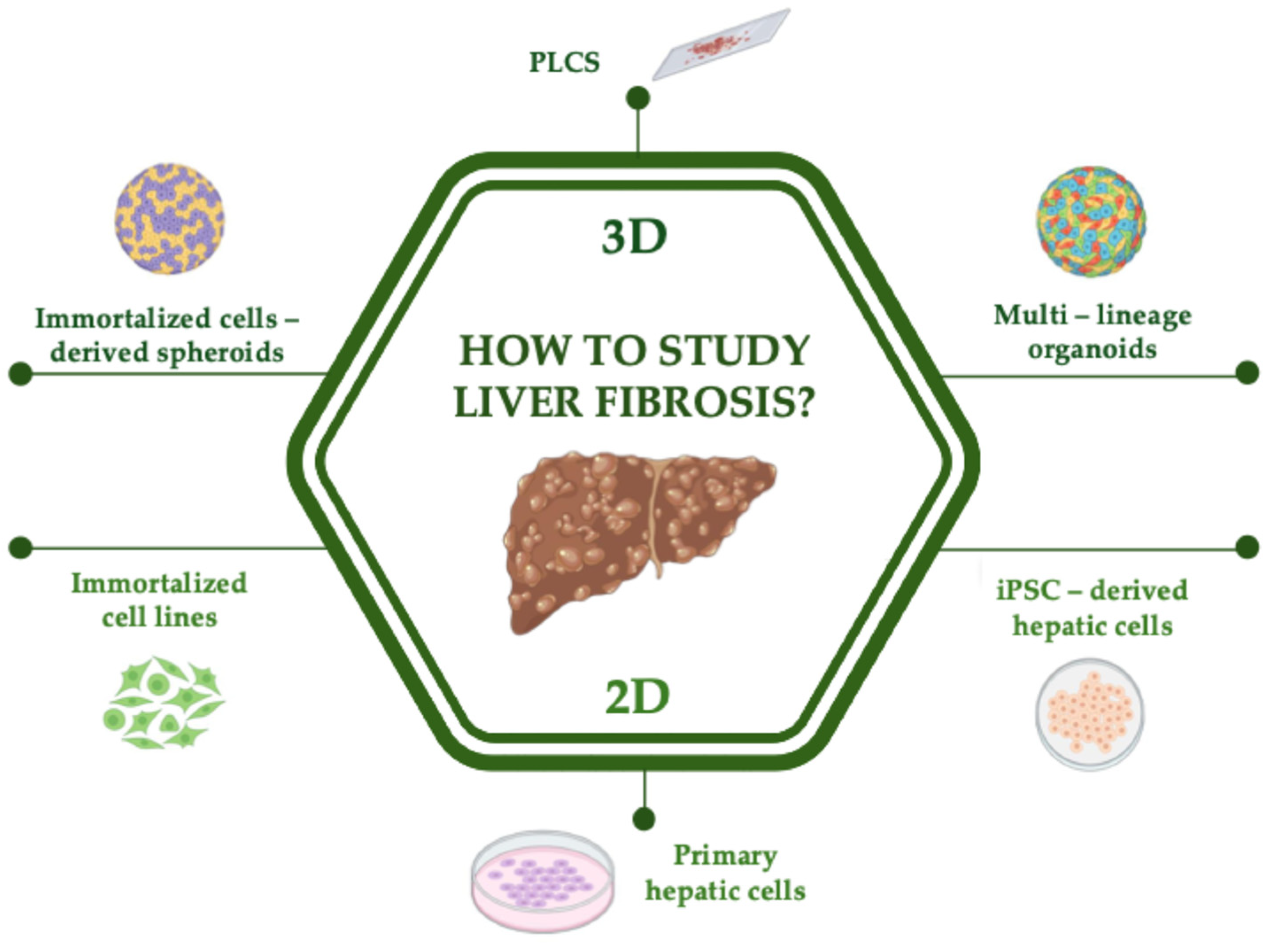
3. Unraveling the Molecular Mechanisms Involved in Liver Fibrosis: Gold Standard and Emerging 2D In Vitro Models
3.1. Primary and Immortalized Cell Lines
3.2. iPSC-Derived Hepatic Cells
4. Unraveling the Molecular Mechanisms Involved in Liver Fibrosis: Gold Standard and Emerging 3D In Vitro Models
4.1. Immortalized Cell Lines Derived 3D-Models
4.2. iPSC-Derived Multilineage Organoids
5. Unraveling the Structural and Architectural Features Involved in Liver Fibrosis: Modeling Liver Fibrosis with Ex Vivo Models
Precision-Cut Liver Slices
6. Future Perspectives: Organoids and Omics as a Combined Tool for Precision and Translational Medicine
7. Conclusions
Funding
Conflicts of Interest
Abbreviations
| CLD | Chronic liver disease |
| iPSC | Induced pluripotent stem cells |
| ECM | Extra-cellular matrix |
| KCs | Kupffer cells |
| HSCs | Hepatic Stellate cells |
| SAGE | Serial analysis of gene expression |
| RNA-seq | RNA-sequencing |
| scRNA-seq | Single-cell RNA sequencing |
| snRNA-seq | Single-nucleus RNA sequencing |
| ST | Spatial Transcriptomics |
| MASLD | Metabolic dysfunction-associated steatotic liver disease |
| CaHSCs | Portal vein-associated HSCs |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| ICM | Inner cell mass |
| ESCs | Embryonic stem cells |
| hiPSC | Human induced pluripotent stem cells |
| HO | Hepatobiliary organoid |
| FFAs | Free fatty acids |
| HLOs | Human liver organoids |
| AFM | Atomic force microscopy |
| ARPKD | Autosomal recessive polycystic kidney disease |
| PCLS | Precision-cut liver slice |
| MAIT | mucosal-associated invariant T |
| NRG | Naringenin |
| AA | Asiatic acid |
| ICA | Icariin |
| mPCLS | Mouse precision-cut liver slice |
| chPCLS | Cirrohotic human precision-cut liver slice |
| DC | Dyskeratosis congenita |
References
- Younossi, Z.M.; Wong, G.; Anstee, Q.M.; Henry, L. The Global Burden of Liver Disease. Clin. Gastroenterol. Hepatol. 2023, 21, 1978–1991. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, Y.; Sun, B. The Molecular Mechanisms of Liver Fibrosis and Its Potential Therapy in Application. Int. J. Mol. Sci. 2022, 23, 12572. [Google Scholar] [CrossRef]
- Kisseleva, T.; Brenner, D. Molecular and Cellular Mechanisms of Liver Fibrosis and Its Regression. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 151–166. [Google Scholar] [CrossRef] [PubMed]
- Roehlen, N.; Crouchet, E.; Baumert, T.F. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 2020, 9, 875. [Google Scholar] [CrossRef] [PubMed]
- Tacke, F.; Zimmermann, H.W. Macrophage Heterogeneity in Liver Injury and Fibrosis. J. Hepatol. 2014, 60, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.L.; Nakib, D.; Perciani, C.T.; MacParland, S.A. The Immune Niche of the Liver. Clin. Sci. 2021, 135, 2445–2466. [Google Scholar] [CrossRef]
- Chen, G.; Xu, W.; Long, Z.; Chong, Y.; Lin, B.; Jie, Y. Single-Cell Technologies Provide Novel Insights into Liver Physiology and Pathology. J. Clin. Transl. Hepatol. 2024, 12, 79–90. [Google Scholar] [CrossRef]
- Ramachandran, P.; Matchett, K.P.; Dobie, R.; Wilson-Kanamori, J.R.; Henderson, N.C. Single-Cell Technologies in Hepatology: New Insights into Liver Biology and Disease Pathogenesis. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 457–472. [Google Scholar] [CrossRef]
- Chu, A.L.; Schilling, J.D.; King, K.R.; Feldstein, A.E. The Power of Single-Cell Analysis for the Study of Liver Pathobiology. Hepatology 2021, 73, 437–448. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, B.; Zhao, G.; Lee, Y.; Buzdin, A.; Mu, X.; Zhao, J.; Chen, H.; Li, X. Spatial Transcriptomics: Technologies, Applications and Experimental Considerations. Genomics 2023, 115, 110671. [Google Scholar] [CrossRef]
- Porat-Shliom, N. Compartmentalization, Cooperation, and Communication: The 3Cs of Hepatocyte Zonation. Curr. Opin. Cell Biol. 2024, 86, 102292. [Google Scholar] [CrossRef] [PubMed]
- Rosenberger, F.A.; Thielert, M.; Strauss, M.T.; Schweizer, L.; Ammar, C.; Mädler, S.C.; Metousis, A.; Skowronek, P.; Wahle, M.; Madden, K.; et al. Spatial Single-Cell Mass Spectrometry Defines Zonation of the Hepatocyte Proteome. Nat. Methods 2023, 20, 1530–1536. [Google Scholar] [CrossRef] [PubMed]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Toth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-Cell Spatial Reconstruction Reveals Global Division of Labour in the Mammalian Liver. Nature 2017, 542, 352–356. [Google Scholar] [CrossRef]
- Dobie, R.; Wilson-Kanamori, J.R.; Henderson, B.E.P.; Smith, J.R.; Matchett, K.P.; Portman, J.R.; Wallenborg, K.; Picelli, S.; Zagorska, A.; Pendem, S.V.; et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 2019, 29, 1832–1847.e8. [Google Scholar] [CrossRef]
- Andrews, T.S.; Atif, J.; Liu, J.C.; Perciani, C.T.; Ma, X.-Z.; Thoeni, C.; Slyper, M.; Eraslan, G.; Segerstolpe, A.; Manuel, J.; et al. Single-Cell, Single-Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol. Commun. 2022, 6, 821–840. [Google Scholar] [CrossRef]
- Chung, B.K.; Øgaard, J.; Reims, H.M.; Karlsen, T.H.; Melum, E. Spatial Transcriptomics Identifies Enriched Gene Expression and Cell Types in Human Liver Fibrosis. Hepatol. Commun. 2022, 6, 2538–2550. [Google Scholar] [CrossRef]
- Li, J.-Z.; Yang, L.; Xiao, M.-X.; Li, N.; Huang, X.; Ye, L.-H.; Zhang, H.-C.; Liu, Z.-Q.; Li, J.-Q.; Liu, Y.-Y.; et al. Spatial and Single-Cell Transcriptomics Reveals the Regional Division of the Spatial Structure of MASH Fibrosis. Liver Int. 2024, 45, e16125. [Google Scholar] [CrossRef]
- Bao, Y.-L.; Wang, L.; Pan, H.-T.; Zhang, T.-R.; Chen, Y.-H.; Xu, S.-J.; Mao, X.-L.; Li, S.-W. Animal and Organoid Models of Liver Fibrosis. Front. Physiol. 2021, 12, 666138. [Google Scholar] [CrossRef] [PubMed]
- Shang, L.; Hosseini, M.; Liu, X.; Kisseleva, T.; Brenner, D.A. Human Hepatic Stellate Cell Isolation and Characterization. J. Gastroenterol. 2018, 53, 6–17. [Google Scholar] [CrossRef]
- Rutt, L.N.; Orlicky, D.J.; McCullough, R.L. Investigating the Role of Wnt3a and Wnt5a as Critical Factors of Hepatic Stellate Cell Activation in Acute Toxicant-Induced Liver Injury. Cell Biol. Toxicol. 2024, 41, 5. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Seki, E. In Vivo and In Vitro Models to Study Liver Fibrosis: Mechanisms and Limitations. Cell Mol. Gastroenterol. Hepatol. 2023, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Anfuso, B.; Tiribelli, C.; Adorini, L.; Rosso, N. Obeticholic Acid and INT-767 Modulate Collagen Deposition in a NASH in Vitro Model. Sci. Rep. 2020, 10, 1699. [Google Scholar] [CrossRef]
- da Silva Lima, N.; Cabaleiro, A.; Novoa, E.; Riobello, C.; Knerr, P.J.; He, Y.; Esquinas-Román, E.M.; González-García, I.; Prevot, V.; Schwaninger, M.; et al. GLP-1 and GIP Agonism Has No Direct Actions in Human Hepatocytes or Hepatic Stellate Cells. Cell Mol. Life Sci. 2024, 81, 468. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hui, A.Y.; Albanis, E.; Arthur, M.J.; O’Byrne, S.M.; Blaner, W.S.; Mukherjee, P.; Friedman, S.L.; Eng, F.J. Human Hepatic Stellate Cell Lines, LX-1 and LX-2: New Tools for Analysis of Hepatic Fibrosis. Gut 2005, 54, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Prior, N.; Inacio, P.; Huch, M. Liver Organoids: From Basic Research to Therapeutic Applications. Gut 2019, 68, 2228–2237. [Google Scholar] [CrossRef]
- Gao, X.; Li, R.; Cahan, P.; Zhao, Y.; Yourick, J.J.; Sprando, R.L. Hepatocyte-like Cells Derived from Human Induced Pluripotent Stem Cells Using Small Molecules: Implications of a Transcriptomic Study. Stem Cell Res. Ther. 2020, 11, 393. [Google Scholar] [CrossRef]
- Park, S.J.; Kim, Y.Y.; Han, J.Y.; Kim, S.W.; Kim, H.; Ku, S.-Y. Advancements in Human Embryonic Stem Cell Research: Clinical Applications and Ethical Issues. Tissue Eng. Regen. Med. 2024, 21, 379–394. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a Dish: Modeling Development and Disease Using Organoid Technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Coll, M.; Perea, L.; Boon, R.; Leite, S.B.; Vallverdú, J.; Mannaerts, I.; Smout, A.; El Taghdouini, A.; Blaya, D.; Rodrigo-Torres, D.; et al. Generation of Hepatic Stellate Cells from Human Pluripotent Stem Cells Enables In Vitro Modeling of Liver Fibrosis. Cell Stem Cell 2018, 23, 101–113.e7. [Google Scholar] [CrossRef]
- Pingitore, P.; Sasidharan, K.; Ekstrand, M.; Prill, S.; Lindén, D.; Romeo, S. Human Multilineage 3D Spheroids as a Model of Liver Steatosis and Fibrosis. Int. J. Mol. Sci. 2019, 20, 1629. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Lee, W.; Liu, X.; Jang, H.; Sakane, S.; Carvalho-Gontijo Weber, R.; Diggle, K.; Kerk, S.A.; Metallo, C.M.; Kisseleva, T.; et al. Protocol to Generate Human Liver Spheroids to Study Liver Fibrosis Induced by Metabolic Stress. STAR Protoc. 2024, 5, 103111. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Thompson, W.L.; Takebe, T. Human Liver Model Systems in a Dish. Dev. Growth Differ. 2021, 63, 47–58. [Google Scholar] [CrossRef]
- Ramli, M.N.B.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e12. [Google Scholar] [CrossRef]
- Ouchi, R.; Togo, S.; Kimura, M.; Shinozawa, T.; Koido, M.; Koike, H.; Thompson, W.; Karns, R.A.; Mayhew, C.N.; McGrath, P.S.; et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 2019, 30, 374–384.e6. [Google Scholar] [CrossRef] [PubMed]
- Al Reza, H.; Santangelo, C.; Al Reza, A.; Iwasawa, K.; Sachiko, S.; Glaser, K.; Bondoc, A.; Merola, J.; Takebe, T. Self-Assembled Generation of Multi-Zonal Liver Organoids from Human Pluripotent Stem Cells. bioRxiv, 2024; preprint. [Google Scholar] [CrossRef]
- Guan, Y.; Xu, D.; Garfin, P.M.; Ehmer, U.; Hurwitz, M.; Enns, G.; Michie, S.; Wu, M.; Zheng, M.; Nishimura, T.; et al. Human Hepatic Organoids for the Analysis of Human Genetic Diseases. JCI Insight 2017, 2, e94954. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Enejder, A.; Wang, M.; Fang, Z.; Cui, L.; Chen, S.-Y.; Wang, J.; Tan, Y.; Wu, M.; Chen, X.; et al. A Human Multi-Lineage Hepatic Organoid Model for Liver Fibrosis. Nat. Commun. 2021, 12, 6138. [Google Scholar] [CrossRef]
- de Graaf, I.A.M.; Olinga, P.; de Jager, M.H.; Merema, M.T.; de Kanter, R.; van de Kerkhof, E.G.; Groothuis, G.M.M. Preparation and Incubation of Precision-Cut Liver and Intestinal Slices for Application in Drug Metabolism and Toxicity Studies. Nat. Protoc. 2010, 5, 1540–1551. [Google Scholar] [CrossRef]
- Palma, E.; Doornebal, E.J.; Chokshi, S. Precision-Cut Liver Slices: A Versatile Tool to Advance Liver Research. Hepatol. Int. 2019, 13, 51–57. [Google Scholar] [CrossRef]
- van de Bovenkamp, M.; Groothuis, G.M.M.; Meijer, D.K.F.; Slooff, M.J.H.; Olinga, P. Human Liver Slices as an in Vitro Model to Study Toxicity-Induced Hepatic Stellate Cell Activation in a Multicellular Milieu. Chem. Biol. Interact. 2006, 162, 62–69. [Google Scholar] [CrossRef] [PubMed]
- van de Bovenkamp, M.; Groothuis, G.M.M.; Meijer, D.K.F.; Olinga, P. Precision-Cut Fibrotic Rat Liver Slices as a New Model to Test the Effects of Anti-Fibrotic Drugs in Vitro. J. Hepatol. 2006, 45, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Mabire, M.; Hegde, P.; Hammoutene, A.; Wan, J.; Caër, C.; Sayegh, R.A.; Cadoux, M.; Allaire, M.; Weiss, E.; Thibault-Sogorb, T.; et al. MAIT Cell Inhibition Promotes Liver Fibrosis Regression via Macrophage Phenotype Reprogramming. Nat. Commun. 2023, 14, 1830. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Leaker, B.; Qiao, G.; Sojoodi, M.; Eissa, I.R.; Epstein, E.T.; Eddy, J.; Dimowo, O.; Lauer, G.M.; Qadan, M.; et al. Precision-Cut Liver Slices as an Ex Vivo Model to Evaluate Antifibrotic Therapies for Liver Fibrosis and Cirrhosis. Hepatol. Commun. 2024, 8, e0558. [Google Scholar] [CrossRef]
- Luo, K.; Geng, Y.; Oosterhuis, D.; de Meijer, V.E.; Olinga, P. Evaluating the Antifibrotic Potential of Naringenin, Asiatic Acid, and Icariin Using Murine and Human Precision-Cut Liver Slices. Physiol. Rep. 2024, 12, e16136. [Google Scholar] [CrossRef]
- Kimura, M.; Takebe, T. Cellotype-Phenotype Associations Using “Organoid Villages”. Trends Endocrinol. Metab. 2024, 35, 462–465. [Google Scholar] [CrossRef]
- Osonoi, S.; Takebe, T. Organoid-Guided Precision Hepatology for Metabolic Liver Disease. J. Hepatol. 2024, 80, 805–821. [Google Scholar] [CrossRef]
- Teriyapirom, I.; Batista-Rocha, A.S.; Koo, B.-K. Genetic Engineering in Organoids. J. Mol. Med. 2021, 99, 555–568. [Google Scholar] [CrossRef]
- Choi, Y.-J.; Kim, M.S.; Rhoades, J.H.; Johnson, N.M.; Berry, C.T.; Root, S.; Chen, Q.; Tian, Y.; Fernandez, R.J.; Cramer, Z.; et al. Patient-Induced Pluripotent Stem Cell–Derived Hepatostellate Organoids Establish a Basis for Liver Pathologies in Telomeropathies. Cell. Mol. Gastroenterol. Hepatol. 2023, 16, 451–472. [Google Scholar] [CrossRef]
- Ramakrishna, G.; Babu, P.E.; Singh, R.; Trehanpati, N. Application of CRISPR-Cas9 Based Gene Editing to Study the Pathogenesis of Colon and Liver Cancer Using Organoids. Hepatol. Int. 2021, 15, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Iguchi, T.; Iwasawa, K.; Dunn, A.; Thompson, W.L.; Yoneyama, Y.; Chaturvedi, P.; Zorn, A.M.; Wintzinger, M.; Quattrocelli, M.; et al. En Masse Organoid Phenotyping Informs Metabolic-Associated Genetic Susceptibility to NASH. Cell 2022, 185, 4216–4232.e16. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Kimura, M.; Li, X.; Sulc, J.; Wang, Q.; Rodríguez-López, S.; Scantlebery, A.M.L.; Strotjohann, K.; Gallart-Ayala, H.; Vijayakumar, A.; et al. ACMSD Inhibition Corrects Fibrosis, Inflammation, and DNA Damage in MASLD/MASH. J. Hepatol. 2025, 82, 174–188. [Google Scholar] [CrossRef]
- Giraudi, P.J.; Laraño, A.A.; Monego, S.D.; Pravisani, R.; Bonazza, D.; Gondolesi, G.; Tiribelli, C.; Baralle, F.; Baccarani, U.; Licastro, D. Genome-Wide DNA Methylation and Transcriptomic Analysis of Liver Tissues Subjected to Early Ischemia/Reperfusion Injury upon Human Liver Transplantation. Ann. Hepatol. 2024, 29, 101506. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Song, Q.; Ren, J.; Zhang, W.; Wang, Y.; Zhou, L.; Wang, D.; Chen, K.; Jiang, L.; Zhang, B.; et al. Simultaneous Analysis of Mutations and Methylations in Circulating Cell-Free DNA for Hepatocellular Carcinoma Detection. Sci. Transl. Med. 2022, 14, eabp8704. [Google Scholar] [CrossRef]
- Angelini, G.; Panunzi, S.; Castagneto-Gissey, L.; Pellicanò, F.; De Gaetano, A.; Pompili, M.; Riccardi, L.; Garcovich, M.; Raffaelli, M.; Ciccoritti, L.; et al. Accurate Liquid Biopsy for the Diagnosis of Non-Alcoholic Steatohepatitis and Liver Fibrosis. Gut 2023, 72, 392–403. [Google Scholar] [CrossRef]
| Cell Model | Potentials | Limitations | Applications | References |
|---|---|---|---|---|
| Primary hepatic stellate cells | Gold standard to study the activation stages of hepatic fibrosis | Single cell type | Study of molecular mechanism of hepatic fibrosis | [19,20] |
| Limited availability | Drug testing | |||
| Can be cultured for few days | ||||
| Cell lines | Single cell type | Derived from transformed or immortalized tumoral cell lines | Study of molecular mechanisms of hepatic fibrosis | [22,23] |
| CRISPR—Cas9 genome editing | Genetic and chromosomal aberration | |||
| Resemble the activated phenotype of myofibroblasts | Single cell type | |||
| Reproduce liver fibrosis mechanisms | ||||
| induced pluripotent stem cells (iPSC)—derived hepatic cells | Maintain cell functional activities and genetic identity | Lack full maturity | Study of molecular mechanism of hepatic fibrosis | [30] |
| Starting material easily available | Challenging technology | |||
| Genetic reprogramming through CRISPR—Cas9 | Drug testing | |||
| Immortalized cell lines—derived 3D models | Assembled, not developed Easy to generate Long-term culture | Lack 3D structure | Study of molecular mechanism of hepatic fibrosis | [31,32] |
| Genetic and genomic aberrations | Drug testing | |||
| induced pluripotent stem cells (iPSC)—derived multilineage organoids | Cell heterogeneity | Lack full maturity | Study of molecular mechanism of hepatic fibrosis | [35,36,38,39] |
| Multi-cell type | Cell line dependency | Drug testing | ||
| Long-term culture Genetic reprogramming (CRISPR-Cas9 technology) | Complexity in generation and maintenance | Metabolic studies | ||
| Precision—cut liver slices | Tissue architecture maintained | Short-term culture | Liver architecture study | [40,42,43] |
| Drug testing | ||||
| Cell heterogeneity maintained | ||||
| Same genetic background of the patient |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Codotto, G.; Blarasin, B.; Tiribelli, C.; Bellarosa, C.; Licastro, D. Decoding Liver Fibrosis: How Omics Technologies and Innovative Modeling Can Guide Precision Medicine. Int. J. Mol. Sci. 2025, 26, 2658. https://doi.org/10.3390/ijms26062658
Codotto G, Blarasin B, Tiribelli C, Bellarosa C, Licastro D. Decoding Liver Fibrosis: How Omics Technologies and Innovative Modeling Can Guide Precision Medicine. International Journal of Molecular Sciences. 2025; 26(6):2658. https://doi.org/10.3390/ijms26062658
Chicago/Turabian StyleCodotto, Gabriele, Benedetta Blarasin, Claudio Tiribelli, Cristina Bellarosa, and Danilo Licastro. 2025. "Decoding Liver Fibrosis: How Omics Technologies and Innovative Modeling Can Guide Precision Medicine" International Journal of Molecular Sciences 26, no. 6: 2658. https://doi.org/10.3390/ijms26062658
APA StyleCodotto, G., Blarasin, B., Tiribelli, C., Bellarosa, C., & Licastro, D. (2025). Decoding Liver Fibrosis: How Omics Technologies and Innovative Modeling Can Guide Precision Medicine. International Journal of Molecular Sciences, 26(6), 2658. https://doi.org/10.3390/ijms26062658







