Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants
Abstract
:1. Introduction
2. Results
2.1. Genomic Identification and Evolutionary Analysis of TaCOBL6A2
2.2. Subcellular Localization of TaCOBL6A2 Reveals Tripartite Targeting of the Plasma Membrane, Cell Wall, and Nucleus
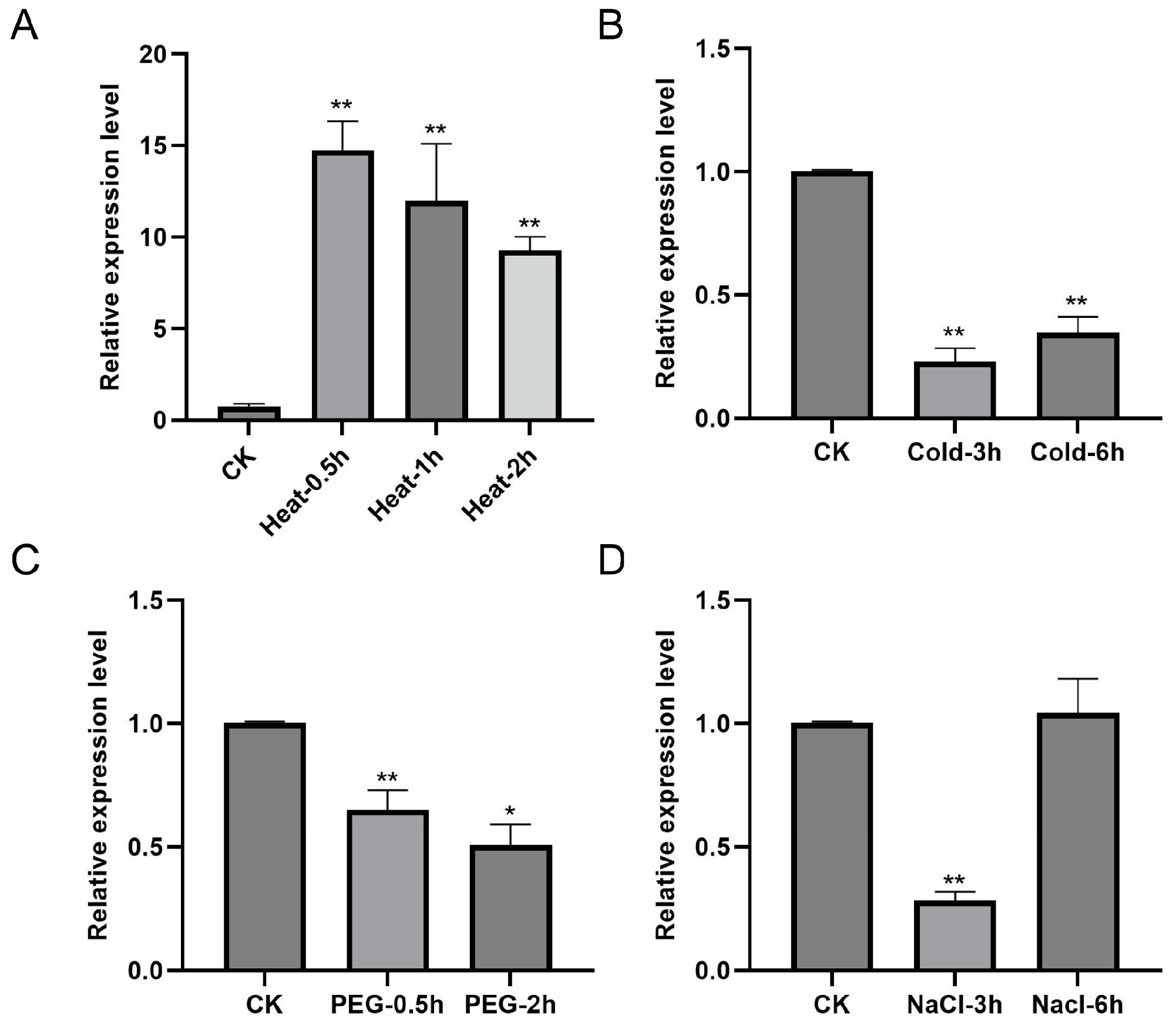
2.3. Expression Profiling and Stress Response Analysis of TaCOBL6A2
2.4. Overexpression of TaCOBL6A2 Enhances Thermotolerance in Arabidopsis
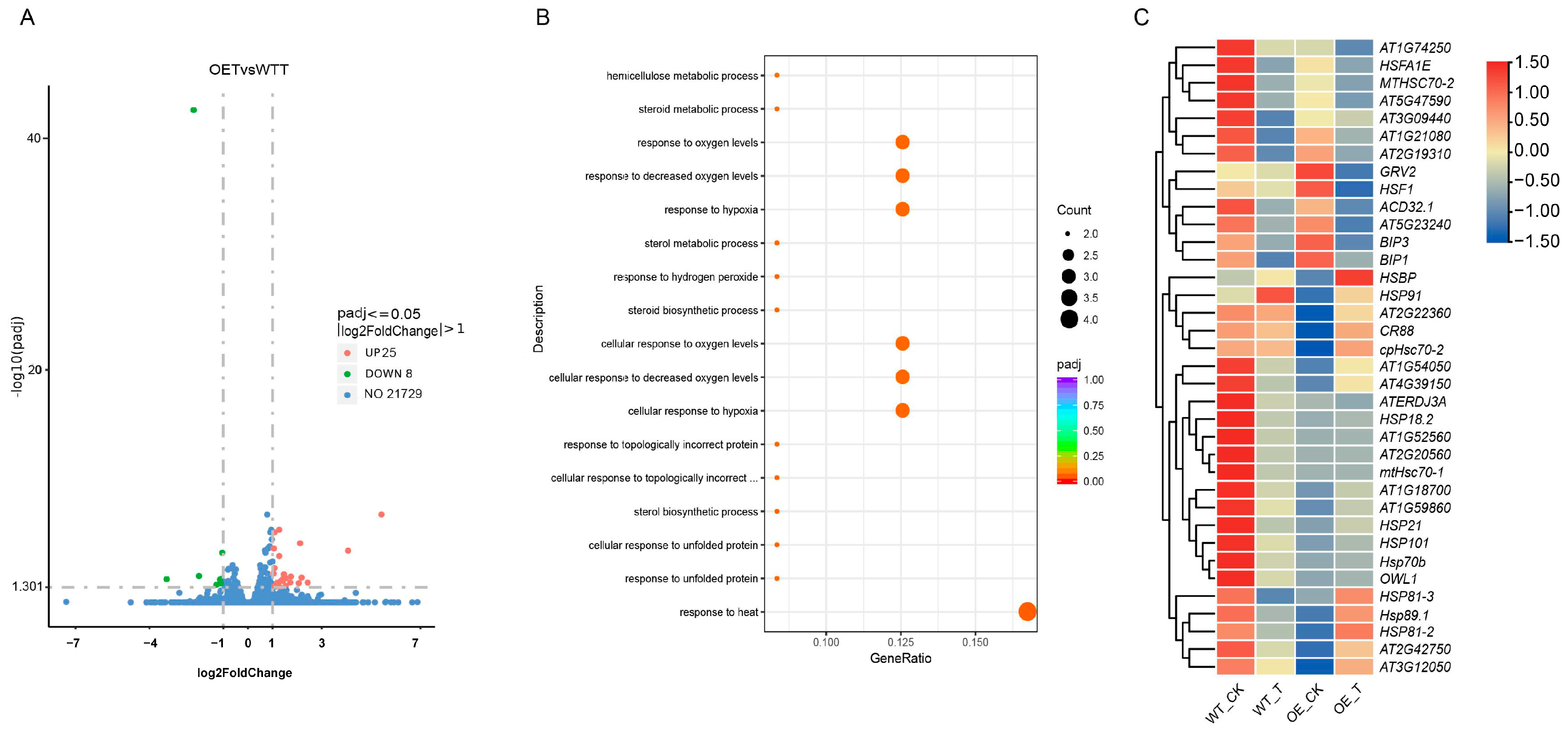
2.5. Comparative Transcriptomic Analysis of TaCOBL6A2 Overexpression Lines
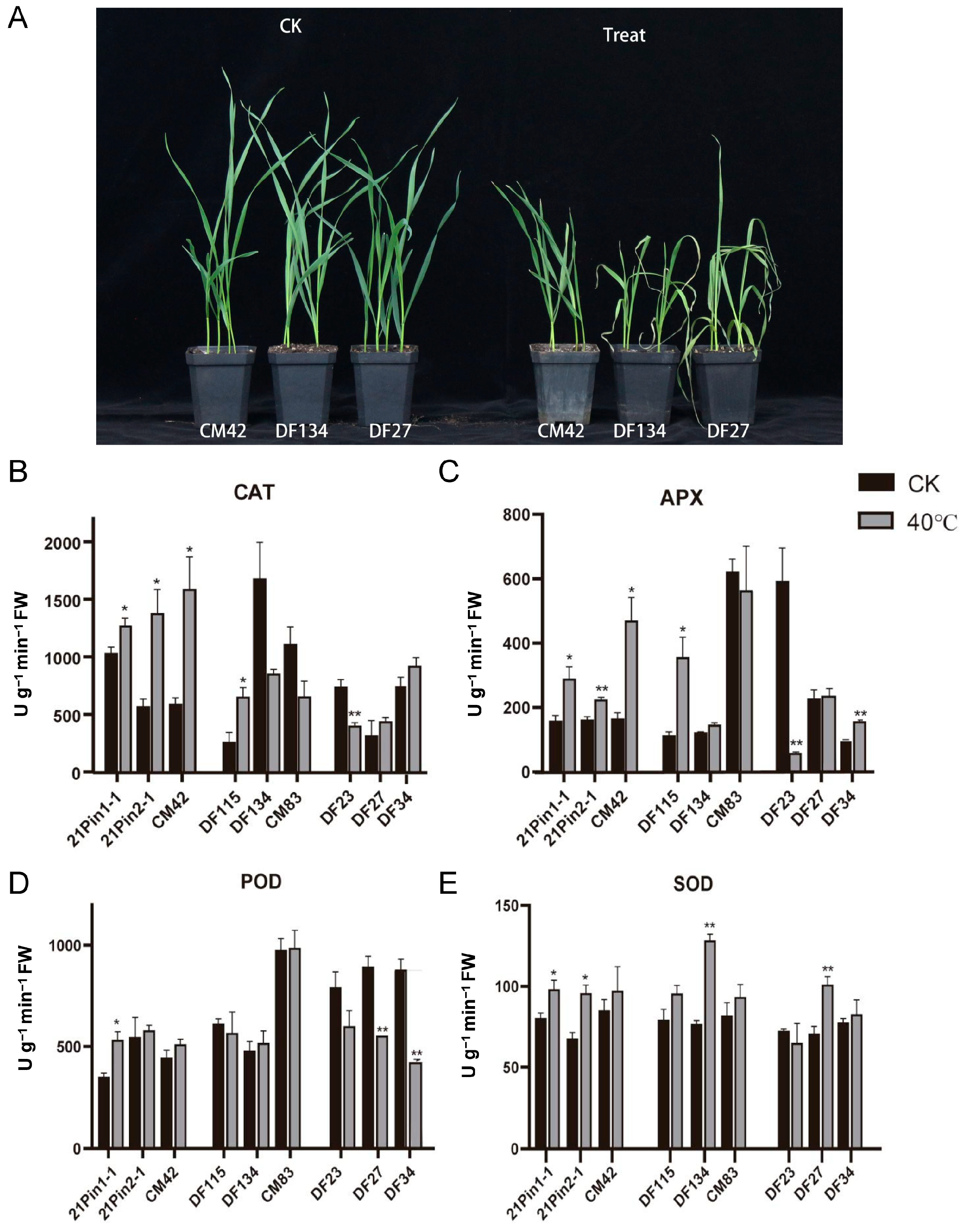
2.6. Natural Variation and Elite Haplotypes of TaCOBL6A2 Enhance Thermotolerance in Wheat
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions and Heat Stress Treatment
4.2. Arabidopsis Thaliana Transformation
4.3. RNA-Seq Analysis
4.4. Protein Feature and Phylogenetic Analyses
4.5. Subcellular Localization
4.6. Haplotype Analysis
4.7. RNA Isolation and Real-Time PCR Analysis
4.8. Determination of Antioxidant Enzyme Activity
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Feldman, M.; Levy, A.A. Evolution of wheat under cultivation. In Wheat Evolution and Domestication; Springer International Publishing: Cham, Germany, 2023; pp. 605–663. [Google Scholar]
- Mishra, D.; Shekhar, S.; Chakraborty, S.; Chakraborty, N. High temperature stress responses and wheat: Impacts and alleviation strategies. Environ. Exp. Bot. 2021, 190, 104589. [Google Scholar] [CrossRef]
- Abasi, F.; Raja, N.I.; Mashwani, Z.-u.-R.; Ehsan, M.; Ali, H.; Shahbaz, M. Heat and wheat: Adaptation strategies with respect to heat shock proteins and antioxidant potential; an era of climate change. Int. J. Biol. Macromol. 2024, 256, 128379. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, K.; Senthilkumar, K.M.; Mamrutha, H.M.; Singh, G.; Singh, G.P. Chapter 8—High-temperature stress in wheat under climate change scenario, effects and mitigation strategies. In Climate Change and Crop Stress; Shanker, A.K., Shanker, C., Anand, A., Maheswari, M., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 209–229. [Google Scholar]
- Mishra, D.; Shekhar, S.; Agrawal, L.; Chakraborty, S.; Chakraborty, N. Cultivar-specific high temperature stress responses in bread wheat (Triticum aestivum L.) associated with physicochemical traits and defense pathways. Food Chem. 2017, 221, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Fonseca De Lima, C.F.; De Smet, I.; Penfield, S. The heat is on: How crop growth, development, and yield respond to high temperature. J. Exp. Bot. 2021, 72, 7359–7373. [Google Scholar] [CrossRef]
- Muthusamy, S.K.; Dalal, M.; Chinnusamy, V.; Bansal, K.C. Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J. Plant Physiol. 2017, 211, 100–113. [Google Scholar] [CrossRef]
- Wang, Y.X.; Yu, T.F.; Wang, C.X.; Wei, J.T.; Zhang, S.X.; Liu, Y.W.; Chen, J.; Zhou, Y.B.; Chen, M.; Ma, Y.Z.; et al. Heat shock protein TaHSP17.4, a TaHOP interactor in wheat, improves plant stress tolerance. Int. J. Biol. Macromol. 2023, 246, 125694. [Google Scholar] [CrossRef]
- Tabusam, J.; Shi, Q.; Feng, D.; Zulfiqar, S.; Shen, S.; Ma, W.; Zhao, J. HSP70 gene family in Brassica rapa: Genome-wide identification, characterization, and expression patterns in response to heat and cold stress. Cells 2022, 11, 2316. [Google Scholar] [CrossRef]
- Guo, X.L.; Yuan, S.N.; Zhang, H.N.; Zhang, Y.Y.; Zhang, Y.J.; Wang, G.Y.; Li, Y.Q.; Li, G.L. Heat-response patterns of the heat shock transcription factor family in advanced development stages of wheat (Triticum aestivum L.) and thermotolerance-regulation by TaHsfA2-10. BMC Plant Biol. 2020, 20, 364. [Google Scholar] [CrossRef]
- Bi, H.; Miao, J.; He, J.; Chen, Q.; Qian, J.; Li, H.; Xu, Y.; Ma, D.; Zhao, Y.; Tian, X.; et al. Characterization of the wheat heat shock factor TaHsfA2e-5D conferring heat and drought tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 2784. [Google Scholar] [CrossRef]
- Wen, J.; Qin, Z.; Sun, L.; Zhang, Y.; Wang, D.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Alternative splicing of TaHSFA6e modulates heat shock protein–mediated translational regulation in response to heat stress in wheat. New Phytol. 2023, 239, 2235–2247. [Google Scholar] [CrossRef]
- Niu, X.; Luo, T.; Zhao, H.; Su, Y.; Ji, W.; Li, H. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liang, Y.; Qin, Q.; Zhao, Y.; Yang, C.; Liu, R.; Gan, Y.; Zhou, H.; Qiu, Z.; Chen, L.; et al. Transcription cofactor CsMBF1c enhances heat tolerance of cucumber and interacts with heat-related proteins CsNFYA1 and CsDREB2. J. Agric. Food Chem. 2024, 72, 15586–15600. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Qin, Z.; Zhao, Y.; Wen, J.; Lan, T.; Zhang, L.; Wang, F.; Qin, D.; Yu, K.; Zhao, A.; et al. Stress granule-associated TaMBF1c confers thermotolerance through regulating specific mRNA translation in wheat (Triticum aestivum). New Phytol. 2022, 233, 1719–1731. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Zang, X.; Li, H.; Liu, Z.; Zhao, A.; Liu, J.; Peng, H.; Yao, Y.; Hu, Z.; Ni, Z.; et al. Unconventional splicing of wheat TabZIP60 confers heat tolerance in transgenic Arabidopsis. Plant Sci. 2018, 274, 252–260. [Google Scholar] [CrossRef]
- Zang, X.; Geng, X.; He, K.; Wang, F.; Tian, X.; Xin, M.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and Arabidopsis. Front. Plant Sci. 2018, 9, 1710. [Google Scholar] [CrossRef]
- Zhao, Y.; Du, H.; Wang, Y.; Wang, H.; Yang, S.; Li, C.; Chen, N.; Yang, H.; Zhang, Y.; Zhu, Y.; et al. The calcium-dependent protein kinase ZmCDPK7 functions in heat-stress tolerance in maize. J. Integr. Plant Biol. 2021, 63, 510–527. [Google Scholar] [CrossRef]
- Wu, H.-C.; Bulgakov, V.P.; Jinn, T.-L. Pectin methylesterases: Cell wall remodeling proteins are required for plant response to heat stress. Front. Plant Sci. 2018, 9, 1612. [Google Scholar] [CrossRef]
- Xu, J.; Tian, J.; Belanger, F.C.; Huang, B. Identification and characterization of an expansin gene AsEXP1 associated with heat tolerance in C3 Agrostis grass species. J. Exp. Bot. 2007, 58, 3789–3796. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Xu, Q.; Xu, X.; Shi, Y.; Xu, J.; Huang, B. Transgenic tobacco plants overexpressing a grass PpEXP1 gene exhibit enhanced tolerance to heat stress. PLoS ONE 2014, 9, e100792. [Google Scholar] [CrossRef]
- Kwon, Y.; Kim, S.H.; Jung, M.S.; Kim, M.S.; Oh, J.E.; Ju, H.W.; Kim, K.I.; Vierling, E.; Lee, H.; Hong, S.W. Arabidopsis hot2 encodes an endochitinase-like protein that is essential for tolerance to heat, salt and drought stresses. Plant J. 2007, 49, 184–193. [Google Scholar] [CrossRef]
- Huang, Y.-C.; Wu, H.-C.; Wang, Y.-D.; Liu, C.-H.; Lin, C.-C.; Luo, D.-L.; Jinn, T.-L. PECTIN METHYLESTERASE34 contributes to heat tolerance through its role in promoting stomatal movement. Plant Physiol. 2017, 174, 748–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-T.; Ru, J.-N.; Liu, Y.-W.; Li, M.; Zhao, D.; Yang, J.-F.; Fu, J.-D.; Xu, Z.-S. Maize WRKY transcription factor ZmWRKY106 confers drought and heat tolerance in transgenic plants. Int. J. Mol. Sci. 2018, 19, 3046. [Google Scholar] [CrossRef] [PubMed]
- Schindelman, G.; Morikami, A.; Jung, J.; Baskin, T.I.; Carpita, N.C.; Derbyshire, P.; McCann, M.C.; Benfey, P.N. COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 2001, 15, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qian, Q.; Zhou, Y.; Yan, M.; Sun, L.; Zhang, M.; Fu, Z.; Wang, Y.; Han, B.; Pang, X.; et al. BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 2003, 15, 2020–2031. [Google Scholar] [CrossRef]
- Dai, X.; You, C.; Wang, L.; Chen, G.; Zhang, Q.; Wu, C. Molecular characterization, expression pattern, and function analysis of the OsBC1L family in rice. Plant Mol. Biol. 2009, 71, 469–481. [Google Scholar] [CrossRef]
- Julius, B.T.; McCubbin, T.J.; Mertz, R.A.; Baert, N.; Knoblauch, J.; Grant, D.G.; Conner, K.; Bihmidine, S.; Chomet, P.; Wagner, R.; et al. Maize Brittle Stalk2-Like3, encoding a COBRA protein, functions in cell wall formation and carbohydrate partitioning. Plant Cell 2021, 33, 3348–3366. [Google Scholar] [CrossRef]
- Xue, J.Y.; McNair, G.; Watanabe, Y.; Kaplen, M.V.; Guevara-Rozo, S.; Schuetz, M.; Schneider, R.; Mansfield, S.D.; Samuels, A.L. COBRA-LIKE4 modulates cellulose synthase velocity and facilitates cellulose deposition in the secondary cell wall. Plant Physiol. 2024, 196, 2531–2548. [Google Scholar] [CrossRef]
- Liu, L.; Shang-Guan, K.; Zhang, B.; Liu, X.; Yan, M.; Zhang, L.; Shi, Y.; Zhang, M.; Qian, Q.; Li, J.; et al. Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet. 2013, 9, e1003704. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, T.; Sun, P.; Chen, X.; Gong, L.; Sun, P.; Ge, S.; Liang, Y.K. COBL9 and COBL7 synergistically regulate root hair tip growth via controlling apical cellulose deposition. Biochem. Biophys. Res. Commun. 2022, 596, 6–13. [Google Scholar] [CrossRef]
- Ben-Tov, D.; Abraham, Y.; Stav, S.; Thompson, K.; Loraine, A.; Elbaum, R.; de Souza, A.; Pauly, M.; Kieber, J.J.; Harpaz-Saad, S. COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol. 2015, 167, 711–724. [Google Scholar] [CrossRef]
- Li, H.; Yang, Y.; Zhang, H.; Li, C.; Du, P.; Bi, M.; Chen, T.; Qian, D.; Niu, Y.; Ren, H.; et al. The Arabidopsis GPI-anchored protein COBL11 is necessary for regulating pollen tube integrity. Cell Rep. 2023, 42, 113353. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.M.; Song, S.; Dhugga, K.S.; Rafalski, J.A.; Benfey, P.N. Combining expression and comparative evolutionary analysis. The COBRA gene family. Plant Physiol. 2007, 143, 172–187. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Kang, B.-g.; Osburn, L.D.; Cheng, Z.-M. The COBRA gene family in Populus and gene expression in vegetative organs and in response to hormones and environmental stresses. Plant Growth Regul. 2009, 58, 211–223. [Google Scholar] [CrossRef]
- Zaheer, M.; Rehman, S.U.; Khan, S.H.; Shahid, S.; Rasheed, A.; Naz, R.; Sajjad, M. Characterization of new COBRA like (COBL) genes in wheat (Triticum aestivum) and their expression analysis under drought stress. Mol. Biol. Rep. 2022, 49, 1379–1387. [Google Scholar] [CrossRef]
- Sun, X.; Xiong, H.; Jiang, C.; Zhang, D.; Yang, Z.; Huang, Y.; Zhu, W.; Ma, S.; Duan, J.; Wang, X.; et al. Natural variation of DROT1 confers drought adaptation in upland rice. Nat. Commun. 2022, 13, 4265. [Google Scholar] [CrossRef]
- Liu, S.; Fukumoto, T.; Gena, P.; Feng, P.; Sun, Q.; Li, Q.; Matsumoto, T.; Kaneko, T.; Zhang, H.; Zhang, Y.; et al. Ectopic expression of a rice plasma membrane intrinsic protein (OsPIP1;3) promotes plant growth and water uptake. Plant J. 2020, 102, 779–796. [Google Scholar] [CrossRef]
- Guo, S.; Xu, Y.; Liu, H.; Mao, Z.; Zhang, C.; Ma, Y.; Zhang, Q.; Meng, Z.; Chong, K. The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat. Commun. 2013, 4, 1566. [Google Scholar] [CrossRef]
- Adak, S.; Agarwal, T.; Das, P.; Ray, S.; Lahiri Majumder, A. Characterization of myo-inositol oxygenase from rice (OsMIOX): Influence of salinity stress in different indica rice cultivars. Physiol. Mol. Biol. Plants 2023, 29, 927–945. [Google Scholar] [CrossRef]
- Deng, M.; Wang, Y.; Kuzma, M.; Chalifoux, M.; Tremblay, L.; Yang, S.; Ying, J.; Sample, A.; Wang, H.M.; Griffiths, R.; et al. Activation tagging identifies Arabidopsis transcription factor AtMYB68 for heat and drought tolerance at yield determining reproductive stages. Plant J. 2020, 104, 1535–1550. [Google Scholar] [CrossRef]
- Kesten, C.; Menna, A.; Sánchez-Rodríguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef]
- Dai, X.; You, C.; Chen, G.; Li, X.; Zhang, Q.; Wu, C. OsBC1L4 encodes a COBRA-like protein that affects cellulose synthesis in rice. Plant Mol. Biol. 2011, 75, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Midorikawa, K.; Kuroda, M.; Yamashita, H.; Tamura, T.; Abe, K.; Asakura, T. Oryza sativa Brittle Culm 1-like 6 modulates β-glucan levels in the endosperm cell wall. PLoS ONE 2019, 14, e0217212. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Complex plant responses to drought and heat stress under climate change. Plant J. 2024, 117, 1873–1892. [Google Scholar] [CrossRef] [PubMed]
- Fakheri, B.A.; Abouzari, A. Reactive oxygen species: Generation, oxidative damage, and signal transduction. Int. J. Life Sci. 2015, 9, 3–17. [Google Scholar] [CrossRef]
- Rurek, M.; Smolibowski, M. Variability of plant transcriptomic responses under stress acclimation: A review from high throughput studies. Acta Biochim. Pol. 2024, 71, 13585. [Google Scholar] [CrossRef]
- Bita, C.E.; Gerats, T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front. Plant Sci. 2013, 4, 273. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, V.; Tanwar, H.; Mor, V.S.; Kumar, M.; Punia, R.C.; Dalal, M.S.; Khan, M.; Sangwan, S.; Bhuker, A.; et al. Impact of high temperature on germination, seedling growth and enzymatic activity of wheat. Agriculture 2022, 12, 1500. [Google Scholar] [CrossRef]
- Rahman, M.A.; Woo, J.H.; Song, Y.; Lee, S.-H.; Hasan, M.M.; Azad, M.A.K.; Lee, K.-W. Heat shock proteins and antioxidant genes involved in heat combined with drought stress responses in perennial rye grass. Life 2022, 12, 1426. [Google Scholar] [CrossRef]
- Cao, D.; Froehlich, J.E.; Zhang, H.; Cheng, C.L. The chlorate-resistant and photomorphogenesis-defective mutant cr88 encodes a chloroplast-targeted HSP90. Plant J. 2003, 33, 107–118. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Silva-Correia, J.; Freitas, S.; Tavares, R.M.; Lino-Neto, T.; Azevedo, H. Phenotypic analysis of the Arabidopsis heat stress response during germination and early seedling development. Plant Methods 2014, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.-Y.; Luo, J.-T.; Zheng, J.-M.; Tan, W.-F.; Pu, Z.-J.; Wang, F. Genome-wide systematic characterization of the NRT2 gene family and its expression profile in wheat (Triticum aestivum L.) during plant growth and in response to nitrate deficiency. BMC Plant Biol. 2023, 23, 353. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Roudier, F.; Schindelman, G.; DeSalle, R.; Benfey, P.N. The COBRA family of putative GPI-anchored proteins in Arabidopsis. A new fellowship in expansion. Plant Physiol. 2002, 130, 538–548. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Li, A.; Hao, C.; Wang, Z.; Geng, S.; Jia, M.; Wang, F.; Han, X.; Kong, X.; Yin, L.; Tao, S.; et al. Wheat breeding history reveals synergistic selection of pleiotropic genomic sites for plant architecture and grain yield. Mol. Plant. 2022, 15, 504–519. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Ma, S.; Wang, M.; Wu, J.; Guo, W.; Chen, Y.; Li, G.; Wang, Y.; Shi, W.; Xia, G.; Fu, D.; et al. WheatOmics: A platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol. Plant 2021, 14, 1965–1968. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.-Q.; Sorrells, M.E. Genetic analysis of fertility restoration in wheat using restriction fragment length polymorphisms. Crop Sci. 1995, 35, 1137–1143. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Liu, J.; Dai, T.; Jing, Q.; Cao, W.; Jiang, D. Alterations in photosynthesis and antioxidant enzyme activity in winter wheat subjected to post-anthesis water-logging. Photosynthetica 2008, 46, 21–27. [Google Scholar] [CrossRef]
- Patra, H.K.; Kar, M.; Mishra, D. Catalase activity in leaves and cotyledons during plant development and senescence1)1) Part X of the series “Studies on Leaf Senescence”. Supported in part by a Grant from the Council of Scientific and Industrial Research, Government of India to D.M. Biochem. Und Physiol. Der Pflanz. 1978, 172, 385–390. [Google Scholar] [CrossRef]
- Nakano, Y.; Asada, K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar] [CrossRef]
- Maehly, A.C.; Chance, B. The assay of catalases and peroxidases. Methods Biochem. Anal. 1954, 1, 357–424. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, Q.; Luo, J.; Zheng, J.; Liu, P.; Wang, D.; Pu, Z. Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants. Int. J. Mol. Sci. 2025, 26, 4101. https://doi.org/10.3390/ijms26094101
Deng Q, Luo J, Zheng J, Liu P, Wang D, Pu Z. Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants. International Journal of Molecular Sciences. 2025; 26(9):4101. https://doi.org/10.3390/ijms26094101
Chicago/Turabian StyleDeng, Qingyan, Jiangtao Luo, Jianmin Zheng, Peixun Liu, Dejun Wang, and Zongjun Pu. 2025. "Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants" International Journal of Molecular Sciences 26, no. 9: 4101. https://doi.org/10.3390/ijms26094101
APA StyleDeng, Q., Luo, J., Zheng, J., Liu, P., Wang, D., & Pu, Z. (2025). Wheat COBRA-like Gene TaCOBL6A2 Confers Heat Tolerance in Plants. International Journal of Molecular Sciences, 26(9), 4101. https://doi.org/10.3390/ijms26094101





