Transcriptome Dynamics of an Oyster Larval Response to a Conspecific Cue-Mediated Settlement Induction in the Pacific Oyster Crassostrea gigas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Larval Culture and Shell Extract Preparation
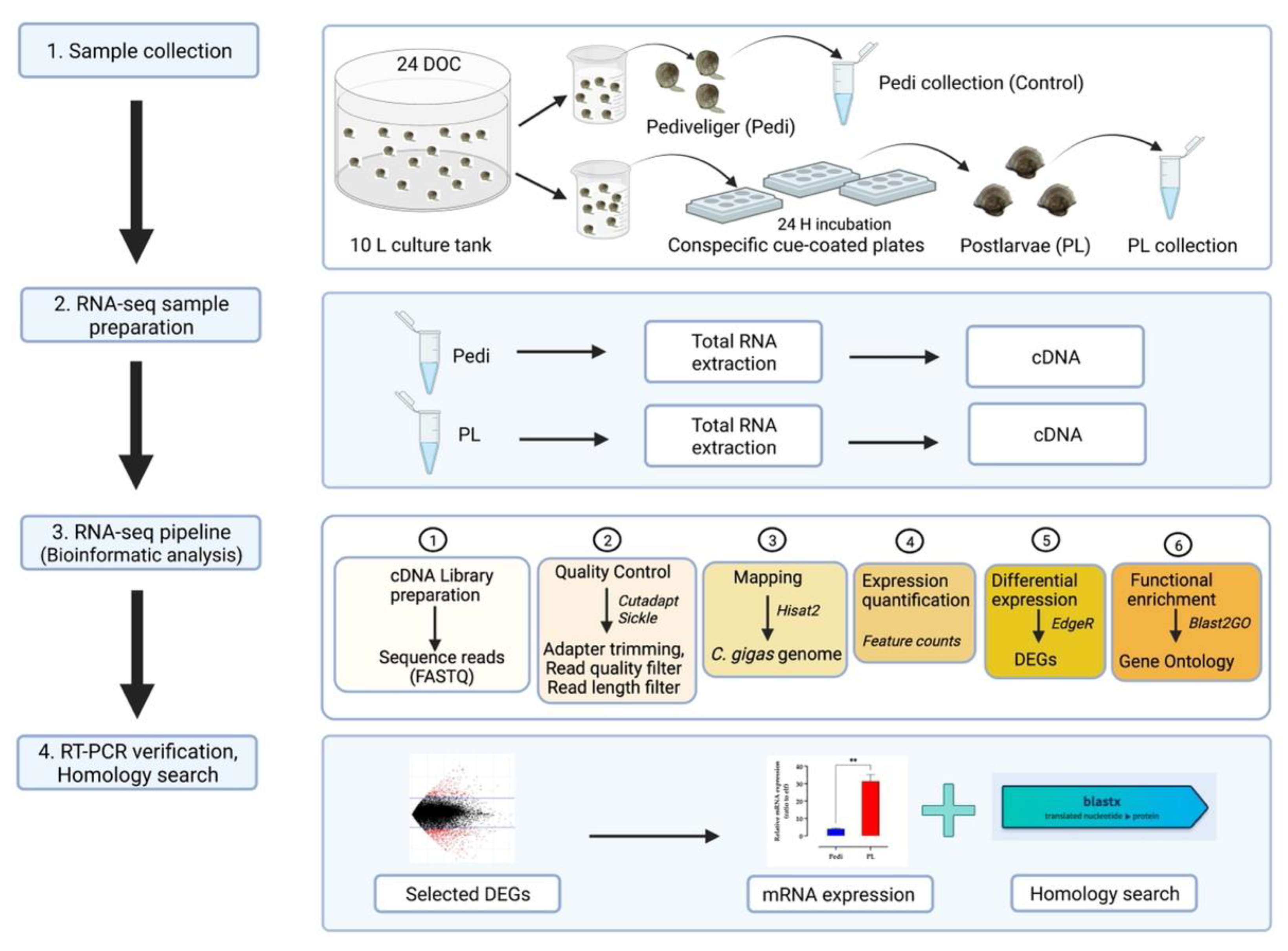
2.2. Sample Collection and Research Design
2.3. RNA Extraction, cDNA Library Preparation, Sequencing, and Bioinformatic Analysis
2.4. Quantitative Real-Time Polymerase Chain Reaction (qPCR)
3. Results
3.1. Transcriptome Sequencing and Mapping
3.2. Identification and Analysis of the Differentially Expressed Genes (DEGs) in the Pediveliger (Pedi) and Postlarvae (PL)
3.2.1. Pre-Settlement Pediveliger Larvae
3.2.2. Postlarvae
3.3. Gene Ontology (GO) Enrichment Analysis
3.4. Validation of Transcriptomic Data via qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedgecock, D.; Gaffney, P.M.; Goulletquer, P.; Guo, X.; Reece, K.; Warr, G.W. The Case for Sequencing the Pacific Oyster Genome. J. Shellfish Res. 2005, 24, 429–441. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; et al. The Oyster Genome Reveals Stress Adaptation and Complexity of Shell Formation. Nature 2012, 490, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Coon, S.L.; Bonar, D.B.; Weiner, R.M. Induction of Settlement and Metamorphosis of the Pacific Oyster, Crassostrea gigas (Thunberg), by L-DOPA and Catecholamines. J. Exp. Mar. Bio. Ecol. 1985, 94, 211–221. [Google Scholar] [CrossRef]
- Bonar, D.B.; Coon, S.L.; Walch, M.; Weiner, R.M.; Fitt, W. Control of Oyster Settlement and Metamorphosis by Endogenous and Exogenous Chemical Cues. Bull. Mar. Sci. 1990, 46, 484–498. [Google Scholar]
- Huan, P.; Wang, H.; Liu, B. A Label-Free Proteomic Analysis on Competent Larvae and Juveniles of the Pacific Oyster (Crassostrea gigas). PLoS ONE 2015, 10, e0135008. [Google Scholar] [CrossRef] [PubMed]
- Laing, I. Bivalve Molluscs: Biology, Ecology and Culture. Aquaculture 2004, 229, 507–508. [Google Scholar] [CrossRef]
- Gosling, E. Reproduction, Settlement and Recruitment. In Marine Bivalve Molluscs; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 157–202. [Google Scholar]
- Joyce, A.; Vogeler, S. Molluscan Bivalve Settlement and Metamorphosis: Neuroendocrine Inducers and Morphogenetic Responses. Aquaculture 2018, 487, 64–82. [Google Scholar] [CrossRef]
- Heyland, A.; Moroz, L.L. Signaling Mechanisms Underlying Metamorphic Transitions in Animals. Integr. Comp. Biol. 2006, 46, 743–759. [Google Scholar] [CrossRef] [Green Version]
- Fitt, W.K.; Labare, M.P.; Fuqua, W.C.; Walch, M.; Coon, S.L.; Bonar, D.B.; Colwell, R.R.; Weiner, R.M. Factors Influencing Bacterial Production of Inducers of Settlement Behavior of Larvae of the Oyster Crassostrea gigas. Microb. Ecol. 1989, 17, 287–298. [Google Scholar] [CrossRef]
- Fitt, W.K.; Coon, S.L.; Walch, M.; Weiner, R.M.; Colwell, R.R.; Bonar, D.B. Settlement Behavior and Metamorphosis of Oyster Larvae (Crassostrea gigas) in Response to Bacterial Supernatants. Mar. Biol. 1990, 106, 389–394. [Google Scholar] [CrossRef]
- Tamburri, M.N.; Zimmer-Faust, R.K.; Tamplin, M.L. Natural Sources and Properties of Chemical Inducers Mediating Settlement of Oyster Larvae: A Re-Examination. Biol. Bull. 1992, 183, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Vasquez, H.E.; Kitamura, H.; Satuito, C.G. Larval Settlement of the Pacific Oyster Crassostrea gigas in Response to Marine Microbial Films and Monospecies Bacterial Films. Sess. Org. 2017, 34, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Swanson, R.L.; De Nys, R.; Huggett, M.J.; Green, J.K.; Steinberg, P.D. In Situ Quantification of a Natural Settlement Cue and Recruitment of the Australian Sea Urchin Holopneustes purpurascens. Mar. Ecol. Prog. Ser. 2006, 314, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Huggett, M.J.; Williamson, J.E.; De Nys, R.; Kjelleberg, S.; Steinberg, P.D. Larval Settlement of the Common Australian Sea Urchin Heliocidaris erythrogramma in Response to Bacteria from the Surface of Coralline Algae. Oecologia 2006, 149, 604–619. [Google Scholar] [CrossRef]
- Huggett, M.J.; Crocetti, G.R.; Kjelleberg, S.; Steinberg, P.D. Recruitment of the Sea Urchin Heliocidaris erythrogramma and the Distribution and Abundance of Inducing Bacteria in the Field. Aquat. Microb. Ecol. 2008, 53, 161–171. [Google Scholar] [CrossRef]
- Hadfield, M.; Paul, V. Natural Chemical Cues for Settlement and Metamorphosis of Marine-Invertebrate Larvae. In Marine Chemical Ecology; McClintock, J.B., Baker, B.J., Eds.; CRC Press: Boca Raton, FL, USA, 2001; pp. 431–461. ISBN 9781420036602. [Google Scholar]
- Hay, M.E. Marine Chemical Ecology: Chemical Signals and Cues Structure Marine Populations, Communities, and Ecosystems. Ann. Rev. Mar. Sci. 2009, 1, 193–212. [Google Scholar] [CrossRef] [Green Version]
- Bayne, B.L. The Gregarious Behaviour of the Larvae of Ostrea edulis L. at Settlement. J. Mar. Biol. Assoc. U. K. 1969, 49, 327–356. [Google Scholar] [CrossRef]
- Crisp, D.J. Chemical Factors Inducing Settlement in Crassostrea virginica (Gmelin). J. Anim. Ecol. 1967, 36, 329–335. [Google Scholar] [CrossRef]
- Hidu, H. Gregarious Setting in the American Oyster Crassostrea virginica Gmelin. Chesap. Sci. 1969, 10, 85–92. [Google Scholar] [CrossRef]
- Hirata, Y.; Tamura, Y.; Nagasawa, K. Influence of Presoaking Conditions of Spat Collectors in Seawater Containing Adult Pacific Oysters (Crassostrea gigas) on Their Larval Settlement. Nippon Suisan Gakkaishi (Jpn.) 2008, 74, 1017–1023. [Google Scholar] [CrossRef] [Green Version]
- Pascual, M.S.; Zampatti, E.A. Evidence of a Chemically Mediated Adult-Larval Interaction Triggering Settlement in Ostrea puelchana: Applications in Hatchery Production. Aquaculture 1995, 133, 33–44. [Google Scholar] [CrossRef]
- Dworjanyn, S.A.; Pirozzi, I. Induction of Settlement in the Sea Urchin Tripneustes gratilla by Macroalgae, Biofilms and Conspecifics: A Role for Bacteria? Aquaculture 2008, 274, 268–274. [Google Scholar] [CrossRef]
- Vasquez, H.E.; Hashimoto, K.; Yoshida, A.; Hara, K.; Imai, C.C.; Kitamura, H.; Satuito, C.G. A Glycoprotein in Shells of Conspecifics Induces Larval Settlement of the Pacific Oyster Crassostrea gigas. PLoS ONE 2013, 8, e82358. [Google Scholar] [CrossRef] [Green Version]
- Vasquez, H.E.; Hashimoto, K.; Kitamura, H.; Satuito, C.G. Wheat Germ Agglutinin-Binding Glycoprotein Extract from Shells of Conspecifics Induces Settlement of Larvae of the Pacific Oyster Crassostrea gigas (Thunberg). J. Shellfish Res. 2014, 33, 415–423. [Google Scholar] [CrossRef]
- Sedanza, M.G.; Kim, H.-J.; Seposo, X.; Yoshida, A.; Yamaguchi, K.; Satuito, C.G. Regulatory Role of Sugars on the Settlement Inducing Activity of a Conspecific Cue in Pacific Oyster Crassostrea gigas. Int. J. Mol. Sci. 2021, 22, 3273. [Google Scholar] [CrossRef] [PubMed]
- Poirier, L.A.; Clements, J.C.; Davidson, J.D.P.; Miron, G.; Davidson, J.; Comeau, L.A. Sink before You Settle: Settlement Behaviour of Eastern Oyster (Crassostrea virginica) Larvae on Artificial Spat Collectors and Natural Substrate. Aquac. Rep. 2019, 13, 100181. [Google Scholar] [CrossRef]
- Tamburri, M.N.; Luckenbach, M.W.; Breitburg, D.L.; Bonniwell, S.M. Settlement of Crassostrea ariakensis Larvae: Effects of Substrate, Biofilms, Sediment and Adult Chemical Cues. J. Shellfish Res. 2008, 27, 601–608. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Hao, R.; Xiong, X.; Jiao, Y.; Deng, Y.; Du, X. Developmental Characteristics of Pearl Oyster Pinctada fucata martensii: Insight into Key Molecular Events Related to Shell Formation, Settlement and Metamorphosis. BMC Genom. 2019, 20, 122. [Google Scholar] [CrossRef]
- Di, G.; Xiao, X.; Tong, M.H.; Chen, X.; Li, L.; Huang, M.; Zhou, L.; Ke, C. Proteome of Larval Metamorphosis Induced by Epinephrine in the Fujian Oyster Crassostrea angulata. BMC Genom. 2020, 21, 675. [Google Scholar] [CrossRef]
- Li, H.; Zhang, B.; Huang, G.; Liu, B.; Fan, S.; Zhang, D.; Yu, D. Differential Gene Expression during Larval Metamorphic Development in the Pearl Oyster, Pinctada fucata, Based on Transcriptome Analysis. Int. J. Genom. 2016, 2016, 2895303. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Zhang, G. Transcriptomic and Proteomic Dynamics during Metamorphosis of Pacific Oyster Crassostrea gigas. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Ji, P.; Xu, F.; Huang, B.; Li, Y.; Li, L.; Zhang, G. Molecular Characterization and Functional Analysis of a Putative Octopamine/Tyramine Receptor during the Developmental Stages of the Pacific Oyster, Crassostrea gigas. PLoS ONE 2016, 11, e0168574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulon, V.; Boudry, P.; Artigaud, S.; Guérard, F.; Hellio, C. In Silico Analysis of Pacific Oyster (Crassostrea gigas) Transcriptome over Developmental Stages Reveals Candidate Genes for Larval Settlement. Int. J. Mol. Sci. 2019, 20, 197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogeler, S.; Bean, T.P.; Lyons, B.P.; Galloway, T.S. Dynamics of Nuclear Receptor Gene Expression during Pacific Oyster Development. BMC Dev. Biol. 2016, 16, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beiras, R.; Widdows, J. Induction of Metamorphosis in Larvae of the Oyster Crassostrea gigas Using Neuroactive Compounds. Mar. Biol. 1995, 123, 327–334. [Google Scholar] [CrossRef]
- Murthy, P.S.; Venugopalan, V.P.; Nair, K.V.K.; Subramoniam, T. Larval Settlement and Surfaces: Implications in Development of Antifouling Strategies. In Marine and Industrial Biofouling; Springer: Berlin/Heidelberg, Germany, 2008; pp. 233–263. [Google Scholar] [CrossRef]
- Fiedler, T.J.; Hudder, A.; McKay, S.J.; Shivkumar, S.; Capo, T.R.; Schmale, M.C.; Walsh, P.J. The Transcriptome of the Early Life History Stages of the California Sea Hare Aplysia californica. Comp. Biochem. Physiol. Part D Genom. Proteom. 2010, 5, 165–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huan, P.; Wang, H.; Liu, B. Transcriptomic Analysis of the Clam Meretrix meretrix on Different Larval Stages. Mar. Biotechnol. 2012, 14, 69–78. [Google Scholar] [CrossRef]
- Qin, J.; Huang, Z.; Chen, J.; Zou, Q.; You, W.; Ke, C. Sequencing and de Novo Analysis of Crassostrea angulata (Fujian Oyster) from 8 Different Developing Phases Using 454 GSFlx. PLoS ONE 2012, 7, e43653. [Google Scholar] [CrossRef] [Green Version]
- Bassim, S.; Tanguy, A.; Genard, B.; Moraga, D.; Tremblay, R. Identification of Mytilus edulis Genetic Regulators during Early Development. Gene 2014, 551, 65–78. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, X.; Deng, Y.; Zheng, Z.; Yang, C.; Du, X. Integrated Application of Transcriptomics and Metabolomics Provides Insights into the Larval Metamorphosis of Pearl Oyster (Pinctada fucata martensii). Aquaculture 2021, 532, 736067. [Google Scholar] [CrossRef]
- Xiong, X.; Cao, Y.; Li, Z.; Huang, R.; Du, X.; Zheng, Z. Ecdysone Signal Pathway Participates in Shell Formation in Pearl Oysters Pinctada fucata martensii. J. Steroid Biochem. Mol. Biol. 2022, 217, 106045. [Google Scholar] [CrossRef] [PubMed]
- Allen, S.K.; Bushek, D. Large-Scale Production of Triploid Oysters, Crassostrea virginica (Gmelin), Using “Stripped” Gametes. Aquaculture 1992, 103, 241–251. [Google Scholar] [CrossRef]
- Sugawara, Y.; Koganezawa, A. Oysters. In Oyster, Scallop and Abalone—Advances in Culture of Molluscs and Related Fields of Research; Nomura, T., Sugawara, Y., Mori, K.M., Takeuchi, M., Nomachi, K., Matsutani, T., Eds.; Kouseisha Kouseikaku: Tokyo, Japan, 1991; pp. 1–17. (In Japanese) [Google Scholar]
- Sugawara, Y. Oysters. In Settlement Mechanism of Marine Organisms; Kajiwara, T., Research Institute of Marine Invertebrates, Eds.; Kouseisha Kouseikaku: Tokyo, Japan, 1991; pp. 62–75. ISBN 978-4-7699-1205-7w. (In Japanese) [Google Scholar]
- Coon, S.L.; Fitt, W.K.; Bonar, D.B. Competence and Delay of Metamorphosis in the Pacific Oyster Crassostrea gigas. Mar. Biol. 1990, 106, 379–387. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Joshi, N.; Fass, J. Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files (Version 1.33) [Software], 2011. Available online: https://github.com/najoshi/sickle (accessed on 25 February 2022).
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-Level Expression Analysis of RNA-Seq Experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; McCarthy, D.; Ritchie, M.; Robinson, M.; Smyth, G. EdgeR: Differential Analysis of Sequence Read Count Data. R Package 2008, 1–119. Available online: https://www.bioconductor.org/packages/devel/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed on 25 February 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; ISBN 3-900051-07-0. Available online: http//www.R-project.org (accessed on 25 February 2022).
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A Universal Tool for Annotation, Visualization and Analysis in Functional Genomics Research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential Analysis of Gene Regulation at Transcript Resolution with RNA-Seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Madden, T. The BLAST Sequence Analysis Tool. In The NCBI Handbook [Internet], 2nd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013; pp. 1–17. [Google Scholar]
- Gao, Y.-L.; Yoshida, A.; Liu, J.-Y.; Shimizu, T.; Shirota, K.; Shiina, Y.; Osatomi, K. Molecular Cloning and Expression Dynamics of UNC-45B upon Heat Shock in the Muscle of Yellowtail. Aquaculture 2021, 541, 736827. [Google Scholar] [CrossRef]
- Marie, B.; Zanella-Cléon, I.; Guichard, N.; Becchi, M.; Marin, F. Novel Proteins from the Calcifying Shell Matrix of the Pacific Oyster Crassostrea gigas. Mar. Biotechnol. 2011, 13, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Mouchi, V.; Lartaud, F.; Guichard, N.; Immel, F.; de Rafélis, M.; Broussard, C.; Crowley, Q.G.; Marin, F. Chalky versus Foliated: A Discriminant Immunogold Labelling of Shell Microstructures in the Edible Oyster Crassostrea gigas. Mar. Biol. 2016, 163, 256. [Google Scholar] [CrossRef]
- Johnstone, M.B.; Wheeler, A.P.; Falwell, E.P.; Staton, M.E.; Saski, C.A.; Mount, A.S. Folian-Cv1 Is a Member of a Highly Acidic Phosphoprotein Class Derived from the Foliated Layer of the Eastern Oyster (Crassostrea virginica) Shell and Identified in Hemocytes and Mantle. Front. Mar. Sci. 2019, 6, 366. [Google Scholar] [CrossRef]
- Coon, S.L.; Bonar, D.B. Pharmacological Evidence That Alpha1.-Adrenoceptors Mediate Metamorphosis of the Pacific Oyster, Crassostrea gigas. Neuroscience 1987, 23, 1169–1174. [Google Scholar] [CrossRef]
- Wang, G.; Liu, B.; Tang, B.; Zhang, T.; Xiang, J. Pharmacological and Immunocytochemical Investigation of the Role of Catecholamines on Larval Metamorphosis by β-Adrenergic-like Receptor in the Bivalve Meretrix meretrix. Aquaculture 2006, 258, 611–618. [Google Scholar] [CrossRef]
- Chen, Z.F.; Matsumura, K.; Wang, H.; Arellano, S.M.; Yan, X.; Alam, I.; Archer, J.A.C.; Bajic, V.B.; Qian, P.Y. Toward an Understanding of the Molecular Mechanisms of Barnacle Larval Settlement: A Comparative Transcriptomic Approach. PLoS ONE 2011, 6, e22913. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.X.; Gong, Y.J.; Gu, J.; Zeng, B.J.; Huang, L.H.; Feng, Q.L. Expression, Subcellular Localization and Protein-Protein Interaction of Four Isoforms of EcR/USP in the Common Cutworm. Insect Sci. 2015, 22, 95–105. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the EdgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar] [CrossRef] [Green Version]
- Burke, R.D. The Induction of Metamorphosis of Marine Invertebrate Larvae: Stimulus and Response. Can. J. Zool. 1983, 61, 1701–1719. [Google Scholar] [CrossRef]
- Morse, D.E. Neurotransmitter-Mimetic Inducers of Larval Settlement and Metamorphosis. Bull. Mar. Sci. 1985, 37, 697–706. [Google Scholar]
- Trapido-Rosenthal, H.; Morse, D. Regulation of Receptor-Mediated Settlement and Metamorphosis in Larvae of a Gastropod Mollusc (Haliotis rufescens). Bull. Mar. Sci. 1986, 39, 383–392. [Google Scholar]
- Hirata, K.Y.; Hadfield, M.G. The Role of Choline in Metamorphic Induction of Phestilla (Gastropoda, Nudibranchia). Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1986, 84, 15–21. [Google Scholar] [CrossRef]
- Satuito, C.G.; Natoyama, K.; Yamazaki, M.; Shimizu, K.; Fusetani, N. Induction of Metamorphosis in the Pediveliger Larvae of the Mussel Mytilus galloprovincialis by Neuroactive Compounds. Fish. Sci. 1999, 65, 384–389. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.L.; Li, Y.F.; Bao, W.Y.; Satuito, C.G.; Kitamura, H. Larval Metamorphosis of the Mussel Mytilus galloprovincialis Lamarck, 1819 in Response to Neurotransmitter Blockers and Tetraethylammonium. Biofouling 2011, 27, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Coon, S.L.; Bonar, D.B.; Weiner, R.M. Chemical Production of Cultchless Oyster Spat Using Epinephrine and Norepinephrine. Aquaculture 1986, 58, 255–262. [Google Scholar] [CrossRef]
- Croll, R.P. Developing Nervous Systems in Molluscs: Navigating the Twists and Turns of a Complex Life Cycle. Brain. Behav. Evol. 2009, 74, 164–176. [Google Scholar] [CrossRef]
- Chandramouli, K.H.; Sun, J.; Mok, F.S.; Liu, L.; Qiu, J.W.; Ravasi, T.; Qian, P.Y. Transcriptome and Quantitative Proteome Analysis Reveals Molecular Processes Associated with Larval Metamorphosis in the Polychaete Pseudopolydora vexillosa. J. Proteome Res. 2013, 12, 1344–1358. [Google Scholar] [CrossRef]
- Song, X.; Liu, Z.; Wang, L.; Song, L. Recent Advances of Shell Matrix Proteins and Cellular Orchestration in Marine Molluscan Shell Biomineralization. Front. Mar. Sci. 2019, 6, 16. [Google Scholar] [CrossRef] [Green Version]
- Chan, V.B.S.; Johnstone, M.B.; Wheeler, A.P.; Mount, A.S. Chitin Facilitated Mineralization in the Eastern Oyster. Front. Mar. Sci. 2018, 5, 12. [Google Scholar] [CrossRef] [Green Version]
- De Wit, P.; Durland, E.; Ventura, A.; Langdon, C.J. Gene Expression Correlated with Delay in Shell Formation in Larval Pacific Oysters (Crassostrea gigas) Exposed to Experimental Ocean Acidification Provides Insights into Shell Formation Mechanisms. BMC Genom. 2018, 19, 160. [Google Scholar] [CrossRef]
- Chandramouli, K.H.; Al-Aqeel, S.; Ryu, T.; Zhang, H.; Seridi, L.; Ghosheh, Y.; Qian, P.Y.; Ravasi, T. Transcriptome and Proteome Dynamics in Larvae of the Barnacle Balanus amphitrite from the Red Sea. BMC Genom. 2015, 16, 1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandramouli, K.H.; Mok, F.S.; Wang, H.; Qian, P.Y. Phosphoproteome Analysis during Larval Development and Metamorphosis in the Spionid Polychaete Pseudopolydora vexillosa. BMC Dev. Biol. 2011, 11, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cranfield, H.J. Observations on the Function of the Glands of the Foot of the Pediveliger of Ostrea edulis during Settlement. Mar. Biol. 1973, 22, 211–223. [Google Scholar] [CrossRef]
- Calado, R.; Leal, M.C. Trophic Ecology of Benthic Marine Invertebrates with Bi-Phasic Life Cycles: What Are We Still Missing? Adv. Mar. Biol. 2015, 71, 1–70. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, M.G.; Carpizo-Ituarte, E.J.; Del Carmen, K.; Nedved, B.T. Metamorphic Competence, a Major Adaptive Convergence in Marine Invertebrate Larvae. Am. Zool. 2001, 41, 1123–1131. [Google Scholar] [CrossRef]
- Lafont, R.; Koolman, J. Diversity of Ecdysteroids in Animal Species. In Ecdysone: Structures and Functions; Smagghe, G., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 47–71. [Google Scholar] [CrossRef]
- Huang, W.; Xu, F.; Qu, T.; Zhang, R.; Li, L.; Que, H.; Zhang, G. Identification of Thyroid Hormones and Functional Characterization of Thyroid Hormone Receptor in the Pacific Oyster Crassostrea gigas Provide Insight into Evolution of the Thyroid Hormone System. PLoS ONE 2015, 10, e0144991. [Google Scholar] [CrossRef]
- Gilbert, L.I.; Tata, J.R.; Atkinson, B.G. Metamorphosis: Postembryonic Reprogramming of Gene Expression in Amphibian and Insect Cells. Gen. Comp. Endocrinol. 1996, 103, 231. [Google Scholar] [CrossRef]



| Accession No. | Annotation | Primer Sequence (5’−3’) | Product Size | Reference |
|---|---|---|---|---|
| LOC105332577 | Chit1-SP | AGGAAATGGGAACAGAAGTTAG | 139 | - |
| Chit1-AP | AGGACGGTTTGTCAATGGGAGG | |||
| LOC105327998 | Calm A-SP | GAGTATTCCGTGTTTCCAATG | 150 | - |
| Calm A-AP | TCTGTTAGCTGTTCTGCCATGT | |||
| LOC105335791 | A-A4-SP | ATTATACAGGTCTTGGGAAAC | 155 | - |
| A-A4-AP | GCAGGGGGTCTCATACAA | |||
| LOC105331104 | CA1-XII-SP | GACGAAATACGGGCACTCAAAG | 145 | - |
| CA1-XII-AP | TTGTCTCCAATGTGACTC | |||
| LOC105327922 | VWC2-SP | TTGCTGTCCAGTGTGTCCTAAC | 114 | - |
| VWC2-AP | CCGAAATGTGAACAATGAC | |||
| LOC105335286 | Wif-1-SP | CGAATGAACGACCACTGCAACAAGTC | 133 | Huan et al. [5] |
| Wif-1-AP | TGTCTGTCTGTCCAGGCTGTAGGC | |||
| LOC105338957 | Elf-SP | CGAGAAGGAAGCTGCTGAGATGG | 208 | Huan et al. [5] |
| Elf-AP | ACAGTCAGCCTGTGAAGTTCCTGTA |
| Sample | Sequencing Results | Quality Filter Results | Summary of Mapping Results | ||||
|---|---|---|---|---|---|---|---|
| No. of Pair Reads | >=Q20 (%) | >=Q30 (%) | High- Quality Pair Reads | No. of Reads Not Mapped to the Reference Array | No. of Reads Mapped to the Reference Array (One Site) | No. of Reads Mapped to the Reference Array (Multiple Sites) | |
| Pediveliger (Pedi) | 23,141,856 | 97.0 | 89.0 | 22,628,121 | 8,184,886 (36.17%) | 13,473,074 (59.54%) | 970,161 (4.29%) |
| Post larvae (PL) | 22,833,100 | 97.2 | 89.7 | 22,362,414 | 8,126,324 (36.34%) | 13,440,437 (60.10%) | 795,653 (3.56%) |
| Average | 22,987,478 | 97.1 | 89.4 | 22,495,268 | 8,155,605 (36.25%) | 13,456,756 (59.82%) | 882,907 (3.93%) |
| Accession No. a | Annotation | Reference Species | Evalue b | logFC c | FDR d |
|---|---|---|---|---|---|
| XP_011437564.2 | Abscisic acid and environmental stress-inducible protein (AESIP) | Crassostrea gigas | 0 | 9 | 1.05 × 10−26 |
| XP_034329440.1 | Coadhesin | Crassostrea gigas | 0 | −6.56 | 3.83 × 10−18 |
| XP_034330544.1 | SCO-spondin | Crassostrea gigas | 0 | −6.29 | 3.83 × 10−18 |
| XP_045201952.1 | General transcriptional corepressor trfA-like, 1 (trfA-like, 1) | Mercenaria mercenaria | 3 × 10−29 | 6.44 | 3.83 × 10−18 |
| XP_034329445.1 | A disintegrin and metalloproteinase with thrombospondin motifs adt-1 (ADAM-1) | Crassostrea gigas | 0 | −6.23 | 1.07 × 10−17 |
| XP_011427257.2 | Calcium-dependent protein kinase 7 (CPK7) | Crassostrea gigas | 2 × 10−37 | −6.28 | 1.44 × 10−17 |
| XP_034323067.1 | Spidroin-1-like, 1 | Crassostrea gigas | 2 × 10−08 | 6.12 | 2.88 × 10−17 |
| OWF40357.1 | Stereocilin | Mizuhopecten yessoensis | 0 | −6.02 | 2.90 × 10−17 |
| XP_011422608.2 | IgGFc-binding protein | Crassostrea gigas | 6 × 10−135 | −6.05 | 4.55 × 10−17 |
| XP_022339848.1 | SCO-spondin-like, 1 | Crassostrea virginica | 0 | −5.99 | 7.63 × 10−17 |
| XP_022339848.1 | SCO-spondin-like, 2 | Crassostrea virginica | 0 | −5.99 | 7.63 × 10−17 |
| XP_045201952.1 | General transcriptional corepressor trfA-like, 2 (trfA-like, 2) | Mercenaria mercenaria | 9 × 10−31 | 5.96 | 7.63 × 10−17 |
| XP_034323070.1 | Spidroin-1-like, 2 | Crassostrea gigas | 7 × 10−13 | 5.97 | 7.63 × 10−17 |
| XP_034322956.1 | uncharacterized LOC117688748 | Crassostrea gigas | 0 | 5.95 | 7.71 × 10−17 |
| XP_034322955.1 | Uncharacterized LOC117688749 | Crassostrea gigas | 0 | 5.85 | 1.81 × 10−16 |
| XP_022339848.1 | SCO-spondin-like, 3 | Crassostrea virginica | 0 | −5.79 | 2.11 × 10−16 |
| XP_034313686.1 | ADP-ribosylation factor 1-like | Crassostrea gigas | 3 × 10−136 | 5.93 | 2.28 × 10−16 |
| XP_011426386.2 | Mediator of DNA damage checkpoint protein 1 (MDC1) | Crassostrea gigas | 0 | 5.87 | 5.08 × 10−16 |
| XP_034323479.1 | Glycine-rich cell wall structural protein 1.8 transcript isoform X2 (GRP 1.8- isoform X2) | Crassostrea gigas | 0 | 5.74 | 7.33 × 10−16 |
| XP_011450501.3 | Glycine-rich cell wall structural protein 1.8 transcript isoform X1 (GRP 1.8-isoform X1) | Crassostrea gigas | 0 | 5.74 | 7.33 × 10−16 |
| APC92582.1 | Tyrp-1 | Hyriopsis cumingii | 2 × 10−157 | 5.74 | 7.66 × 10−16 |
| XP_011436430.2 | Collagen alpha-2(I) chain | Crassostrea gigas | 6 × 10−37 | 5.70 | 1.03 × 10−15 |
| XP_011422608.2 | IgGFc-binding protein | Crassostrea gigas | 0 | −5.58 | 1.40 × 10−15 |
| XP_011452538.2 | Leucine-rich repeat-containing protein 4C (LRRC4C) | Crassostrea gigas | 0 | −5.63 | 2.00 × 10−15 |
| XP_011419747.2 | Uncharacterized LOC105322623 | Crassostrea gigas | 0 | −5.56 | 2.75 × 10−15 |
| XP_034323071.1 | Spidroin-1-like, 3 | Crassostrea gigas | 3 × 10−08 | 5.51 | 2.83 × 10−15 |
| CAG2230452.1 | CD283 | Mytilus edulis | 3 × 10−59 | −5.49 | 5.61 × 10−15 |
| XP_011415271.2 | Acanthoscurrin-2 | Crassostrea gigas | 0 | −5.40 | 7.66 × 10−15 |
| XP_011452536.2 | Leucine-rich repeat LGI family member 2 (LGI2) | Crassostrea gigas | 0 | −5.38 | 2.24 × 10−14 |
| XP_021357253.1 | Serine/arginine repetitive matrix protein 1-like (SRRM1-like) | Mizuhopecten yessoensis | 9 × 10−62 | 5.44 | 2.88 × 10−14 |
| Description | Chitin-Binding | Calcium Ion Binding | Extracellular Region |
|---|---|---|---|
| GO ID | GO:0008061 | GO:0005509 | GO:0005576 |
| FDR value | 8.13 × 10−5 | 0.0024 | 5.67 × 10−6 |
| Ontology | Molecular function (MF) | Molecular function (MF) | Cellular component (CC) |
| Count | 9 | 17 | 17 |
| Gene | cgPif97 (↑), Peritrophin-44 (↓), Lactahedrin (↓), Collagen alpha 1 (XII) chain (↓), Peritrophin-1-like,1 (↓), Peritrophin-like,2 (↓), Cleavage and polyadenylation factor 1 subunit 1 (Clip1) (↑), Putative chitinase 1 precursor (↑), Putative chitinase (↑) | DNA ligase 1 (↓), EF-hand domain-containing protein 1 (↓), Calmodulin-4 (↓), Calcium-binding protein E-63-1-like (↓), Calmodulin-A isoforms (↓), Leucine-rich repeat-containing protein 74B (↓), Sarcoplasmic calcium-binding protein (↑), Myosin (↑), EGF-like repeat and discoidin-I domain-containing protein 3 (↓), Mammalian ependymin-related protein 1 (↓), EF-hand domain-containing family member B (↓), Sarcoplasmic calcium-binding protein (↑), Neurocalcin homolog,1,(↓), Neurocalcin homolog,2 (↑), Regucalcin,1 (↓), Regucalcin,2 (↓), Annexin A4 (↓) | Poly(3-hydroxyalkanoate) depolymerase C (↓), cgPif97 (↑), Trithorax group protein osa-like (↑), NPC intracellular cholesterol transporter 2 (↓), Mammalian ependymin-related protein 1 (↓), Peritrophin-44 (↓), Lactadherin (↓), Golgi-associated plant pathogenesis-related protein 1 (↓), Collagen alpha 1 (XII) chain (↓), Dermatopontin (↓), Metalloproteinase inhibitor 3 (↓), Peritrophin-1-like, 1 (↓), Peritrophin-1-like, 2 (↓), inactive pancreatic lipase-related protein 1 (↓), pancreatic lipase-related protein 2 (↓), adenosine deaminase AGSA (↓), von Willebrand factor C domain-containing protein 2-like (VWC2L) (↓) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedanza, M.G.; Alshaweesh, J.; Gao, Y.-L.; Yoshida, A.; Kim, H.-J.; Yamaguchi, K.; Satuito, C.G. Transcriptome Dynamics of an Oyster Larval Response to a Conspecific Cue-Mediated Settlement Induction in the Pacific Oyster Crassostrea gigas. Diversity 2022, 14, 559. https://doi.org/10.3390/d14070559
Sedanza MG, Alshaweesh J, Gao Y-L, Yoshida A, Kim H-J, Yamaguchi K, Satuito CG. Transcriptome Dynamics of an Oyster Larval Response to a Conspecific Cue-Mediated Settlement Induction in the Pacific Oyster Crassostrea gigas. Diversity. 2022; 14(7):559. https://doi.org/10.3390/d14070559
Chicago/Turabian StyleSedanza, Mary Grace, Jalal Alshaweesh, Yi-Li Gao, Asami Yoshida, Hee-Jin Kim, Kenichi Yamaguchi, and Cyril Glenn Satuito. 2022. "Transcriptome Dynamics of an Oyster Larval Response to a Conspecific Cue-Mediated Settlement Induction in the Pacific Oyster Crassostrea gigas" Diversity 14, no. 7: 559. https://doi.org/10.3390/d14070559
APA StyleSedanza, M. G., Alshaweesh, J., Gao, Y.-L., Yoshida, A., Kim, H.-J., Yamaguchi, K., & Satuito, C. G. (2022). Transcriptome Dynamics of an Oyster Larval Response to a Conspecific Cue-Mediated Settlement Induction in the Pacific Oyster Crassostrea gigas. Diversity, 14(7), 559. https://doi.org/10.3390/d14070559







