Bacterial Communities in a Gradient of Abiotic Factors Near a Sulfide Thermal Spring in Northern Baikal
Abstract
:1. Introduction
2. Materials and Methods
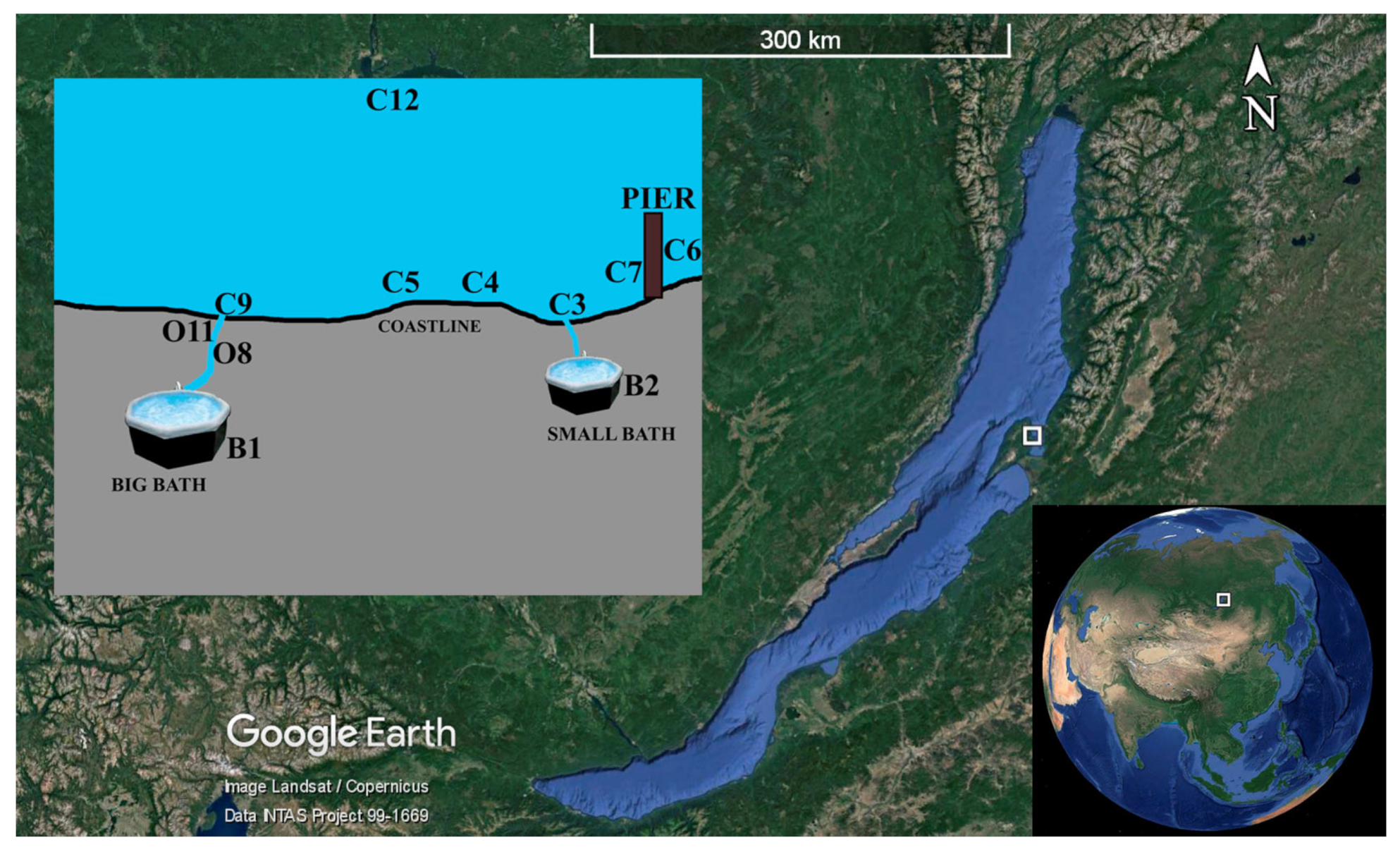
2.1. Characteristics of the Study and Sampling Areas
2.2. Physical and Chemical Parameters of Sites
2.3. DNA Extraction and Sequencing
2.4. Bioinformatics Analysis
3. Results
3.1. Chemical Composition
3.2. Alpha Diversity Differences among Sites
3.3. Beta Diversity Differences between Sites
3.4. Comparative Community Analysis
3.5. Predicted Metabolic Functions
3.6. Interaction between Microbial Diversity and Abiotic Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brock, T.D.; Freeze, H. Thermus aquaticus, a nonsporulating extreme thermophile. J. Bacteriol. 1969, 98, 289–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowen De Leon, K.; Gerlach, R.; Peyton, B.M.; Fields, M.W. Archaeal and bacterial communities in three alkaline hot springs in Heart Lake Geyser Basin, Yellowstone National Park. Front. Microbiol. 2013, 4, 330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, C.M.; Beard, B.L.; Beukes, N.J.; Klein, C.; O’Leary, J.M. Ancient geochemical cycling in the Earth as inferred from Fe isotope studies of banded iron formations from the Transvaal Craton. Contrib. Mineral. Petrol. 2003, 144, 523–547. [Google Scholar] [CrossRef]
- Beam, J.P.; Jay, Z.J.; Schmid, M.C.; Rusch, D.B.; Romine, M.F.; Jennings, R.M.; Kozubal, M.A.; Tringe, S.G.; Wagner, M.; Inskeep, W.P. Ecophysiology of an uncultivated lineage of Aigarchaeota from an oxic, hot spring filamentous ‘streamer’ community. ISME J. 2016, 10, 210–224. [Google Scholar] [CrossRef] [Green Version]
- Kochetkova, T.V.; Podosokorskaya, O.A.; Elcheninov, A.G.; Kublanov, I.V. Diversity of thermophilic prokaryotes inhabiting Russian natural hot springs. Microbiology 2022, 91, 3–31. (In Russian) [Google Scholar] [CrossRef]
- Zavarzin, G.A.; Karpov, G.A.; Gorlenko, V.M.; Golovacheva, R.S.; Gerasimenko, L.M.; Bonch-Osmolovskaya, E.A.; Orleanskii, V.K. Kaldernye Mikroorganizmy (Caldera Microorganisms); Nauka: Moscow, Russia, 1989; p. 120. (In Russian) [Google Scholar]
- Gorlenko, V.M.; Starynin, D.A.; Bonch-Osmolovskaya, E.A.; Kachalkin, V.I. Production processes in microbial cenoses of the Thermofil’nyi Hot Spring. Microbiology 1987, 56, 872–878. (In Russian) [Google Scholar]
- Reigstad, L.J.; Jorgensen, S.L.; Schleper, C. Diversity and abundance of Korarchaeota in terrestrial hot springs of Iceland and Kamchatka. ISME J. 2010, 4, 346–356. [Google Scholar] [CrossRef] [Green Version]
- Bonch-Osmolovskaya, E.A.; Karpov, G.A. Microbial methane formation in Uzon Caldera hydrothermal vents. Microbiology 1986, 56, 516–518. (In Russian) [Google Scholar]
- Lazareva, E.V.; Anisimova, N.S.; Bryanskaya, A.V.; Ogorodnikova, O.L.; Zhmodik, S.M. Features of Mineral Formation in Microbial Communities Developing along the Outflow of Termofil’nyi Vent (Uzon caldera, Kamchatka). Tr. Kronotsk. Gos. Biosfer. Zapovednika (Proc. Kronotsky State Biosphere Reserve); Mosolov, V.I., Ed.; Kamchatpress: Petropavlovsk-Kamchatsky, Russia, 2012; pp. 143–156. (In Russian) [Google Scholar]
- Chernousova, E.Y. Biodiversity of Filamentous Sulfur Bacteria in Sulfide Ecosystems of the North Caucasus Region: Ecological and Molecular Genetic Aspects. Ph.D. Thesis, Voronezh State University, Voronezh, Russia, 2011. [Google Scholar]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef] [Green Version]
- Borisenko, I.M.; Zamana, L.V. Mineral’nye Vody Buryatskoi ASSR (Mineral Waters of Buryat ASSR); Buryat. Izd.: Ulan-Ude, Russia, 1978; p. 162. [Google Scholar]
- Kompantseva, E.I.; Gorlenko, V.M. Phototrophic communities of some thermal springs of Lake Baikal. Microbiology 1988, 57, 841–846. (In Russian) [Google Scholar]
- Namsaraev, Z.B.; Gorlenko, V.M.; Namsaraev, B.B.; Buriukhaev, S.P.; Iurkov, V.V. The structure and biogeochemical activity of the phototrophic communities from the Bol’sherechenskii alkaline hot spring. Microbiology 2003, 72, 228–238. (In Russian) [Google Scholar] [CrossRef] [PubMed]
- Babasanova, O.B.; Barkhutova, D.D.; Namsaraev, Z.B.; Kalashnikova, O.M. Aerobic organothrophic bacteria in hot springs of Baikal region. Bulletin of the Buryat State University. Biol. Geogr. 2007, 3, 112–118. (In Russian) [Google Scholar]
- Koeksoy, E.; Halama, M.; Hagemann, N.; Weigold, P.R.; Laufer, K.; Kleindienst, S.; Byrne, J.M.; Sundman, A.; Hanselmann, K.; Halevy, I.; et al. A case study for late Archean and Proterozoic biogeochemical iron- and sulphur cycling in a modern habitat-the Arvadi Spring. Geobiology 2017, 16, 353–368. [Google Scholar] [CrossRef] [PubMed]
- Sorokovikova, E.G.; Belykh, O.I.; Likhoshvai, E.V. Cyanobacteria of Thermal Springs of the Baikal Rift Zone: Species Composition, Cell Ultrastructure, Silicon Biomineralization; Modern Paleontology, Classical and Modern Methods; Borissiak Paleontological Institute of the Russian Academy of Sciences: Moscow, Russia, 2007; pp. 11–18. [Google Scholar]
- Zelenkina, T.S.; Dagurova, O.P.; Namsaraev, B.B.; Eshinimayev, B.T.; Suzina, N.E.; Trotsenko, Y.A. Aerobic methanotrophs from the coastal thermal springs of Lake Baikal. Microbiology 2009, 78, 492–497. (In Russian) [Google Scholar] [CrossRef]
- Wetzel, R.G.; Likens, G.E. Limnological Analyses; Springer: New York, NY, USA, 1991; p. 391. [Google Scholar]
- Baram, G.I.; Vereshchagin, A.L.; Golobokova, L.P. Microcolumn high-performance liquid chromatography with UV detection for the determination of anions in environmental materials. J. Anal. Chem. 1999, 54, 962–965. (In Russian) [Google Scholar]
- Cline, J.D. Spectrophotometric determination of hydrogen sulphide in natural waters. Limnol. Oceanogr. 1969, 14, 454–458. [Google Scholar] [CrossRef]
- Marmur, J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 1961, 3, 208–218. [Google Scholar] [CrossRef]
- Sahm, K.; John, P.; Nacke, H.; Wemheuer, B.; Grote, R.; Daniel, R.; Antranikian, G. High abundance of heterotrophic prokaryotes in hydrothermal springs of the Azores as revealed by a network of 16S rRNA gene-based methods. Extremophiles 2013, 17, 649–662. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. 2022. Available online: https://www.R-project.org/ (accessed on 4 October 2022).
- Falkner, K.K.; Measures, C.I.; Herbelin, S.E.; Edmond, J.M.; Weiss, R.F. The major and minor element geochemistry of Lake Baikal. Limnol. Oceanogr. 1991, 36, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 10. [Google Scholar] [CrossRef] [PubMed]
- Matheus Carnevali, P.B.; Schulz, F.; Castelle, C.J.; Kantor, R.S.; Shih, P.M.; Sharon, I.; Santini, J.M.; Olm, M.R.; Amano, Y.; Thomas, B.C.; et al. Hydrogen-based metabolism as an ancestral trait in lineages sibling to the Cyanobacteria. Nat. Commun. 2019, 10, 463. [Google Scholar] [CrossRef] [Green Version]
- Yeoh, Y.K.; Sekiguchi, Y.; Parks, D.H.; Hugenholtz, P.B. Comparative genomics of candidate phylum TM6 suggests that parasitism is widespread and ancestral in this lineage. Mol. Biol. Evol. 2016, 33, 915–927. [Google Scholar] [CrossRef] [Green Version]
- Suproniene, S.; Doyeni, M.O.; Viti, C.; Tilvikiene, V.; Pini, F. Characterization of the Soil Prokaryotic community with respect to time and fertilization with animal waste–based digestate in a humid continental climate. Front. Environ. Sci. 2022, 10, 12. [Google Scholar] [CrossRef]
- Barhutova, D.D. Effect of Environmental Parameters on Bacterial Diversity and Functional Activity of Bacteria-Destructors in Sulfide-Rich Springs of the Baikal Region. Ph.D. Thesis, Banzarov Buryat State University, Ulan-Ude, Russia, 2000. [Google Scholar]
- Plyusnin, A.M.; Zamana, L.V.; Shvartsev, S.L.; Tokarenko, O.G.; Chernyavskii, M.K. Hydrogeochemical peculiarities of the composition of nitric thermal waters in the Baikal Rift Zone. Geol. Geophys. 2013, 54, 647–664. [Google Scholar] [CrossRef]
- Chernitsyna, S.M.; Mamaeva, E.V.; Lomakina, A.V.; Pogodaeva, T.V.; Galach’yants, Y.P.; Bukin, S.V.; Khlystov, O.M.; Zemskaya, T.I.; Pimenov, N.V. Phylogenetic diversity of microbial communities of the Posolsk Bank bottom sediments, Lake Baikal. Microbiology 2016, 85, 672–680. [Google Scholar] [CrossRef]
- Yang, T.; Lyons, S.; Aguilar, C.; Cuhel, R.; Teske, A. Microbial communities and chemosynthesis in Yellowstone Lake sublacustrine hydrothermal vent waters. Front. Microbiol. 2011, 2, 130. [Google Scholar] [CrossRef] [Green Version]
- Martínez, A.; Pibernat, I.; Figueras, J.; García-Gil, J. Structure and composition of freshwater microbial mats from a sulfur spring (“Font Pudosa”, NE Spain). Microbiologia 1997, 13, 45–56. [Google Scholar]
- Lan, L.; Zhao, J.; Wang, S.; Li, X.; Qiu, L.; Liu, S.; Bin Nasry, A.A. NO and N2O accumulation during nitrite-based sulfide-oxidizing autotrophic denitrification. Bioresour. Technol. Rep. 2019, 7, 100190. [Google Scholar] [CrossRef]
- Potapova, Z.M. The Species Composition of Cyanobacteria and Their Ecophysiology in Nitrogen-Rich Thermal Springs of Northern Transbaikal Region. Ph.D. Thesis, Banzarov Buryat State University, Ulan-Ude, Russia, 2010. [Google Scholar]
- Yurkov, V.V.; Gorlenko, V.M.; Mityushina, L.L.; Starynin, D.A. The Effect of Limiting Factors on the Structure of Phototrophic Communities in the Bol’sherechenskie Thermal Springs. Microbiology 1991, 60, 129–138. (In Russian) [Google Scholar]
- Namsaraev, B.B.; Barkhutova, D.D.; Danilova, E.V.; Bryanskaya, A.V.; Buryukhaev, S.P.; Garmaev, E.Z.; Gorlenko, V.M.; Dagurova, O.P.; Dambaev, V.B.; Zaitseva, S.V.; et al. The Geochemical Activity of Microorganisms of the Thermal Springs in the Baikal Rift Zone; Weinstein, M.B., Ed.; Academic Publishing House “Geo”: Novosibirsk, Russia, 2011; p. 302. [Google Scholar]
- Chernitsyna, S.M.; Khalzov, I.A.; Sitnikova, T.Y.; Naumova, T.V.; Khabuev, A.V.; Zemskaya, T.I. Microbial Communities Associated with Bentic Invertebrates of Lake Baikal. Curr. Microbiol. 2021, 78, 3020–3031. [Google Scholar] [CrossRef] [PubMed]
- Khalzov, I.A.; Bukin, S.V.; Zakharenko, A.S.; Chernitsyna, S.M.; Galachyants, Y.P.; Sitnikova, T.Y.; Zemskaya, T.I. Microbial communities associated with the ostracods Candona sp. inhabiting the area of the methane seep Goloustnoye (Lake Baikal). Symbiosis 2021, 85, 163–174. [Google Scholar] [CrossRef]
- Boeuf, D.; Eppley, J.M.; Mende, D.R.; Malmstrom, R.R.; Woyke, T.; DeLong, E.F. Metapangenomics reveals depth-dependent shifts in metabolic potential for the ubiquitous marine bacterial SAR324 lineage. Microbiome 2021, 9, 18. [Google Scholar] [CrossRef]
- Anantharaman, K.; Brown, C.T.; Hug, L.A.; Sharon, I.; Castelle, C.J.; Probst, A.J.; Thomas, B.C.; Singh, A.; Wilkins, M.J.; Karaoz, U.; et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat. Commun. 2016, 7, 11. [Google Scholar] [CrossRef]
- Van Veen, W.L.; Mulder, E.G.; Deinema, M.H. The Sphaerotilus−Leptothrix group of bacteria. Microbiol. Rev. 1978, 42, 162–329. [Google Scholar] [CrossRef]
- Chernousova, E.Y.; Gridneva, E.V.; Grabovich, M.Y.; Akimov, V.N.; Dubinina, G.A. Phylogenetic in situ/ex situ analysis of a sulfur mat microbial community from a thermal sulfide spring in the North Caucasus. Microbiology 2008, 77, 219–223. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, C.; Li, W.; Weng, S.; Song, W.; Li, J.; Wang, Y. Functional diversity of microbial communities in inactive seafloor sulfide deposits. FEMS Microbiol. Ecol. 2021, 97, 14. [Google Scholar] [CrossRef]
- Cabello-Yeves, P.J.; Zemskaya, T.I.; Zakharenko, A.S.; Sakirko, M.V.; Ivanov, V.G.; Ghai, R.; Rodriguez-Valera, F. Microbiome of the deep Lake Baikal, a unique oxic bathypelagic habitat. Limnol. Oceanogr. 2020, 65, 1471–1488. [Google Scholar] [CrossRef] [Green Version]
- Yuriev, D.A.; Zaitseva, S.V.; Zhdanova, G.O.; Tolstoy, M.Y.; Barkhutova, D.D.; Vyatchina, O.F.; Konovalova, E.Y.; Stom, D.I. Microbial mat of the thermal springs Kuchiger Republic of Buryatia: Species composition, biochemical properties and electrogenic activity in biofuel cell. IOP Conf. Ser. Earth Environ. Sci. 2018, 121, 022012. [Google Scholar] [CrossRef]
- Andreeva, I.S.; Pechurkina, N.I.; Morozova, O.V.; Ryabchikova, E.I.; Belikov, S.I.; Puchkova, L.I.; Emel’yanova, E.K.; Torok, T.; Repin, V.E. The new eubacterium Roseomonas baikalica sp. nov. isolated from core samples collected by deep-hole drilling of the bottom of Lake Baikal. Microbiology 2007, 76, 487–493. [Google Scholar] [CrossRef]
- Sukhanova, E.V.; Shtykova, Y.R.; Suslova, M.Y.; Pestunova, O.S.; Kostornova, T.Y.; Khanaev, I.V.; Zimens, E.A.; Podlesnaya, G.V.; Parfenova, V.V. Diversity and Physiological and Biochemical Properties of Heterotrophic Bacteria Isolated from Lake Baikal Epilithic Biofilms. Microbiology 2019, 88, 345–357. [Google Scholar] [CrossRef]





| Factor | R Value | p Value |
|---|---|---|
| T° | 0.741 | 1 × 10−4 |
| pH | 0.398 | 6.7× 10−3 |
| Eh | 0.350 | 1.4 × 10−2 |
| HCO3− | 0.457 | 8.5 × 10−3 |
| Cl− | 0.432 | 2.6 × 10−3 |
| NO3− | 0.604 | 8 × 10−4 |
| SO42− | 0.786 | 1 × 10−4 |
| Na+ | 0.814 | 1 × 10−4 |
| K+ | 0.515 | 1 × 10−3 |
| Ca2+ | 0.711 | 1 × 10−4 |
| Mg2+ | 0.509 | 8 × 10−4 |
| H2S | 0.664 | 1 × 10−4 |
| O2 | 0.790 | 1 × 10−4 |
| Ʃions | 0.799 | 1 × 10−4 |
| total | 0.867 | 1 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chernitsyna, S.; Elovskaya, I.; Pogodaeva, T.; Bukin, S.; Zakharenko, A.; Zemskaya, T. Bacterial Communities in a Gradient of Abiotic Factors Near a Sulfide Thermal Spring in Northern Baikal. Diversity 2023, 15, 298. https://doi.org/10.3390/d15020298
Chernitsyna S, Elovskaya I, Pogodaeva T, Bukin S, Zakharenko A, Zemskaya T. Bacterial Communities in a Gradient of Abiotic Factors Near a Sulfide Thermal Spring in Northern Baikal. Diversity. 2023; 15(2):298. https://doi.org/10.3390/d15020298
Chicago/Turabian StyleChernitsyna, Svetlana, Irina Elovskaya, Tatyana Pogodaeva, Sergei Bukin, Aleksandra Zakharenko, and Tamara Zemskaya. 2023. "Bacterial Communities in a Gradient of Abiotic Factors Near a Sulfide Thermal Spring in Northern Baikal" Diversity 15, no. 2: 298. https://doi.org/10.3390/d15020298
APA StyleChernitsyna, S., Elovskaya, I., Pogodaeva, T., Bukin, S., Zakharenko, A., & Zemskaya, T. (2023). Bacterial Communities in a Gradient of Abiotic Factors Near a Sulfide Thermal Spring in Northern Baikal. Diversity, 15(2), 298. https://doi.org/10.3390/d15020298






