Genetic Variability and Kinship Analyses of Seized Red-Browed Amazon, Amazona rhodocorytha (Aves, Psittacidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and DNA Extraction
2.2. UCE Sequence Processing, SNP Calling and Filtering
2.3. Genetic Diversity and Kinship Analysis
2.4. Structure Analysis
3. Results
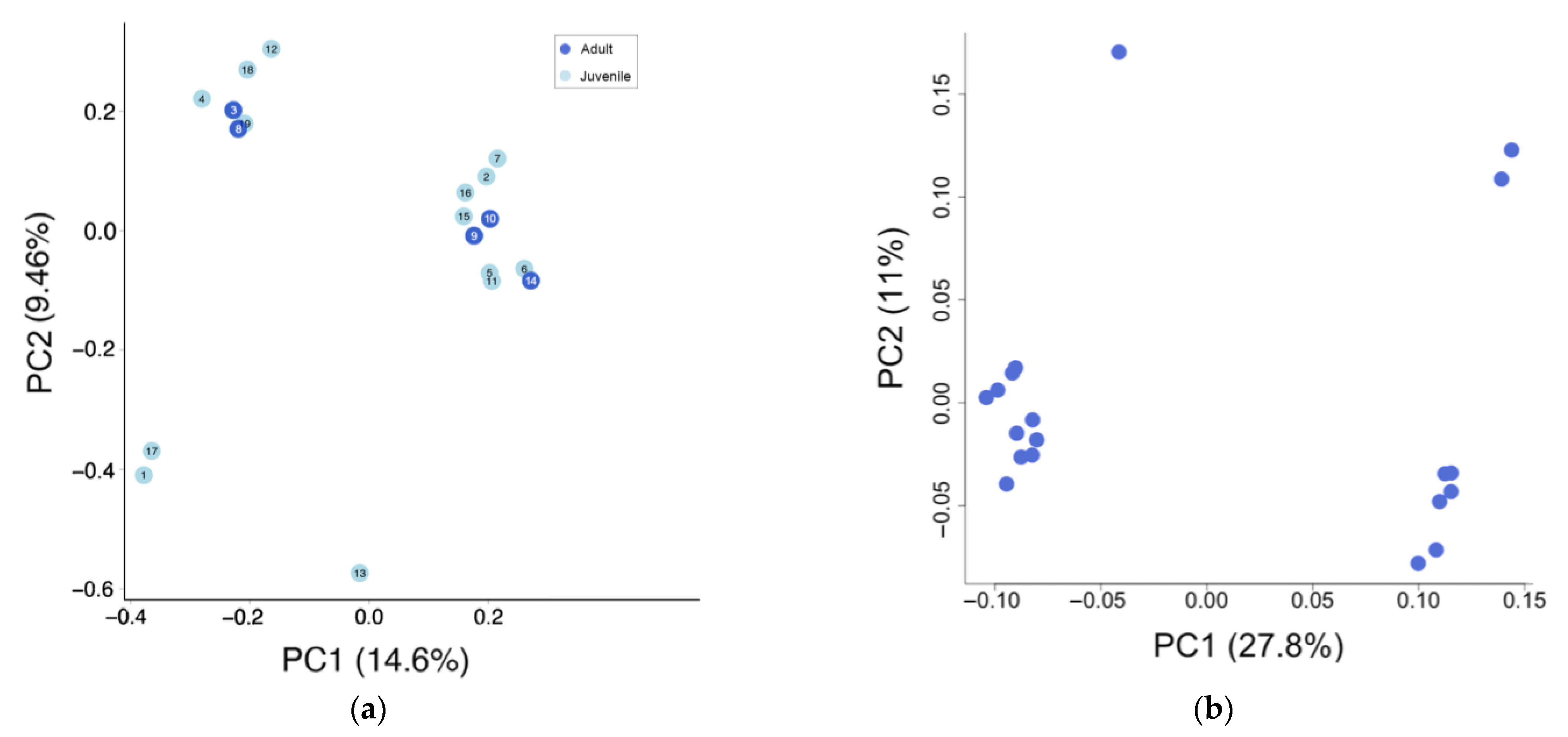
3.1. Genetic Diversity
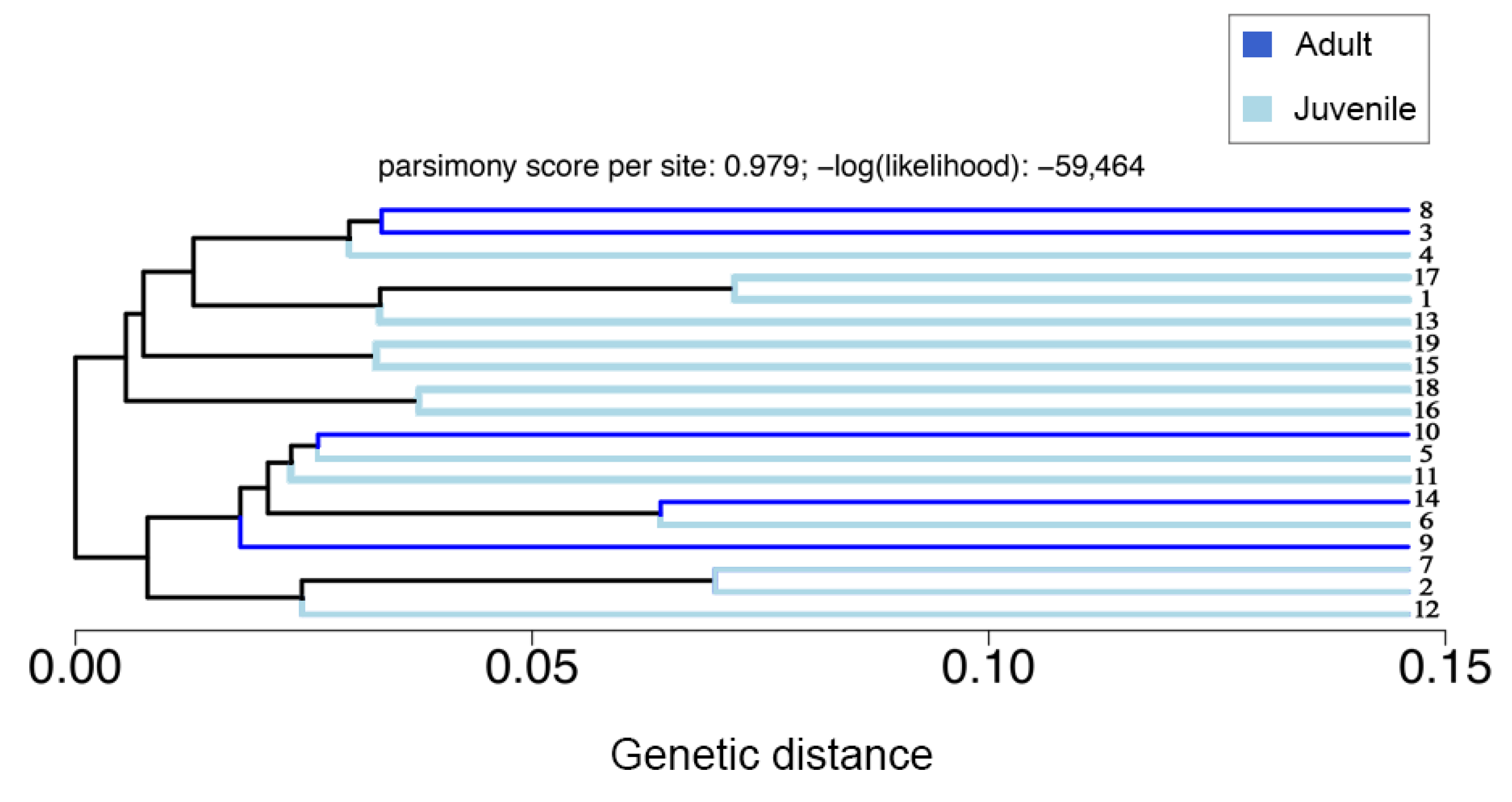
3.2. Population Structure
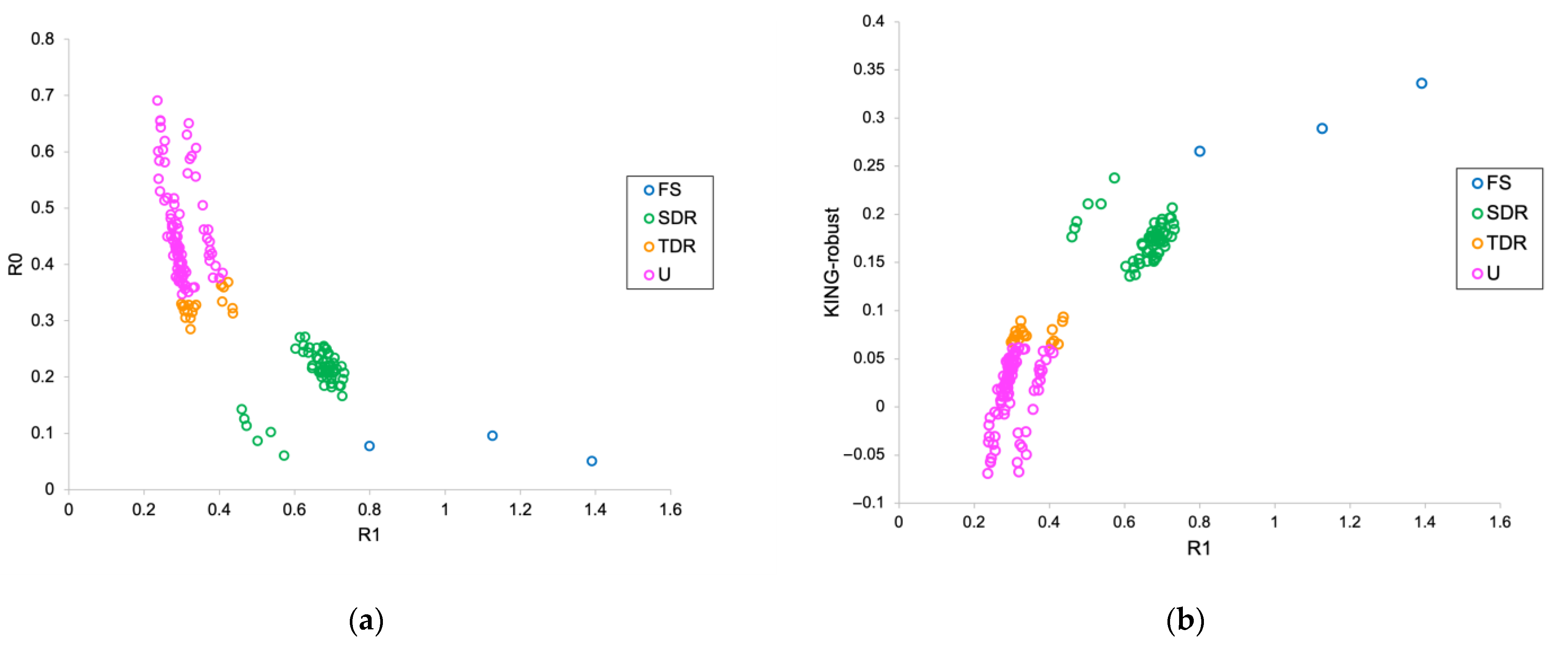
3.3. Pairwise Relatedness
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Winkler, D.W.; Billerman, S.M.; Lovette, I.J. New World and African Parrots (Psittacidae), version 1.0. In Birds of the World; Billerman, S.M., Keeney, B.K., Rodewald, P.G., Schulenberg, T.S., Eds.; Cornell Lab of Ornithology: Ithaca, NY, USA, 2020. [Google Scholar] [CrossRef]
- Forshaw, J.M. Parrots of the World; Lansdowne Editions: Sidney, Australia, 1989; p. 573. [Google Scholar]
- Berkunsky, I.; Quillfeldt, P.; Brightsmith, D.J.; Abbud, M.C.; Aguilar, J.M.R.E.; Alemán-Zelaya, U.; Aramburú, R.M.; Arce Arias, A.; Balas McNab, R.; Balsby, T.J.S.; et al. Current threats faced by Neotropical parrot populations. Biol. Conserv. 2017, 214, 278–287. [Google Scholar] [CrossRef] [Green Version]
- Feliciano, R.; Leite, A.B.; Garbeloto, M.C.; Silveira, L.F.; Francisco, M.R. A critical assessment of ex situ conservation based on the Brazilian avifauna: Are we focusing on what is easier? Perspect. Ecol. Conserv. 2023, 21, 62–70. [Google Scholar] [CrossRef]
- Berkunsky, I.; Daniele, G.; Kacoliris, F.P.; Díaz-Luque, J.A.; Silva Frias, C.P.; Aramburu, R.M.; Gilardi, J.D. Reproductive Parameters in the Critically Endangered Blue-Throated Macaw: Limits to the Recovery of a Parrot under Intensive Management. PLoS ONE 2014, 9, e99941. [Google Scholar] [CrossRef]
- Birdlife International IUCN Red List of Threatened Species. The IUCN Red List of Threatened Species. 2022. Available online: https://www.iucnredlist.org (accessed on 13 May 2023).
- Vergara-Tabares, D.L.; Cordier, J.M.; Landi, M.A.; Olah, G.; Nori, J. Global trends of habitat destruction and consequences for parrot conservation. Glob. Change Biol. 2020, 26, 4251–4262. [Google Scholar] [CrossRef] [PubMed]
- Olah, G.; Butchart, S.H.; Symes, A.; Guzmán, I.M.; Cunningham, R.; Brightsmith, D.J.; Heinsohn, R. Ecological and socio-economic factors affecting extinction risk in parrots. Biodivers. Conserv. 2016, 25, 205–223. [Google Scholar]
- ICMBio. Livro Vermelho da Fauna Brasileira Ameaçada de Extinção: Volume III—Aves. In Livro Vermelho da Fauna Brasileira Ameaçada de Extinção; Instituto Chico Mendes de Conservação da Biodiversidade. (Org.): Brasília, Brazil, 2018; Volume 3, pp. 299–301. [Google Scholar]
- Tella, J.L.; Blanco, G.; Carrete, M. Recent Advances in Parrot Research and Conservation. Diversity 2022, 14, 419. [Google Scholar] [CrossRef]
- Pacífico, E.C.; Barbosa, E.A.; Filadelfo, T.; Oliveira, K.G.; Silveira, L.F.; Tella, J.L. Breeding to non-breeding population ratio and breeding performance of the globally Endangered Lear’s Macaw Anodorhynchus leari: Conservation and monitoring implications. Bird Conserv. Int. 2014, 24, 466–476. [Google Scholar]
- Olah, G.; Vigo, G.; Heinsohn, R.; Brightsmith, D.J. Nest site selection and efficacy of artificial nests for breeding success of Scarlet Macaws Ara macao macao in lowland Peru. J. Nat. Conserv. 2014, 22, 176–185. [Google Scholar] [CrossRef]
- Scherer-Neto, P.; Guedes, N.M.R.; Toledo, M.C.B. Long-term monitoring of a hyacinth macaw Anodorhynchus hyacinthinus (Psittacidae) roost in the Pantanal, Brazil. Endanger. Species Res. 2019, 39, 25–34. [Google Scholar]
- Gautschi, D.; Heinsohn, R.; Crates, R.; Macgregor, N.A.; Wilson, M.; Stojanovic, D. Utilization of modified and artificial nests by endemic and introduced parrots on Norfolk Island. Restor. Ecol. 2022, 30, e13586. [Google Scholar] [CrossRef]
- Kanaan, V. Re-introduction of the vinaceous-breasted Amazon at the Araucárias National Park, Santa Catarina, Brazil. Glob. Re-Introd. Perspect. Case-Stud. Around Globe 2016, 106, 106–110. [Google Scholar]
- Lopes, A.R.; Rocha, M.S.; Mesquita, W.U.; Drumond, T.; Ferreira, N.F.; Camargos, R.A.; Vilela, D.A.R.; Azevedo, C.S. Translocation and post-release monitoring of captive-raised Blue-fronted Amazons (Amazona aestiva). Acta Ornithol. 2018, 53, 37–48. [Google Scholar] [CrossRef]
- Vilarta, M.R.; Wittkoff, W.; Lobato, C.; Oliveira, R.D.A.; Pereira, N.G.P.; Silveira, L.F. Reintroduction of the Golden Conure (Guaruba guarouba) in Northern Brazil: Establishing a Population in a Protected Area. Diversity 2021, 13, 198. [Google Scholar] [CrossRef]
- Donald, P.; Collar, N.; Marsden, S.; Pain, D. Facing Extinction: The World’s Rarest Birds and the Race to Save Them; T&AD Poyser: London, UK, 2010. [Google Scholar]
- White, T.H., Jr.; Abreu, W.; Benitez, G.; Jhonson, A.; Lopez, M.; Ramirez, L.; Rodriguez, I.; Toledo, M.; Torres, P.; Velez, J. Minimizing potential allee effects in psittacine reintroductions: An example from Puerto Rico. Diversity 2021, 13, 13. [Google Scholar] [CrossRef]
- Fraga, R.E.; Schiavetti, A.; Santos, C.M.R.; Silva, R.S.; Teixeira, R.E.R.; Tomazi, L.; dos Santos, C.Z. Reintroduction and monitoring of the bird Amazona aestiva (Psittaciformes: Psittacidae) in Brazil. Rev. Biol. Trop. 2021, 71, e53145. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A. A Primer of Conservation Genetics; Cambridge University Press: Cambridge, UK, 2004; p. 236. [Google Scholar]
- Frankham, R. Challenges and opportunities of genetic approaches to biological conservation. Biol Conserv. 2010, 143, 1919–1927. [Google Scholar] [CrossRef]
- Camargo, C.; Gibbs, H.L.; Costa, M.C.; Del-Rio, G.; Silveira, L.F.; Wasko, A.P.; Francisco, M.R. Marshes as “Mountain Tops”: Genetic analyses of the critically endangered São Paulo Marsh Antwren (Aves: Thamnophilidae). PLoS ONE 2015, 10, e0140145. [Google Scholar] [CrossRef] [Green Version]
- Oliveira-Jr, P.R.R.; Costa, M.C.; Silveira, L.F.; Francisco, M.R. Genetic guidelines for captive breeding and reintroductions of the endangered Black-fronted Piping Guan, Aburria jacutinga (Galliformes, Cracidae), an Atlantic Forest Endemic. Zoo Biol. 2016, 35, 313–318. [Google Scholar] [CrossRef]
- Costa, M.C.; Oliveira Jr, P.R.R.; Davanço, P.V.; Camargo, C.; Laganaro, N.M.; Azeredo, R.A.; Simpson, J.; Silveira, L.F.; Francisco, M.R. Recovering the genetic identity of an Extinctin-the-Wild species: The puzzling case of the Alagoas Curassow. PLoS ONE 2017, 12, e0169636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moritz, C. Defining “Evolutionarily Significant Units” for conservation. Trends Ecol. Evol. 1994, 9, 373–375. [Google Scholar] [CrossRef]
- Caparroz, R.; Miyaki, C.Y.; Baker, A.J. Contrasting phylogeographic patterns in mitochondrial DNA and microsatellites: Evidence of female philopatry and male-biased gene flow among regional populations of the Blue-and-yellow Macaw (Psittaciformes: Ara ararauna) in Brazil. Auk 2009, 126, 359–370. [Google Scholar] [CrossRef]
- Presti, F.T.; Guedes, N.M.R.; Antas, P.T.Z.; Miyaki, C.Y. Population genetic structure in Hyacinth Macaws (Anodorhynchus hyacinthinus) and identification of the probable origin of confiscated individuals. J. Hered. 2015, 106, 491–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fundação SOS Mata Atlântica; INPE. Atlas dos Remanescentes Florestais da Mata Atlântica: Período 2019/2020, Relatório Técnico; Fundação SOS Mata Atlântica: São Paulo, Brazil, 2021; 73p. [Google Scholar]
- Vale, M.M.; Tourinho, L.; Lorini, M.L.; Rajão, H.; Figueiredo, M.S. Endemic birds of the Atlantic Forest: Traits, conservation status, and patterns of biodiversity. J. Field Ornithol. 2018, 89, 193–206. [Google Scholar] [CrossRef]
- Sargatal, J.; del Hoyo, J.; Llobet, T.; Christie, D.A.; Collar, N. Family Psittacidae (parrots). In Handbook of the Birds of the World Sandgrouse to Cuckoos; Lynx Edicions: Barcelona, Spain, 1977; Volume 4, pp. 280–477. [Google Scholar]
- Juniper, T.; Parr, M. Parrots—A Guide to the Parrots of the World; Pica Press/Yale University Press: East Sussex, UK; New Haven, CT, USA, 1998. [Google Scholar]
- Pinto, O.M.D.O. 1ª Parte aves não Passeriformes e Passeriformes não Oscines, com exclusão da família Tyrannidae. In Novo catálogo das Aves do Brasil; Biodiversity Heritage Library: Washington, DC, USA, 1978. [Google Scholar]
- Martinez, J.; Prestes, N.P. Biologia da Conservação: Programa Nacional para a Conservação do Papagaio-de-Peito-Roxo e Outras Iniciativas; Editora LEW: Tapera, Brasil, 2021. [Google Scholar]
- Klemann-Júnior, L.; Shimakura, S.E.; Ribeiro Junior, P.J.; Scherer-Neto, P.; Passos, F.C. Different land use types influence Red-browed Amazon (Amazona rhodocorytha) ccurrence in Southeastern Brazil. An. Acad. Bras. Ciências 2022, 94, e20191227. [Google Scholar]
- Marsden, S.J.; Whiffin, M.; Sadgrove, L.; Guimarães, P., Jr. Parrot populations and habitat use in and around two lowland Atlantic Forest reserves, Brazil. Biol. Conserv. 2000, 96, 209–217. [Google Scholar] [CrossRef] [Green Version]
- Klemann-Júnior, L.; Neto, P.S.; Monteiro, T.V.; Ramos, F.M.; de Almeida, R. Mapeamento da distribuição e conservação do chauá (Amazona rhodocorytha) no estado do Espírito Santo, Brasil. Ornitol. Neotrop. 2008, 19, 183–196. [Google Scholar]
- Zulian, V.; Miller, D.A.; Ferraz, G. Endemic and Threatened Amazona Parrots of the Atlantic Forest: An Overview of Their Geographic Range and Population Size. Diversity 2021, 13, 416. [Google Scholar]
- Destro, G.F.G.; Pimentel, T.L.; Sabaini, R.M.; Borges, R.C.; Barreto, R. Efforts to combat wild animals trafficking in Brazil. Biodivers. Enrich. Divers. World 2012, 1, 421–436. [Google Scholar]
- MMA, Ministério do Meio Ambiente. Operação AVEAS CORPUS PF/IBAMA (Relatório de Fiscalização Ambiental No. I397QQB); Ministério do Meio Ambiente: Vitória, Spain, 2021. [Google Scholar]
- Faircloth, B.C.; McCormack, J.E.; Crawford, N.G.; Harvey, M.G.; Brumfield, R.T.; Glenn, T.C. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 2012, 61, 717–726. [Google Scholar] [CrossRef]
- Faircloth, B.C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 2016, 32, 786–788. [Google Scholar] [CrossRef] [Green Version]
- Faircloth, B.C. Illumiprocessor: A Trimmomatic Wrapper for Parallel Adapter and Quality Trimming. 2013. Available online: https://github.com/faircloth-lab/illumiprocessor (accessed on 3 January 2023).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocalini, F.; Bolivar-Leguizamon, S.D.; Silveira, L.F.; Bravo, G.A. Comparative phylogeographic and demographic analyses reveal a congruent pattern of sister relationships between bird populations of the northern and south-central Atlantic Forest. Mol. Phylogenet. Evol. 2021, 154, 106973. [Google Scholar] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- De Jong, M.J.; de Jong, J.F.; Hoelzel, A.R.; Janke, A. SambaR: An R package for fast, easy and reproducible population-genetic analyses of biallelic SNP data sets. Mol. Ecol. Resour. 2021, 21, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jombart, T.; Ahmed, I. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 2011, 27, 3070–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Leary, S.J.; Puritz, J.B.; Willis, S.C.; Hollenbeck, C.M.; Portnoy, D.S. These aren’t the loci you’re looking for: Principles of effective SNP filtering for molecular ecologists. Mol. Ecol. 2018, 27, 3193–3206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kardos, M.; Luikart, G.; Allendorf, F.W. Measuring individual inbreeding in the age of genomics: Marker-based measures are better than pedigrees. Heredity 2015, 115, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.K.; Albrechtsen, A.; Moltke, I. Allele frequency-free inference of close familial relationships from genotypes or low-depth sequencing data. Mol. Ecol. 2019, 28, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef] [PubMed]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Berkunsky, I.; Reboreda, J.C. Nest-site fidelity and cavity reoccupation by Blue-fronted Parrots Amazona aestiva in the dry Chaco of Argentina. Ibis 2009, 151, 145–150. [Google Scholar] [CrossRef]
- Francisco, M.R.; Gibbs, H.L.; Galetti, M.; Lunardi, V.O.; Galetti, P.M. Genetic structure in a tropical lek-breeding bird, the Blue Manakin (Chiroxiphia caudata) in the Brazilian Atlantic Forest. Mol. Ecol. 2007, 16, 4908–4918. [Google Scholar] [CrossRef] [PubMed]
- Medolago, C.A.B.; Costa, M.C.; Silveira, L.F.; Francisco, M.R. Hybridization between two recently diverged Neotropical passerines: The Pearly-bellied Seedeater Sporophila pileata, and the Copper Seedeater S. bouvreuil (Aves, Passeriformes, Thraupidae). PLoS ONE 2020, 15, e0229714. [Google Scholar] [CrossRef] [PubMed]
| Individual | Name | Age | Sex | Ho | He | F | p-Value |
|---|---|---|---|---|---|---|---|
| 1 | 707_MZUSP_a2572_Ama_rho | Juvenile | F | 0.363 | 0.243 | 0.037 | 0.096166 |
| 2 | 708_MZUSP_a2554_Ama_rho | Juvenile | F | 0.533 | 0.243 | −0.416 | 0 |
| 3 | 709_MZUSP_a2563_Ama_rho | Subadult | M | 0.337 | 0.243 | 0.106 | 0.000002 |
| 4 | 710_MZUSP_a2564_Ama_rho | Juvenile | M | 0.262 | 0.243 | 0.304 | 0 |
| 5 | 711_MZUSP_a2556_Ama_rho | Juvenile | F | 0.476 | 0.243 | −0.263 | 0 |
| 6 | 712_MZUSP_a2557_Ama_rho | Juvenile | F | 0.470 | 0.243 | −0.248 | 0 |
| 7 | 713_MZUSP_a2560_Ama_rho | Juvenile | F | 0.521 | 0.243 | −0.381 | 0 |
| 8 | 714_MZUSP_a235_Ama_rho | Adult | M | 0.347 | 0.243 | 0.080 | 0.000335 |
| 9 | 715_MZUSP_a2568_Ama_rho | Subadult | F | 0.462 | 0.243 | −0.227 | 0 |
| 10 | 716_MZUSP_a2570_Ama_rho | Adult | F | 0.472 | 0.243 | −0.254 | 0 |
| 11 | 717_MZUSP_a2553_Ama_rho | Juvenile | F | 0.475 | 0.243 | −0.260 | 0 |
| 12 | 718_MZUSP_a2562_Ama_rho | Juvenile | M | 0.333 | 0.243 | 0.115 | 0 |
| 13 | 719_MZUSP_a2575_Ama_rho | Juvenile | M | 0.510 | 0.243 | −0.354 | 0 |
| 14 | 720_MZUSP_a2568_Ama_rho | Adult | F | 0.464 | 0.243 | −0.232 | 0 |
| 15 | 721_MZUSP_a2567_Ama_rho | Juvenile | M | 0.503 | 0.243 | −0.334 | 0 |
| 16 | 722_MZUSP_a2558_Ama_rho | Juvenile | M | 0.497 | 0.243 | −0.319 | 0 |
| 17 | 723_MZUSP_a2573_Ama_rho | Juvenile | M | 0.344 | 0.243 | 0.088 | 0.000079 |
| 18 | 724_MZUSP_a2559_Ama_rho | Juvenile | M | 0.337 | 0.243 | 0.107 | 0.000002 |
| 19 | 725_MZUSP_a2574_Ama_rho | Juvenile | M | 0.350 | 0.243 | 0.070 | 0.001761 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agazzi Migotto, A.; Bocalini, F.; Francisco, M.R.; Reillo, P.; Silveira, L.F. Genetic Variability and Kinship Analyses of Seized Red-Browed Amazon, Amazona rhodocorytha (Aves, Psittacidae). Diversity 2023, 15, 923. https://doi.org/10.3390/d15080923
Agazzi Migotto A, Bocalini F, Francisco MR, Reillo P, Silveira LF. Genetic Variability and Kinship Analyses of Seized Red-Browed Amazon, Amazona rhodocorytha (Aves, Psittacidae). Diversity. 2023; 15(8):923. https://doi.org/10.3390/d15080923
Chicago/Turabian StyleAgazzi Migotto, Anna, Fernanda Bocalini, Mercival Roberto Francisco, Paul Reillo, and Luís Fábio Silveira. 2023. "Genetic Variability and Kinship Analyses of Seized Red-Browed Amazon, Amazona rhodocorytha (Aves, Psittacidae)" Diversity 15, no. 8: 923. https://doi.org/10.3390/d15080923
APA StyleAgazzi Migotto, A., Bocalini, F., Francisco, M. R., Reillo, P., & Silveira, L. F. (2023). Genetic Variability and Kinship Analyses of Seized Red-Browed Amazon, Amazona rhodocorytha (Aves, Psittacidae). Diversity, 15(8), 923. https://doi.org/10.3390/d15080923









