Abstract
The genetic structure and dispersal dynamics of reptile populations are profoundly influenced by natural processes and human activities. While natural dispersal is shaped by species’ characteristics and paleogeographical features, human-mediated translocations have become increasingly prevalent, posing ecological challenges. Mitochondrial genetic markers have been pivotal in untangling invasion pathways for various species. Our study focuses on the Dalmatian Algyroides, Algyroides nigropunctatus (Duméril & Bibron, 1839), a lizard species endemic to the Balkan Peninsula, where recent observations in the Apulian region of Italy suggest an introduced population. Genetic analyses employing two mtDNA markers (16S and ND4 genes) elucidate the phylogenetic relationships of the Dalmatian Algyroides and trace the geographic origin of the introduced population. Our findings reveal areas in western Greece and southwestern Albania as the most probable areas of the source population, while we identify two previously undetected geographical lineages in the native range, highlighting the complex evolutionary history of the species in the region. Additionally, indications of potential glacial refugia and post-glacial dispersal patterns shed more light on the species’ demographic dynamics.
1. Introduction
Past geological and climatic events, including ice ages, have shaped the distribution of multiple European species and acted as pivotal factors in genetic differentiation and speciation events that took place during periods of isolation within glacial refugia [1]. However, with the onset of the Anthropocene, increased human-mediated translocations of species beyond their native range have been altering the results of such processes by eliminating barriers among allopatric taxa. Introductions of non-native species have triggered negative ecological impacts including increased competition, altered predator-prey relationships, hybridization events and the spread of pathogens, compromising conservation efforts at the expense of native biodiversity [2,3,4,5,6,7]. Among the most widespread and translocated species are reptiles, often unintentionally moved by human activities due to their habit of living in modified environments and finding refuge in small cavities and crevices. This passive transportation has significant direct and indirect impacts on ecosystems and native species, including native reptiles [8,9]. Currently, the origin of introduced species can be effectively investigated through the employment of genetic tools facilitating the identification of the source populations and assisting the reconstruction of the history and route of an invasion [10,11,12]. Cases of biological invasions for various lizard species have been disentangled using mitochondrial genetic markers, due to their rapid evolutionary rate, lack of recombination and maternal inheritance, and such markers have been used to monitor introduced species along different invasion pathways [13,14,15]. For instance, analyses of mitochondrial genes have revealed several independent introductions of the Italian wall lizard (Podarcis siculus) outside its native range, which is restricted to the Italian Peninsula and Sicily, to multiple locations either via the pet trade or unintentional translocations through shipments [16,17,18]. These studies took advantage of previous knowledge and strong phylogeographic structure in the species’ native range [19]. It is therefore evident that in order to understand human-mediated dispersal patterns, the use of genetic markers is of utmost importance for the accurate identification and assignment to likely sources and introduction pathways of alien species and populations.
Recently, a population of another lizard species, the Dalmatian Algyroides Algyroides nigropunctatus (Duméril & Bibron, 1839), has been observed in the Apulian region in Italy, far outside its native range [20]. Interestingly, another allochthonous population has recently been reported from a suburb region in Athens, suggesting some invasive capacity in this species, since it is considered—among other lacertid lizards as well—as being capable to establish viable populations, even far outside its native range [21]. The Dalmatian Algyroides is a lacertid lizard sub-endemic to the Balkan peninsula, and its native distribution expands from north-eastern Italy down to the Gulf of Corinth in Greece, including the Ionian islands of Corfu, Diapontia Islets, Paxoi, Lefkada, Kefalonia and Ithaca [22,23,24,25,26]. The Balkan peninsula has long been recognized as an important center of diversification and represents the origin of postglacial dispersal movements towards central and northern Europe, with several species forming distinct lineages and exhibiting complex evolutionary histories [27,28,29,30]. The Dalmatian Algyroides is no exception, with molecular studies indicating the existence of three major phylogenetic clades, the Adria-Pindos clade showing the widest distribution, the northern Ionian clade from Parga and the Island of Corfu, and the southern Ionian clade where the A. n. kephalithacius is found, highlighting the importance of the Balkan peninsula as a glacial refugium [31,32]. The species is considered highly differentiated within Greece, showing high genetic diversity in the southern parts of its distribution while, furthermore, spatial distribution analysis indicates the existence of a glacial refugium in southwestern Greece [32]. However, a more intensive sampling scheme throughout the species’ distribution, especially in the western parts of its current range, is needed to fully resolve the phylogeographic patterns in this species.
To this end, we aim to elucidate the phylogeographic patterns of Algyroides nigropunctatus populations in the Balkan Peninsula along the species’ southwestern distribution using mitochondrial genetic markers, commonly used in resolving intra-specific phylogenies, and determine the origin of the likely introduced Italian population outside its native range.
2. Materials and Methods
2.1. Sample Collection and Laboratory Procedures
In total, 95 A. nigropunctatus specimens from the whole distribution range of the species and 1 A. moreoticus specimen were collected between 2013 and 2019 (Figure 1, Table S1).
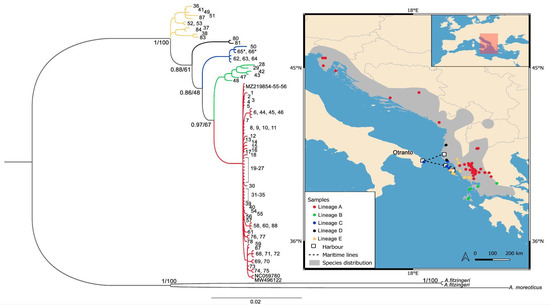
Figure 1.
Bayesian tree based on the concatenated mitochondrial DNA dataset (16S + ND4). Bayesian posterior probabilities followed by Maximum Likelihood bootstrap support values are shown on nodes. Lineages C and D are newly detected in the present study. Asterisks indicate the samples from the introduced Italian population belonging to lineage C. Distribution map and sample localities (numbers) are shown according to Table S1. Previously published sequences are indicated by GenBank accession numbers. Colors represent mitochondrial lineages. The map was created using QGIS v. 3.22 software [33].
Geographic data were recorded with GPS for each caught animal while tail tissues were collected and stored in ethanol. Total genomic DNA was extracted from the tissue samples following standard extraction protocols [34]. Two mitochondrial gene fragments, commonly used in resolving intra-specific phylogenies, the ND4 (NADH dehydrogenase subunit 4) mtDNA gene with the adjacent tRNAs (ND4 + His, Ser, Leu) and the ribosomal 16S rRNA gene, were amplified using the primers described in [35] and [36], respectively. PCR amplifications were performed in a total volume of 20 μL, with 4 μL of 5 X reaction Buffer, 2.4 μL of 25 mM MgCl2, 0.4 μL of 10 mM dNTPs, 0.8 μL of 5.0 μM of each primer, and 0.2 μL of Kapa Taq DNA Polymerase (Kapa Biosystems, Cape Town, South Africa). Thermocycling conditions consisted of an initial denaturation at 94 °C for 300 s, 35 cycles of 94 °C for 60 s, primer annealing at 54 °C for 60 s for ND4 and 47 °C for 60 s for 16S, followed by primer extension at 72 °C for 60 s and a final extension of 72 °C for 300 s. PCR products were purified using the NucleoSpin ExtractII (Macherey-Nagel, Düren, Germany) cleanup kit and single-strand sequencing was conducted by CEMIA (Cellular & Molecular Immunological Applications, Larisa, Greece).
2.2. Phylogenetic and Demographic Analyses
Sequences were edited in MEGA v.11 [37] and aligned with CLUSTAL W [38]. Additionally, available ND4 and 16S sequences from previous studies were retrieved through GenBank (Table S1), while four Algyroides fitzingeri sequences were used as outgroups along with sequences from an A. moreoticus (Table S1) [31,39,40,41,42,43]. The number of haplotypes, haplotypic diversity and nucleotide diversity parameters were estimated with DnaSP v.5 [44], and the relationships among the detected haplotypes were visualized with a Median Joining (MJ) network [45] in PopArt [46]. Genetic uncorrected p-distances and nucleotide diversity calculations (Φst) were calculated in MEGA v.11 and Arlequin v.3.5 [47] between each detected lineage. Phylogenetic inference was conducted using a concatenated dataset including both genes, and the best-fit partitioning schemes and nucleotide models were determined according to PartitionFinder v1.1.1 [48] using the Akaike Information Criterion (AIC). Bayesian Inference (BI) analysis was performed with MrBayes v. 3.2.7a [49] under the Markov chain Monte Carlo method (MCMC), and the models for the two partitions were K80 + I for 16S and HKY + I for ND4. Two independent runs with four chains were conducted for 107 generations; sampling was carried out every 500 steps and 25% of the sampled trees were discarded as burn-in. Maximum Likelihood (ML) analysis was carried out using RaxML v. 8.2.12 [50], and the tree was inferred using the GTR + G substitution model, while bootstrap support was assessed by 1000 replicates. The resulting mitogenomic phylogenies were visualized in FigTree v.1.4.4 [51]. In order to reconstruct historical demography and detect possible expansion events and changes in population size, a Mismatch distribution analysis was performed in DnaSP v5 [44] and Arlequin v. 3.5. for the detected lineages consisting of more than 10 sequences in the concatenated dataset.
3. Results
The analysis of the 16S fragment included 494 bp (variable sites = 90, parsimony informative = 60) from 109 individuals excluding the outgroups and revealed 23 haplotypes with high haplotype diversity (Hd ± SD: 0.774 ± 0.002). A total of 17 newly sequenced haplotypes were added to the six existing haplotypes covering 23 different localities across five countries (Figure 1). The MJ network based on the 16S revealed five lineages as the reconstructed phylogenetic tree, allowing to assign each haplotype to the respective geographical clade (Figure 1 and Figure 2).
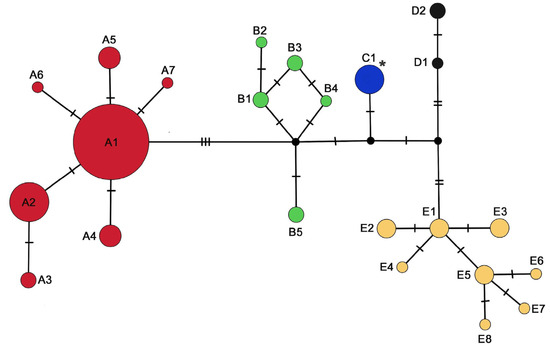
Figure 2.
Median joining network of the 16S gene set. Circles correspond to haplotypes, with size proportional to the number of individuals per haplotype. Vertical lines correspond to mutational steps. The asterisk indicates the haplotype detected in the introduced Italian population. Colors represent mitochondrial lineages according to Figure 1 and Table S1.
The ND4 mtDNA alignments included 702 bp (variable sites = 208, parsimony informative = 147) from 89 individuals excluding the outgroups, revealing 36 haplotypes with high levels of haplotypic diversity (Hd ± SD: 0.805 ± 0.002). A total of 34 newly sequenced haplotypes were detected from 33 different localities covering the species distribution (Figure 3), while both the MJ network and phylogenetic tree revealed five lineages (Figure 1 and Figure 3).
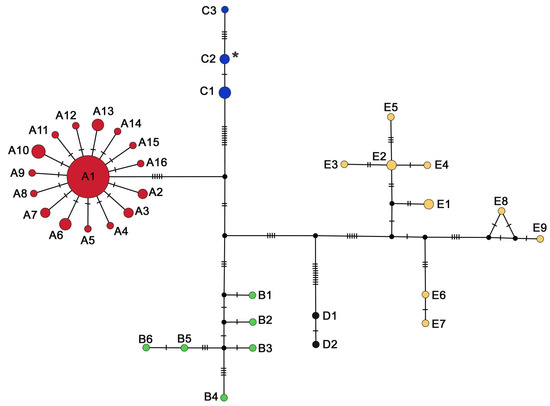
Figure 3.
Median joining network of the ND4 gene set. Circles correspond to haplotypes, with size proportional to the number of individuals per haplotype. Vertical lines correspond to mutational steps. The asterisk indicates the haplotype detected in the introduced Italian population. Colors represent mitochondrial lineages according to Figure 1 and Table S1.
The uncorrected genetic divergences (p-distances) between the detected lineages range from 2.0% to 3.3% for ND4 and from 0.7% to 1.8% for 16S (Table 1), while the respective between-lineage divergence values (Φst) are presented in Table S2.

Table 1.
Average genetic distances (% p-distances) between the main lineages for ND4 (above the diagonal) and 16S (below the diagonal). Genetic distances within lineages are shown diagonally in bold for 16S/ND4, respectively.
The concatenated dataset included 89 individuals comprising a total alignment length of 1196 bp. Both Bayesian Inference and Maximum Likelihood analysis recovered almost identical topologies revealing five mitochondrial lineages (Figure 1). However, most of the nodes display low to moderate support for both approaches (Figure 1). Lineage A (Adria/Pindos Clade) has the widest expansion, ranging across the mainland from Pindos (NW Greece) to Slovenia. Lineage B corresponds to the Southern Ionian Clade and comprises individuals from Aetoloakarnania and the Ionian islands of Kefalonia and Lefkada. The Northern Ionian region includes members of both Lineages C and E. Individuals from north Corfu, Erikoussa Island and southwest Albania belong to the here newly reported Lineage C. Lineage E is further divided in two sub-lineages: the first comprises individuals from south Albania and Corfu Island, and the second comprises individuals from the adjacent coast in Greece (Figure 1, Table S1). The newly detected Lineage D is located in the regions of Llogare and Divjake, in Albania. The recently discovered population in Otranto, Italy [20], belongs to Lineage C along with individuals from Erikousa and northern Corfu. The population belongs to the C1 and C2 haplotypes, according to the 16S and ND4 gene sets, respectively (Table S1).
Both gene sets reveal a star-like haplotypic network for Lineage A (Figure 2 and Figure 3), surrounded by several closely related haplotypes with low nucleotide differences (Hd = 0.729, Pi = 0.002) (Table S3, Figure S1). At the same time, Lineage E shows high haplotype and low nucleotide diversity (Hd = 0.982, Pi = 0.007), while the mismatch distribution analysis and neutrality tests reveal a multimodal distribution (Table S3, Figure S1).
4. Discussion
Our approach identified multiple new haplotypes, allowing us to further elucidate the phylogenetic relationships of the Dalmatian Algyroides in the Balkan peninsula, while the employment of mitochondrial genetic markers untangled the origin and phylogeographic position of the population in question from southern Italy. The population was discovered in southern Italy quite recently; its significant geographic separation from the nearest native population from NE Italy (more than 900 km overland; Figure 1), combined with the absence of any relevant historical records, strongly suggests that these individuals were introduced to the area rather than actively dispersed.
Our phylogenetic analysis assigned the individuals from the Apulia region in Italy to the same lineage with individuals from southwestern Albania, Corfu and Erikousa Island, hence corroborating the introduced status of this population, since the 16S haplotype found is identical to the one found in samples originating from those areas. Regarding the ND4 sequences, the Apulian population bears a different haplotype from northern Corfu and Erikousa, while the respective sequence from the southwestern parts of Albania was not successfully amplified in the present study. More specifically, the northern Corfu population shows a highly divergent haplotype (haplotype C3), therefore it is unlikely that individuals were translocated from Corfu. These areas were not connected even during the marine regression of the last glacial maximum, although the distribution models identified that area as ecologically suitable for the species [32]. As such, we conclude that the population might have originated from either Erikousa or Albania, with known shipment connections to the port of Otranto (Figure 1). These translocations could have been mediated through recreational yachts and fishing vessels towards Italy. A more robust scenario involves translocation events through the regular maritime connections carrying construction materials from Vlore harbor in southwestern Albania to Otranto [20]. Indeed, these shipments could favor the unintentional movements of saxicolous reptiles despite them being typically sedentary with low dispersal capabilities, since their use of small cavities offers high resilience in harsh environments [9]. Indeed, Algyroides nigropunctatus is frequently found in humanized areas [52,53] and is highly resistant to dehydration compared to other members of the genus such as A. moreoticus [54].
Biological invasions are multistep processes encompassing translocation and a successful establishment followed by further dispersal across the new geographical area. The integration of multiple information pathways, monitoring schemes and genetic data is vital in elucidating the success of translocations and alien species distribution and their persistence in changing environments [55].
Regarding the phylogenetic relationships of the Dalmatian Algyroides, along with the previously identified mitochondrial lineages of Adria-Pindos, Southern Ionian and Northern Ionian clades [31,32], our mitochondrial analysis revealed the existence of additional and previously undetected geographic lineages in the southwestern distribution of the species (Lineages C and D; Figure 1), indicative of the complex history of the species in north-western Greece. Albeit their low/moderate node support (e.g., [56]), their clear separation is further supported by the network analyses (Figure 2 and Figure 3). However, this low support might indicate the retention of ancestral polymorphisms during recent cladogenetic events by incomplete lineage sorting [57,58].
Assuming an evolutionary rate for the ND4-tRNA (Leu) fragment of 1.74–2.78% per million years, as proposed by Pinho et al. [59] and applied in Iberian Algyroides by Rato et al. [43], the extent of between-lineage genetic divergence (Table 1), suggests that lineage splitting might have occurred approximately 0.72–1.89 Mya, corresponding to Pleistocene’s climatic oscillations.
During the Last Glacial Maximum, the Balkan Peninsula provided more stable climatic conditions, offering refuge and suitable habitats for several species, supporting diverse ecosystems and serving as a center of biodiversity [29,60,61]. In a recent study, they identified suitable ecological habitats for the Dalmatian Algyroides during the Mid-Holocene and Last Glacial Maximum in southwestern Albania and western Greece, providing evidence of refugia in the area where the species might have occurred and perpetuated itself during the harsh environmental conditions [32]. Indeed, sufficient time must have elapsed in order to accumulate the large number of mutational steps separating the haplotypes among the detected mitochondrial lineages in the present study [62], supporting the existence of multiple refugia in the area and highlighting the importance of the Balkan Peninsula as a center of diversification.
At the end of the glacial oscillations, the Balkan Peninsula served as the basis of northward expansion, providing colonization routes that allowed many animal and plant species to gradually extend their distributions into central and northern Europe [1]. This has led to patterns of higher diversification in southern populations compared to northern ones, a trend observed in various species in the Balkan Peninsula, particularly in Greece [63,64,65,66]. The elevation of sea level at the end of the Pleistocene, however, may have limited that northern expansion to coastal areas and impeded the colonization of the Italian side of the Adriatic, which includes ecologically suitable areas such as the introduction site [32]. Our analysis corroborates this pattern to the Dalmatian Algyroides as well for the Adria/Pindos lineage (Lineage A), which shows the widest distribution while the combination of high haplotype and low nucleotide diversity might reflect a rapid demographic growth from an initially small population in the area of Pindos massif (Table S2, Figure S1), which also acted as a glacial refugium in western Greece [32].
So far, only lineage B has been found to exhibit a distinct phenotype, members of which have been conventionally classified as belonging to subspecies A. n. kephalithacius [31,32]. Future research on the phenotypic distinctiveness of the remaining mitochondrial lineages would assist considerably on the taxonomical resolution of the Dalmatian Algyroides.
5. Conclusions
Our study elucidates the origin of the introduced population of the Dalmatian Algyroides outside its native range and offers new insights into the phylogeography of the species. Specifically, we identified two new mitochondrial lineages with deep phylogenetic splits suggesting the persistence of old populations for the Dalmatian Algyroides in the southwestern parts of its distribution, while at the same time demonstrating that they constitute the origin of the recently reported introduced population in the Italian Peninsula. To further reconstruct the invasion history and assess the processes leading to invasive success, fast-evolving genetic markers at the population level should be employed with extensive sampling efforts in the area of Vlore, which constitutes the probable origin area of the translocated population. The present study highlights the importance of the Balkan Peninsula and particularly the area of western Greece as a refuge for the Dalmatian Algyroides and its diverse phylogenetic relationships.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16070406/s1, Figure S1: Mismatch distribution analysis showing unimodal and multimodal distribution (non-significant) for Clades A and E, respectively, based on the concatenated dataset; Table S1: Sample codes, species, sampling locality and sequences’ GenBank accession numbers used in the present study. Haplotypes for the 16S and ND4 gene fragments according to the present study: no haplotypes were retrieved; Table S2: Mismatch distribution analysis and neutrality tests based on the concatenated datasets for clades with over 10 individuals. Statistically significant results are shown in bold; Table S3: Mismatch distribution analysis and neutrality tests based on the concatenated datasets for lineages with over 10 individuals. Statistically significant results are shown in bold.
Author Contributions
Conceptualization, K.S., P.C. and A.B.; methodology, K.S. and A.B.; software, E.-A.T. and D.S.; validation, K.S.; formal analysis, E.-A.T. and D.S.; investigation, P.C., D.S., E.-A.T., A.B., C.P., M.A.C., R.C., D.J.H., O.S.G.P., L.L. and K.S.; resources, K.S. and P.C.; data curation, D.S., E.-A.T. and A.B.; writing—original draft preparation, E.-A.T. and D.S.; writing—review and editing, E.-A.T., D.S., A.B., P.C., C.P., M.A.C., R.C., D.J.H., O.S.G.P., L.L. and K.S; visualization, E.-A.T., D.S. and A.B.; supervision, K.S.; project administration, K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Sampling was conducted under special permits issued by the Hellenic Ministry of the Environment (ΩKZT4653Π8-422), the National Agency of Protected Areas-Ministry of Tourism and Environment of Albania (481/16-4-2019), the Environmental Protection Agency of Montenegro (UPI-584/4, UPI-101/2-02-824/6) and the Slovenian Environment Agency (no. 35601-47/2011-6).
Data Availability Statement
The novel sequences obtained in the present study are openly available in GenBank (Accession numbers: PP776044–PP776060 and PP947930 for 16S; PP784216–PP784249 and PP955415 for ND4; Table S1).
Acknowledgments
Adnan Zimić (National Museum of Bosnia and Herzegovina) kindly provided the sample from Bosnia. Three anonymous reviewers provided insightful comments on the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hewitt, G.M. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 1996, 58, 247–276. [Google Scholar] [CrossRef]
- Sakai, A.K.; Allendorf, F.W.; Holt, J.S.; Lodge, D.M.; Molofsky, J.; With, K.A.; Baughman, S.; Cabin, R.J.; Cohen, J.E.; Ellstrand, N.C.; et al. The population biology of invasive species. Annu. Rev. Ecol. Syst. 2001, 2, 305–332. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Lundquist, L.L. Introduction: Population biology, evolution, and control of invasive species. Conserv. Biol. 2003, 17, 24–30. [Google Scholar] [CrossRef]
- Storfer, A.; Murphy, M.A.; Spear, S.F.; Holderegger, R.; Waits, L.P. Landscape genetics: Where are we now? Mol. Ecol. 2010, 19, 3496–3514. [Google Scholar] [CrossRef]
- Vila, M.; Basnou, C.; Pysek, P.; Josefsson, M.; Genovesi, P.; Gollasch, S.; Nentwig, W.; Olenin, S.; Roques, A.; Roy, D.; et al. How well do we understand the impacts of alien species on ecosystem services? A pan-European, cross-taxa assessment. Front. Ecol. Environ. 2010, 8, 135–144. [Google Scholar] [CrossRef]
- Crispo, E.; Moore, J.S.; Lee-Yaw, J.A.; Gray, S.M.; Haller, B.C. Broken barriers: Human-induced changes to gene flow and introgression in animals: An examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays 2011, 33, 508–518. [Google Scholar] [CrossRef]
- Capinha, C.; Essl, F.; Seebens, H.; Moser, D.; Pereira, H.M. The dispersal of alien species redefines biogeography in the Anthropocene. Science 2015, 348, 1248–1251. [Google Scholar] [CrossRef]
- Kraus, F. Alien Reptiles and Amphibians: A Scientific Compendium and Analysis; Invading Nature—Springer Series in Invasion Ecology; Springer: Dordrecht, The Netherlands, 2007; Volume 4, pp. 133–369. [Google Scholar] [CrossRef]
- Silva-Rocha, I.R.; Salvi, D.; Carretero, M.A.; Ficetola, G.F. Alien reptiles on Mediterranean Islands: A model for invasion biogeography. Divers. Distrib. 2019, 25, 995–1005. [Google Scholar] [CrossRef]
- Fitzpatrick, B.M.; Fordyce, J.A.; Niemiller, M.L.; Reynolds, R.G. What can DNA tell us about biological invasions? Biol. Invasions 2012, 14, 245–253. [Google Scholar] [CrossRef]
- Cristescu, M.E. Genetic reconstructions of invasion history. In: Invasion Genetics: The Baker and Stebbins Legacy. Mol. Ecol. 2016, 24, 267–282. [Google Scholar] [CrossRef]
- Oficialdegui, F.J.; Clavero, M.; Sánchez, M.I.; Green, A.J.; Boyero, L.; Michot, T.C.; Klose, K.; Kawai, T.; Lejeusne, C. Unravelling the global invasion routes of a worldwide invader, the red swamp crayfish (Procambarus clarkii). Freshw. Biol. 2019, 64, 1382–1400. [Google Scholar] [CrossRef]
- Eales, J.; Thorpe, R.S. Revealing the geographic origin of an invasive lizard: The problem of native population genetic diversity. Biol. Invasions 2010, 12, 77–86. [Google Scholar] [CrossRef]
- Michaelides, S.; While, G.M.; Bell, C.; Uller, T. Human introductions create opportunities for intra-specific hybridization in an alien lizard. Biol. Invasions 2013, 15, 1101–1112. [Google Scholar] [CrossRef]
- Santos, J.L.; Žagar, A.; Drašler, K.; Rato, C.; Ayres, C.; Harris, D.J.; Carretero, M.A.; Salvi, D. Phylogeographic evidence for multiple long-distance introductions of the common wall lizard associated with human trade and transport. Amphib.-Reptil. 2019, 40, 121–127. [Google Scholar] [CrossRef]
- Silva-Rocha, I.R.; Salvi, D.; Carretero, M.A. Genetic data reveal a multiple origin for the populations of the Italian wall lizard Podarcis sicula (Squamata: Lacertidae) introduced in the Iberian Peninsula and Balearic islands. Ital. J. Zool. 2012, 79, 502–510. [Google Scholar] [CrossRef]
- Silva-Rocha, I.R.; Salvi, D.; Harris, D.J.; Freits, S.; Davis, C.; Foster, J.; Deichsel, G.; Adamopoulou, C.; Carretero, M.A. Molecular assessment of Podarcis sicula populations in Britain, Greece and Turkey reinforces a multiple-origin invasion pattern in this species. Acta Herpetol. 2014, 9, 253–258. [Google Scholar] [CrossRef]
- Oskyrko, O.; Sreelatha, L.B.; Hanke, G.F.; Deichsel, G.; Carretero, M.A. Origin of introduced Italian wall lizards, Podarcis siculus (rafinesque-schmaltz, 1810) (Squamata: Lacertidae), in North America. BioInvasions Rec. 2022, 11, 1095–1106. [Google Scholar] [CrossRef]
- Senczuk, G.; Colangelo, P.; De Simone, E.; Aloise, G.; Castiglia, R. A combination of long term fragmentation and glacial persistence drove the evolutionary history of the Italian wall lizard Podarcis siculus. BMC Evol. Biol. 2017, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Carlino, P.; Pauwels, O.S.G. Taxonomic identity of an extralimital population of Algyroides Lizards (Squamata: Lacertidae) from Apulia Region in Southern Italy. Bull. Chic. Herpetol. Soc. 2016, 51, 149–151. [Google Scholar]
- Deimezis-Tsikoutas, A.; Capsalas, G.; Antonopoulos, A.; Strachinis, I.; Pafilis, P. Algyroides nigropunctatus (Squamata: Lacertidae) in the city of Athens: An unexpected finding. Russ. J. Herpetol. 2020, 27, 172–174. [Google Scholar] [CrossRef]
- Arnold, E.N.; Ovenden, D. A Field Guide to the Reptiles and Amphibians of Britain and Europe, 2nd ed.; Collins: London, UK, 1978. [Google Scholar]
- Chondropoulos, B.P. A checklist of the Greek reptiles. I. The lizards. Amphib.-Reptil. 1986, 7, 217–235. [Google Scholar] [CrossRef]
- Valakos, E.; Pafilis, P.; Sotiropoulos, K.; Lymberakis, P.; Maragou, P.; Foufopoulos, J. The Amphibians and Reptiles of Greece; Chimaira: Frankfurt, Germany, 2008. [Google Scholar]
- Wilson, M.; Stille, B.; Stille, M. Herpetofauna of Paxos, Ionian Islands, Greece, including two species new to the island. Herpetozoa 2014, 27, 108–112. [Google Scholar]
- Stille, B.; Stille, M. The Herpetofauna of Corfu and Adjacent Islands; Chimaira: Frankfurt, Germany, 2017. [Google Scholar]
- Griffiths, H.I.; Krystufek, B.; Reed, J.M. Balkan Biodiversity: Pattern and Process in the European Hotspot; Springer: Dordrecht, The Netherlands, 2004. [Google Scholar]
- Lymberakis, P.; Poulakakis, N. Three continents claiming an archipelago: The evolution of Aegean’s herpetofaunal diversity. Diversity 2010, 2, 233–255. [Google Scholar] [CrossRef]
- Hewitt, G.M. Mediterranean peninsulas: The evolution of hotspots. In Biodiversity Hotspots: Distribution and Protection of Conservation Priority Areas; Zachos, F., Habel, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 123–147. [Google Scholar]
- Psonis, N.; Antoniou, A.; Karameta, E.; Leaché, A.D.; Kotsakiozi, P.; Darriba, D.; Kozlov, A.; Stamatakis, A.; Poursanidis, D.; Kukushkin, O.; et al. Resolving complex phylogeographic patterns in the Balkan Peninsula using closely related wall-lizard species as a model system. Mol. Phylogenet. Evol. 2018, 125, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Podnar, M.; Mayer, W. First insights into the mitochondrial DNA diversity of Dalmatian Algyroides, Algyroides nigropunctatus (Lacertidae). Period. Biol. 2006, 108, 85–87. [Google Scholar]
- Strachinis, I.; Poulakakis, N.; Karaiskou, N.; Patronidis, P.; Patramanis, I.; Poursanidis, D.; Jablonski, D.; Triantafyllidis, A. Phylogeography and systematics of Algyroides (Sauria: Lacertidae) of the Balkan Peninsula. Zool. Scr. 2021, 50, 282–299. [Google Scholar] [CrossRef]
- QGIS.org. QGIS Geographic Information System. Open Source Geospatial Foundation Project. 2023. Available online: http://qgis.org (accessed on 12 April 2024).
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Arèvalo, E.; Davis, S.K.; Sites, J.W. Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico. Syst. Biol. 1994, 43, 387–418. [Google Scholar] [CrossRef]
- Palumbi, S.R.; Martin, A.; Romano, S.; McMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, version 2.0; University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Pavlicev, M.; Mayer, W. Fast radiation of the subfamily Lacertinae (Reptilia: Lacertidae): History or methodical artefact? Mol. Phylogenet. Evol. 2009, 52, 727–734. [Google Scholar] [CrossRef]
- Salvi, D.; Harris, D.J.; Perera, A.; Bologna, M.A.; Carretero, M.A. Preliminary survey on genetic variation within the Pygmy Algyroides, Algyroides fitzingeri, across Corsica and Sardinia. Amphib.-Reptil. 2011, 32, 281–286. [Google Scholar]
- Greenbaum, E.; Villanueva, C.O.; Kusamba, C.; Aristote, M.M.; Branch, W.R. A molecular phylogeny of Equatorial African Lacertidae, with the description of a new genus and species from eastern Democratic Republic of the Congo. Zool. J. Linn. Soc. 2011, 163, 913–942. [Google Scholar] [CrossRef] [PubMed]
- Kirchhof, S.; Lyra, M.L.; Rodríguez, A.; Ineich, I.; Müller, J.; Rödel, M.O.; Trape, J.F.; Vences, M.; Boissinot, S. Mitogenome analyses elucidate the evolutionary relationships of a probable Eocene wet tropics relic in the xerophilic lizard genus Acanthodactylus. Sci. Rep. 2021, 11, 4858. [Google Scholar] [CrossRef] [PubMed]
- Rato, C.; Sillero, N.; Ceacero, F.; Muñoz, E.G.; Carretero, M.A. A survival story: Evolutionary history of the Iberian Algyroides (Squamata: Lacertidae), an endemic lizard relict. Biodivers. Conserv. 2021, 30, 2707–2729. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Ho, S.Y.W.; Guindon, S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012, 29, 1695–1701. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post- analysis of large phylogenies. Bioinformatics 2014, 22, 1312–1313. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Bressi, N. Algyroides nigropunctatus nigropunctatus in Italy: Notes on ecology, habitat selection and conservation (Reptilia, Lacertidae). Ital. J. Zool. J. 2004, 71, 113–116. [Google Scholar] [CrossRef]
- Zakkak, S.; Halley, J.M.; Akriotis, T.; Kati, V. Lizards along an agricultural land abandonment gradient in Pindos Mountains, Greece. Amphib.-Reptil. 2015, 36, 253–264. [Google Scholar] [CrossRef]
- Carneiro, D.; García-Muñoz, E.; Žagar, A.; Pafilis, P.; Carretero, M.A. Is ecophysiology congruent with the present-day relictual distribution of a lizard group? Evidence from preferred temperatures and water loss rates. Herpetol. J. 2017, 27, 47–56. [Google Scholar]
- Blackburn, T.M.; Pyšek, P.; Bacher, S.; Carlton, J.T.; Duncan, R.P.; Jarošík, V.; Wilson, J.R.U.; Richardson, D.M. A proposed unified framework for biological invasions. Trends Ecol. Evol. 2011, 26, 333–339. [Google Scholar] [CrossRef]
- Alfaro, M.E.; Zoller, S.; Lutzoni, F. Bayes or Bootstrap? A Simulation Study Comparing the Performance of Bayesian Markov Chain Monte Carlo Sampling and Bootstrapping in Assessing Phylogenetic Confidence. Mol. Biol. Evol. 2003, 20, 255–266. [Google Scholar] [CrossRef]
- Maddison, W.P. Gene trees in species trees. Syst. Biol. 1997, 46, 523–536. [Google Scholar] [CrossRef]
- Maddison, W.P.; Knowles, L.L. Inferring phylogeny despite incomplete lineage sorting. Syst. Biol. 2006, 55, 21–30. [Google Scholar] [CrossRef]
- Pinho, C.; Harris, D.J.; Ferrand, N. Contrasting patterns of population subdivision and historical demography in three western Mediterranean lizard species inferred from mitochondrial DNA variation. Mol. Ecol. 2007, 16, 1191–1205. [Google Scholar] [CrossRef] [PubMed]
- Savić, I.R. Diversification of the Balkan fauna: Its origin, historical development and present status. Adv. Arachnol. Dev. Biol. 2008, 12, 57–78. [Google Scholar]
- Canestrelli, D.; Aloise, G.; Cecchetti, S.; Nascetti, G. Birth of a hotspot of intraspecific genetic diversity: Notes from the underground. Mol. Ecol. 2010, 19, 5432–5451. [Google Scholar] [CrossRef]
- Avise, J.C. Phylogeography: Retrospect and prospect. J. Biogeogr. 2009, 36, 3–15. [Google Scholar] [CrossRef]
- Ursenbacher, S.; Schweiger, S.; Tomović, L.; Crnobrnja-Isailović, J.; Fumagalli, L.; Mayer, W. Molecular phylogeography of the nose-horned viper (Vipera ammodytes, Linnaeus (1758)): Evidence for high genetic diversity and multiple refugia in the Balkan peninsula. Mol. Phylogenet. Evol. 2008, 46, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Kotsakiozi, P.; Parmakelis, A.; Giokas, S.; Papanikolaou, I.; Valakos, E.D. Mitochondrial phylogeny and biogeographic history of the Greek endemic land-snail genus Codringtonia Kobelt 1898 (Gastropoda, Pulmonata, Helicidae). Mol. Phylogenet. Evol. 2012, 62, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Sagonas, K.; Poulakakis, N.; Lymberakis, P.; Parmakelis, A.; Pafilis, P.; Valakos, E.D. Molecular systematics and historical biogeography of the green lizards (Lacerta) in Greece: Insights from mitochondrial and nuclear DNA. Mol. Phylogenet. Evol. 2014, 76, 144–154. [Google Scholar] [CrossRef]
- Kornilios, P.; Thanou, E.; Kapli, P.; Parmakelis, D.; Chatzaki, M. Peeking through the trapdoor: Historical biogeography of the Aegean endemic spider Cyrtocarenum Ausserer, 1871 with an estimation of mtDNA substitution rates for Mygalomorphae. Mol. Phylogenet. Evol. 2016, 98, 300–313. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).