Molecular Phylogeny and Historical Biogeography of Byrsonima (Malpighiaceae) Corroborates the Mid-Miocene Origins of Neotropical Savannas
Abstract
1. Introduction
2. Materials and Methods
2.1. Taxon Sampling and Molecular Protocols
2.2. Phylogenetic Analyses
2.3. Character Selection, Coding, and Morphological Analysis
2.4. Calibration
2.5. Ancestral Range Reconstruction
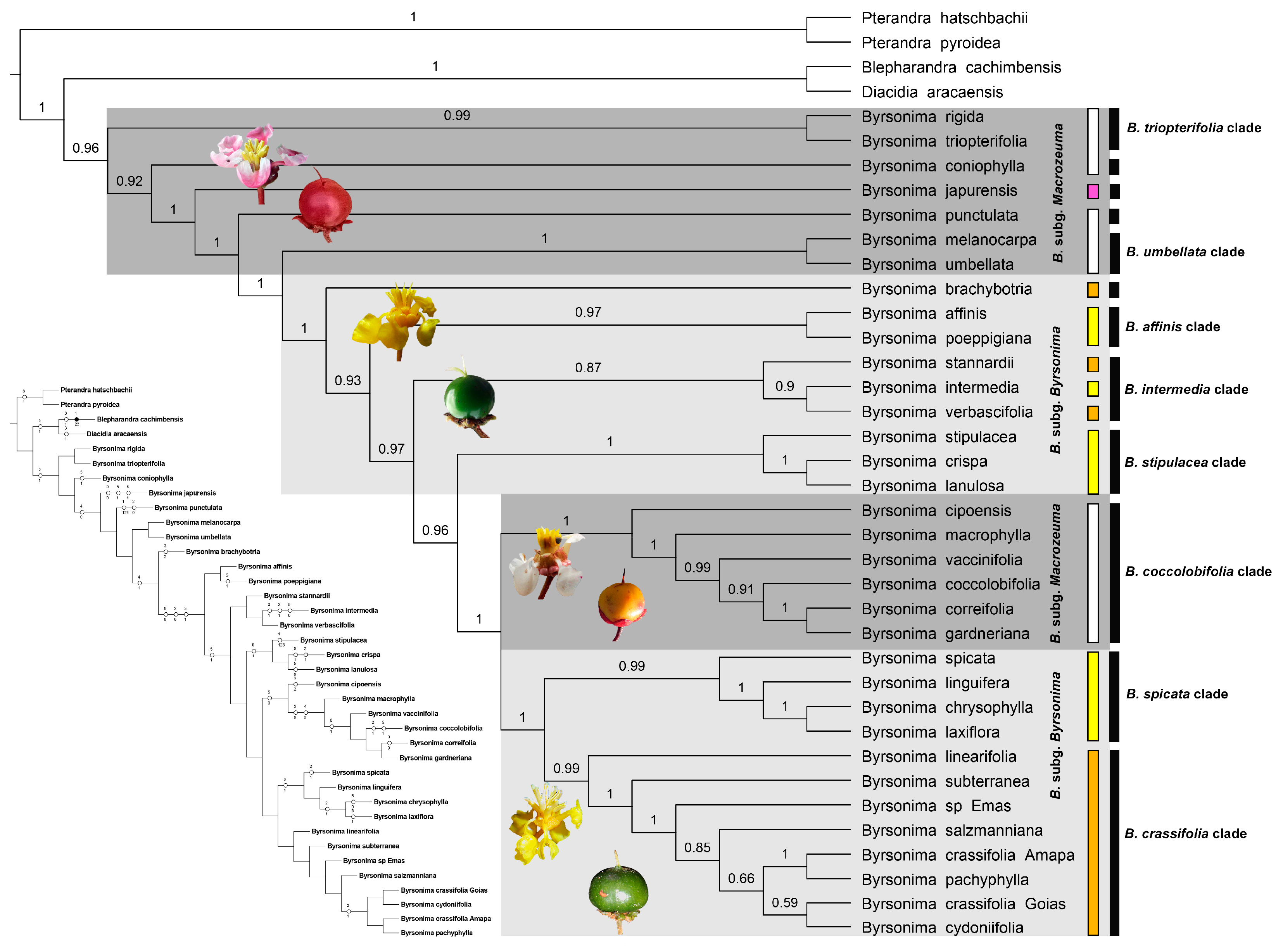
3. Results
3.1. Phylogenetics and Character Mapping
3.2. Divergence Times and Ancestral Ranges of Byrsonima
4. Discussion
4.1. Phylogenetics and Systematics of Byrsonima
4.2. Historical Biogeography of Byrsonima
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Almeida, R.F.; de Morais, I.L.; Alves-Silva, T.; Antonio-Domingues, H.; Pellegrini, M.O.O. A new classification system and taxonomic synopsis for Malpighiaceae (Malpighiales, Rosids) based on molecular phylogenetics, morphology, palynology, and chemistry. PhytoKeys 2024, 242, 69–138. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, R.F.; Francener, A.; Pessoa, C.; Sebastiani, R.; Oliveira, Y.R.; Amorim, A.M.A.; Mamede, M.C.H. Malpighiaceae in Flora do Brasil 2020. Jardim Botânico do Rio de Janeiro. 2020. Available online: https://floradobrasil.jbrj.gov.br/reflora/floradobrasil/FB155 (accessed on 7 December 2023).
- Niedenzu, F. De Genere Byrsonima (Pars Posterior); Arbeiten aus dem botanischen Institut des Kgl; Lyceum Hosianum: Braunsberg, Poland, 1901; pp. 1–45. [Google Scholar]
- Niedenzu, F. Malpighiaceae. In Das Pflanzenreich; Engler, A., Ed.; Wilhelm Engelmann: Leipzig, Germany, 1928; Volume 141, p. 870. [Google Scholar]
- Rolim, S.I.E. Revisão e Redefinição de Byrsonima Rich. ex Kunth Subg. Macrozeugma Nied. (Malpighiaceae). Ph.D. Thesis, University of São Paulo, São Paulo, Brazil, 2004. Available online: https://bdpi.usp.br/item/001449938 (accessed on 7 July 2024).
- Davis, C.C.; Anderson, W.R. A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Amer. J. Bot. 2010, 97, 2031–2048. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J.; Doyle, J.S. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- de Almeida, R.F.; Amorim, A.M.A.; van den Berg, C. Timing the origin and past connections between Andean and Atlantic Seasonally Dry Tropical Forests in South America: Insights from the biogeographical history of Amorimia (Malpighiaceae). Taxon 2018, 67, 739–751. [Google Scholar] [CrossRef]
- Almeida, R.F.; Pellegrini, M.O.O.; de Morais, I.L.; Simão-Bianchini, R.; Rattanakrajang, P.; Cheek, M.; Simões, A.R.G. Barking up the wrong tree: The dangers of taxonomic misidentification in molecular phylogenetic studies. Plant Ecol. Evol. 2023, 156, 146–159. [Google Scholar]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Amer. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Schilling, E.E.; Small, R.L. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. Amer. J. Bot. 2007, 94, 275–288. [Google Scholar] [CrossRef]
- de Almeida, R.F.; Amorim, A.M.A.; Correa, A.M.S.; van den Berg, C. A new infrageneric classification for Amorimia (Malpighiaceae) based on morphological, phytochemical and molecular evidence. Phytotaxa 2017, 313, 231–248. [Google Scholar]
- Paithankar, K.R.; Prasad, K.S.N. Precipitation of DNA by polyethylene glycol and ethanol. Nucl. Acids Res. 1991, 19, 1346. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. Mr Bayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 7 February 2023).
- Thiers, B. Index Herbariorum. Available online: https://sweetgum.nybg.org/science/ih/ (accessed on 7 February 2023).
- Sereno, P.C. Logical basis for morphological characters in phylogenetics. Cladistics 2007, 23, 565–587. [Google Scholar] [CrossRef] [PubMed]
- De Pinna, M.C.C. Concepts and tests of homology in the cladistic paradigm. Cladistics 1991, 7, 367–394. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. Available online: http://www.mesquiteproject.org (accessed on 7 February 2023).
- Nixon, K.C. Winclada (Beta) Ver. 0.9. Published by the Author, Ithaca, NY. Available online: http://www.cladistics.com (accessed on 7 February 2023).
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Davis, C.C.; Schaefer, H.; Xia, Z.; Baum, D.A.; Donoghue, M.J.; Harmon, L.J. Long-term morphological stasis maintained by a plant–pollinator mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 5914–5919. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree Version 1.3.1: Tree Figure Drawing Tool. Computer Program and Documentation Distributed by the Author. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 7 February 2023).
- GBIF. Global Biodiversity Information Facility. Available online: http://gbif.org (accessed on 7 February 2023).
- Matzke, N.J. BioGeoBEARS: Biogeography with Bayesian (And Likelihood) Evolutionary Analysis in R Scripts, CRAN: The Comprehensive R Archive Network, Berkeley, 2013, CA. Available online: http://CRAN.R-project.org/package=BioGeoBEARS (accessed on 7 February 2023).
- Cameron, K.M.; Chase, M.W.; Anderson, W.R.; Hills, H.G. Molecular Systematics of Malpighiaceae: Evidence from plastid rbcL and matK sequences. Amer. J. Bot. 2001, 88, 1847–1862. [Google Scholar] [CrossRef]
- Davis, C.C.; Anderson, W.R.; Donoghue, M. Phylogeny of Malpighiaceae: Evidence from chloroplast ndhF and trnL-F nucleotides sequences. Amer. J. Bot. 2001, 88, 1830–1846. [Google Scholar] [CrossRef]
- Davis, C.C.; Bell, C.D.; Mathews, S.; Donoghue, M.J. Laurasian migration explains Gondwanan disjunctions: Evidence from Malpighiaceae. Proc. Natl. Acad. Sci. USA 2002, 99, 6833–6837. [Google Scholar] [CrossRef]
- Santos-Zanuncio, V.S.; Alves, F.M.; Silva, D.B.; Carollo, C.A. Chemosystematic implications based on metabolic profiling of the genus Byrsonima (Malpighiaceae). Folia Geobot. 2020, 55, 289–300. [Google Scholar] [CrossRef]
- Anderson, W.R. Malpighiaceae. In: The Botany of Guayana Highland—Part IX. Mem. N. Y. Bot. Gard. 1981, 32, 129–131. [Google Scholar]
- Anderson, W.R. Notes on Neotropical Malpighiaceae - I. Contr. Univ. Michigan. Herb. 1982, 16, 55–108. [Google Scholar]
- Jussieu, A. Malpighiacearum synopsis, monographiae mox edendae prodromus. Ann. Sci. Nat. Botanique 1840, 13, 247–291, 321–338. [Google Scholar]
- Aguiar, A.J.C.; Melo, G.A.R.; Vasconcelos, T.N.C.; Gonçalves, R.B.; Giugliano, L.; Martins, A.C. Biogeography and early diversification of Tapinotaspidini oil-bees support presence of Paleocene savannas in South America. Mol. Phylogenetics Evol. 2020, 143, 106692. [Google Scholar] [CrossRef]
- Gonçalves, D.J.P.; Shimizu, G.H.; Ortiz, E.M.; Jansen, R.K.; Simpson, B.B. Historical biogeography of Vochysiaceae reveals an unexpected perspective of plant evolution in the Neotropics. Amer. J. Bot. 2020, 107, 1004–1020. [Google Scholar] [CrossRef]
- Antonelli, A.; Sanmartín, I. Why are there so many plant species in the Neotropics? Taxon 2011, 60, 403–414. [Google Scholar] [CrossRef]
- Carnaval, A.C.; Waltari, E.; Rodrigues, M.T.; Rosauer, D.; VanDerWal, J.; Damasceno, R.; Prates, I.; Strangas, M.; Spanos, Z.; Rivera, D.; et al. Prediction of phylogeographic endemism in an environmentally complex biome. Proc. R. Soc. B 2014, 281, 20141461. [Google Scholar] [CrossRef]
- Amaral, D.T.; Minhós-Yano, I.; Oliveira, J.V.M.; Romeiro-Brito, M.; Bonatelli, I.A.S.; Taylor, N.P.; Zappi, D.C.; Moraes, E.M.; Eaton, D.; Franco, F.F. Tracking the xeric biomes of South America: The spatiotemporal diversification of Mandacaru cactus. J. Biogeogr. 2021, 48, 3085–3103. [Google Scholar] [CrossRef]
- Buzatti, R.S.O.; Pfeilsticker, T.R.; de Magalhães, R.F.; Bueno, M.L.; Lemos-Filho, J.P.; Lovato, M.B. Genetic and historical colonization analyses of an endemic savanna tree, Qualea grandiflora, reveal ancient connections between Amazonian savannas and Cerrado core. Front Plant Sci. 2018, 9, 981. [Google Scholar] [CrossRef]
- Cândido, E.S.; Vatanparast, M.; Vargas, W.; Bezerra, L.M.P.A.; Lewis, G.L.; Mansano, V.F.; Simões, A.O.; Silva, M.J.; Stirton, C.; Tozzi, A.M.G.A.; et al. Molecular phylogenetic insights into the evolution of Eriosema (Fabaceae): A recent tropical savanna-adapted genus. Bot. J. Linn. Soc. 2020, 194, 439–459. [Google Scholar] [CrossRef]
- Fiorini, C.F.; Peres, E.A.; da Silva, M.J.; Araujo, A.O.; Borba, E.L.; Solferini, V.N. Phylogeography of the specialist plant Mandirola hirsuta (Gesneriaceae) suggests ancient habitat fragmentation due to savanna expansion. Flora 2020, 262, 151522. [Google Scholar] [CrossRef]
- Simon, M.F.; Grether, R.; Queiroz, L.P.; Skema, C.; Pennington, R.T.; Hughes, C.E. Recent assembly of the Cerrado, a neotropical plant diversity hotspot, by in situ evolution of adaptations to fire. Proc. Natl. Acad. Sci. USA 2009, 106, 20359–20364. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, T.N.C.; Alcantara, S.; Andrino, C.O.; Forest, F.; Reginato, M.; Simon, M.F.; Pirani, J.R. Fast diversification through a mosaic of evolutionary histories characterizes the endemic flora of ancient Neotropical mountains. Proc. R. Soc. B 2020, 287, 20192933. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, L.C.; Lima-Ribeiro, M.S.; Terribile, L.C.; Collevatti, R.G. Demographical expansion of Handroanthus ochraceus in the Cerrado during the Quaternary: Implications for the genetic diversity of Neotropical trees. Biol. J. Linn. Soc. 2018, 123, 561–577. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Almeida, R.F.; Francener, A.; Mamede, M.C.H.; van den Berg, C. Molecular Phylogeny and Historical Biogeography of Byrsonima (Malpighiaceae) Corroborates the Mid-Miocene Origins of Neotropical Savannas. Diversity 2024, 16, 488. https://doi.org/10.3390/d16080488
de Almeida RF, Francener A, Mamede MCH, van den Berg C. Molecular Phylogeny and Historical Biogeography of Byrsonima (Malpighiaceae) Corroborates the Mid-Miocene Origins of Neotropical Savannas. Diversity. 2024; 16(8):488. https://doi.org/10.3390/d16080488
Chicago/Turabian Stylede Almeida, Rafael Felipe, Augusto Francener, Maria Candida Henrique Mamede, and Cássio van den Berg. 2024. "Molecular Phylogeny and Historical Biogeography of Byrsonima (Malpighiaceae) Corroborates the Mid-Miocene Origins of Neotropical Savannas" Diversity 16, no. 8: 488. https://doi.org/10.3390/d16080488
APA Stylede Almeida, R. F., Francener, A., Mamede, M. C. H., & van den Berg, C. (2024). Molecular Phylogeny and Historical Biogeography of Byrsonima (Malpighiaceae) Corroborates the Mid-Miocene Origins of Neotropical Savannas. Diversity, 16(8), 488. https://doi.org/10.3390/d16080488








