Capacity of Aromatic Compound Degradation by Bacteria from Amazon Dark Earth
Abstract
:1. Introduction
2. Experimental Section
2.1. Sampling
2.2. Enrichment Assay
| DSMZ Standard | AH Metabolism |
|---|---|
| P. putida | toluene dioxygenase |
| P. putida (DSM 291) | |
| P. fluorescens (DSM 8369) | naphthalene dioxygenase |
| P. abietaniphila (DSM 6506) |
2.3. Isolates Identification
2.3.1. 16S rRNA Gene of Bacteria Amplification
2.3.2. Sequencing Reaction
2.3.3. Phylogenetic Analysis
2.4. Biodegradation Assays
2.4.1. Cell Density
2.4.2. The Assays
| No. DSMZ | Microorganism | AH metabolism |
|---|---|---|
| DSM 11192 | Gordonia sp. | n-alkane, naphthalene, toluene, m-xylene, p-xylene |
3. Results and Discussion


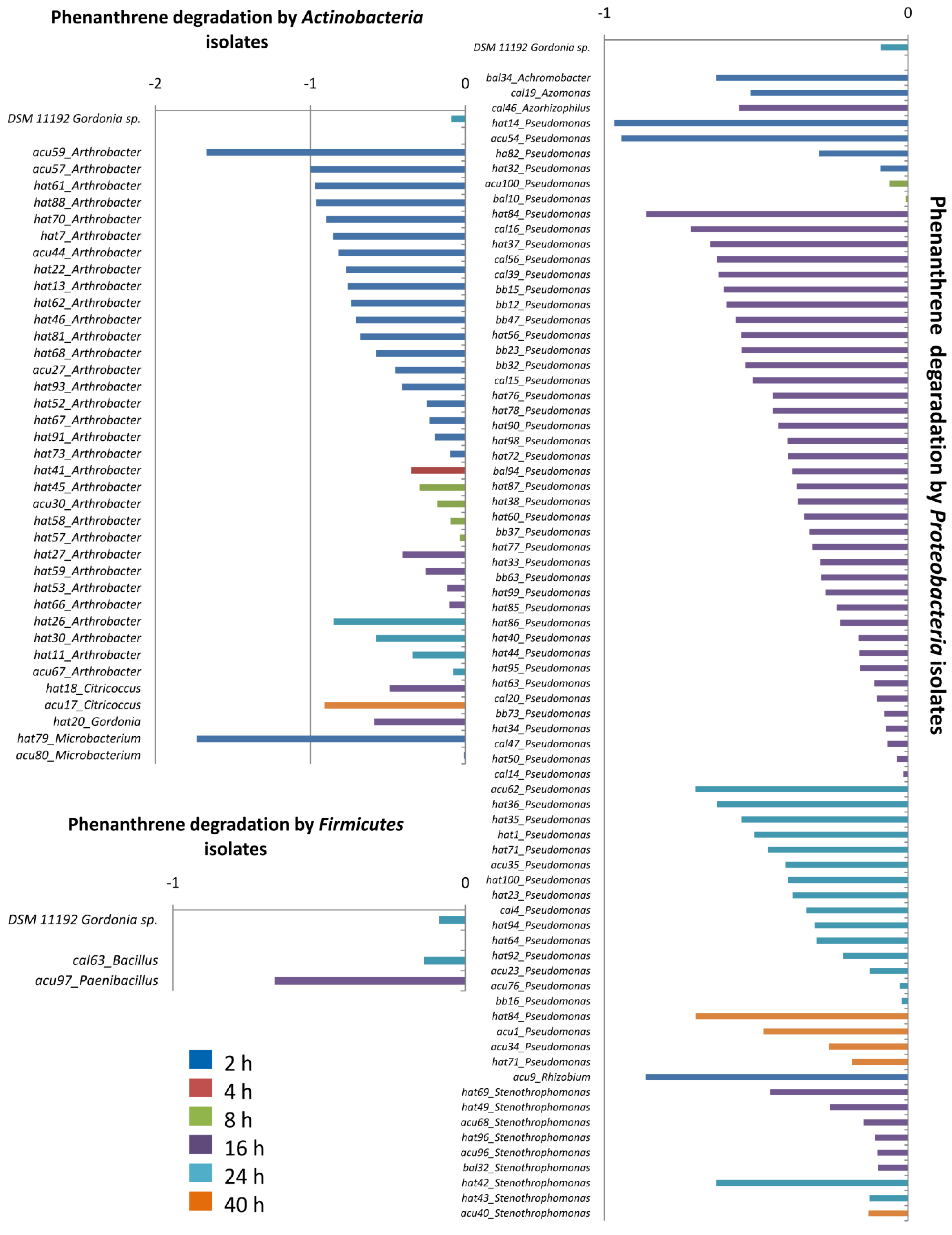

4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Glaser, B.; Haumaier, L.; Guggenberger, G.; Zech, W. The “Terra Preta” phenomenon: A model for sustainable agriculture in the humid tropics. Naturwissenschaften 2001, 88, 37–41. [Google Scholar] [CrossRef]
- Lehmann, J.; Joseph, S. Biochar for Environmental Management; SciTech Earthscan: London, UK, 2009; pp. 1–3. [Google Scholar]
- Brady, N.C.; Weil, R.R. An Introduction to the Nature and Properties of Soils, 14th ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2008; p. 881. [Google Scholar]
- Steiner, C.; Das, K.C.; Garcia, M.; Forster, B.; Zech, W. Charcoal and smoke extract stimulate the soil microbial community in a highly weathered xanthic Ferralsol. Pedobiologia 2008, 51, 359–366. [Google Scholar] [CrossRef]
- Habe, H.; Omori, T. Genetic of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotech. Biochem. 2003, 67, 225–243. [Google Scholar] [CrossRef]
- Volkering, F.; Breure, A.M.; Sterkenburg, A.; van Andel, J.G. Microbial degradation of polycyclic aromatic hydrocarbons: Effect of substrate availability on bacterial growth kinetics. Appl. Microbiol. Biotechnol. 1992, 36, 548–552. [Google Scholar]
- Barkay, T.; Pritchard, H. Adaptation of aquatic microbial communities to pollutant stress. Microbiol. Sci. 1988, 5, 165–169. [Google Scholar]
- Spain, J.C.; van Veld, P.A. Adaptation of natural microbial communities to degradation of xenobiotic compounds: Effects of concentration, exposure time, inoculum, and chemical structure. Appl. Environ. Microbiol. 1983, 54, 428–435. [Google Scholar]
- Peng, R.; Xiong, A.; Xue, Y.; Fu, X.; Gao, F.; Zhao, W.; Tian, Y.; Ya, Q. Microbial biodegradation of polyaromatic hydrocarbons. FEMS Microbiol. Rev. 2008, 32, 927–955. [Google Scholar] [CrossRef]
- Sayler, G.S.; Shields, M.S.; Tedford, E.T.; Breen, A.; Hooper, S.W.; Sirotkin, K. M; Davis, J.W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl. Environ. Microbiol. 1985, 49, 1295–1303. [Google Scholar]
- Park, J.; Crowley, D.E. Dynamic changes in nahAc gene copy numbers during degradation of naphthalene in PAH-contaminated soils. Appl. Microbiol. Biotechnol. 2006, 72, 1322–1329. [Google Scholar] [CrossRef]
- Lynch, J.M.; Hobbie, J.E. Micro-Organisms in Action: Concepts and Applications in Microbial Ecology; Blackwell Science Inc., Osney Mead: Oxford, UK, 1988; p. 363. [Google Scholar]
- Mishra, V.; Lal, R. Srinivasan. Enzymes and operons mediating xenobiotic degradation in bacteria. Crit. Rev. Microbiol. 2001, 27, 133–166. [Google Scholar] [CrossRef]
- Navarrete, A.A.; Cannavan, F.S.; Taketani, R.G.; Tsai, S.M. A molecular survey of the diversity of microbial communities in different Amazonian agricultural model systems. Diversity 2010, 2, 787–809. [Google Scholar] [CrossRef]
- Germano, M.G.; Cannavan, F.S.; Mendes, L.W.; Lima, A.B.; Teixeira, W.G.; Pellizari, V.H.; Tsai, S.M. Functional diversity of bacterial genes associated with aromatic hydrocarbon degradation in anthropogenic dark earth of Amazonia. Pesqui. Agropec. Bras. 2012, 47, 654–664. [Google Scholar] [CrossRef] [Green Version]
- Atlas, R.M. Bioremediation of petroleum pollutants. Int. Biodeterior. Biodegrad. 1995, 35, 317–327. [Google Scholar] [CrossRef]
- Denevan, W. Comments on prehistoric agriculture in Amazônia. Cult. Agric. 1998, 20, 54–59. [Google Scholar]
- Furukawa, K. Modification of PCBs by bacteria and others microorganisms. In PCBs and the Environment; Waid, J.S., Ed.; CRC Press: Boca Raton, FL, USA, 1986; p. 84. [Google Scholar]
- Doyle, J.J. DNA Protocols for Plants. Mol. Tech. Taxon. NATO ASI Ser. 1991, 57, 283–293. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar]
- Kent, A.D.; Triplett, E.W. Microbial communities and their interactions in soil and rhizosphere ecosystems. Ann. Rev. Microbiol. 2002, 56, 211–236. [Google Scholar] [CrossRef]
- Ewing, B.; Green, P. Based-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 1998, 8, 186–194. [Google Scholar]
- Ewing, B.; Hijjier, L.; Wendl, M.C.; Green, P. Based-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 1998, 8, 175–185. [Google Scholar]
- Gordon, D.; Abajian, C.; Green, P. Consed: A geographic tool for sequence finishing. Genome Res. 1998, 8, 195–202. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon: A prokaryotic 16S rRNA Gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Lelliott, R.A.; Stead, D.E. Chapter 5: Host tests methods. In Methods for the Diagnosis of Bacterial Diseases of Plants, 2nd ed.; British Society for Plant Pathology, Blackwell Scientific Publications: Oxford/London, UK, 1987; pp. 167–168. [Google Scholar]
- Hanson, K.G.; Desai, J.D.; Desai, A.J. A rapid and simple screening technique for potential crude oil degrading microorganisms. Biotechnol. Tech. 1993, 7, 745–748. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, L.; Shao, Z. Pseudomonas, the dominant polycyclic aromatic hydrocarbon-degrading bacteria isolated from Antarctic soils and the role of large plasmids in horizontal gene transfer. Environ. Microbiol. 2006, 8, 455–465. [Google Scholar] [CrossRef]
- Leahy, J.F.; Colwell, R.R. Microbial degradation of hydrocarbons in the environment. Microb. Rev. 1990, 54, 305–315. [Google Scholar]
- Pieper, D.H. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 2005, 67, 170–191. [Google Scholar] [CrossRef]
- Guzik, U.; Hupert-Kocurek, K.; Sałek, K.; Wojcieszyn´Ska, D. Influence of metal ions on bioremediation activity of protocatechuate 3,4-dioxygenase from Stenotrophomonas maltophilia KB2. World J. Microbiol. Biotechnol. 2013, 29, 267–273. [Google Scholar] [CrossRef]
- Tuan, N.N.; Hsieh, H.; Lin, Y.; Huang, S. Analysis of bacterial degradation pathways for long-chain alkylphenols involving phenol hydroxylase, alkylphenolmonooxygenase and catechol dioxygenase genes. Bior. Technol. 2011, 102, 4232–4240. [Google Scholar] [CrossRef]
- Reinhold, B.; Hurek, T.; Fendrik, I. Cross-reaction of predominant nitrogen-fixing bacteria with enveloped, round bodies in the root interior of kallar grass. Appl. Environ. Microbiol. 1987, 53, 889–891. [Google Scholar]
- Juhasz, A.L.; Naidu, R. Bioremediation of high molecular weight polycyclic aromatic hydrocarbons: A review of the microbial degradation of benzo[a]pyrene. Int. Biodeterior. Biodegrad. 2000, 45, 57–88. [Google Scholar] [CrossRef]
- Pathak, A.; Green, S.J.; Ogram, A.; Chauhan, A. Draft genome sequence of Rhodococcusopacus strain M213 shows a diverse catabolic potential. Genome. Announc. 2013. [Google Scholar] [CrossRef]
- Mutnuri, S.; Vasudevan, N.; Kaestner, M. Degradation of anthracene and pyrene supplied by microcrystals and non-aqueous-phase liquids. Appl. Microbiol. Biotech. 2005, 67, 569–576. [Google Scholar] [CrossRef]
- Kanashiro, C.H.; Arredondo, D.L.L.; Morales, P.C.; Alcaraz, L.; Olmedo, G.; Gómez, F.B.; Estrella, L.H. First Draft Genome Sequence of a Strain from the Genus Citricoccus. J. Bact. 2011, 193, 6092–6093. [Google Scholar] [CrossRef]
- Van Aken, B.; Yoon, J.M.; Schnoor, J.L. Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoids x nigra DN34). Appl. Environ. Microbiol. 2004, 70, 508–517. [Google Scholar] [CrossRef]
- Daane, L.L.; Harjono, I.; Zylstra, G.J.; Haggblom, M.M. Isolation and characterization of polycyclic aromatic hydrocarbons-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl. Environ. Microbiol. 2011, 67, 2683–2691. [Google Scholar]
- Agency for Toxic Substances and Disease Registry (ATSDR). Toxicological Profile for Fuel Oils; U.S. Department of Health and Human Services, Public Health Service: Atlanta, GA, USA, 1995. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nakamura, F.M.; Germano, M.G.; Tsai, S.M. Capacity of Aromatic Compound Degradation by Bacteria from Amazon Dark Earth. Diversity 2014, 6, 339-353. https://doi.org/10.3390/d6020339
Nakamura FM, Germano MG, Tsai SM. Capacity of Aromatic Compound Degradation by Bacteria from Amazon Dark Earth. Diversity. 2014; 6(2):339-353. https://doi.org/10.3390/d6020339
Chicago/Turabian StyleNakamura, Fernanda Mancini, Mariana Gomes Germano, and Siu Mui Tsai. 2014. "Capacity of Aromatic Compound Degradation by Bacteria from Amazon Dark Earth" Diversity 6, no. 2: 339-353. https://doi.org/10.3390/d6020339
APA StyleNakamura, F. M., Germano, M. G., & Tsai, S. M. (2014). Capacity of Aromatic Compound Degradation by Bacteria from Amazon Dark Earth. Diversity, 6(2), 339-353. https://doi.org/10.3390/d6020339




