Identification of NAC Transcription Factors in Suaeda glauca and Their Responses to Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Physicochemical Characterization of NAC Family Members in S. glauca
2.2. Motif, Phylogenetic Tree, and Protein–Protein Interaction Analyses
2.3. Expression Profile Analysis of NAC Genes
2.4. Subcellular Localization
2.5. qRT-PCR Verification
3. Results
3.1. Identification and Physicochemical Properties of the NAC Transcription Factor Family in S. glauca
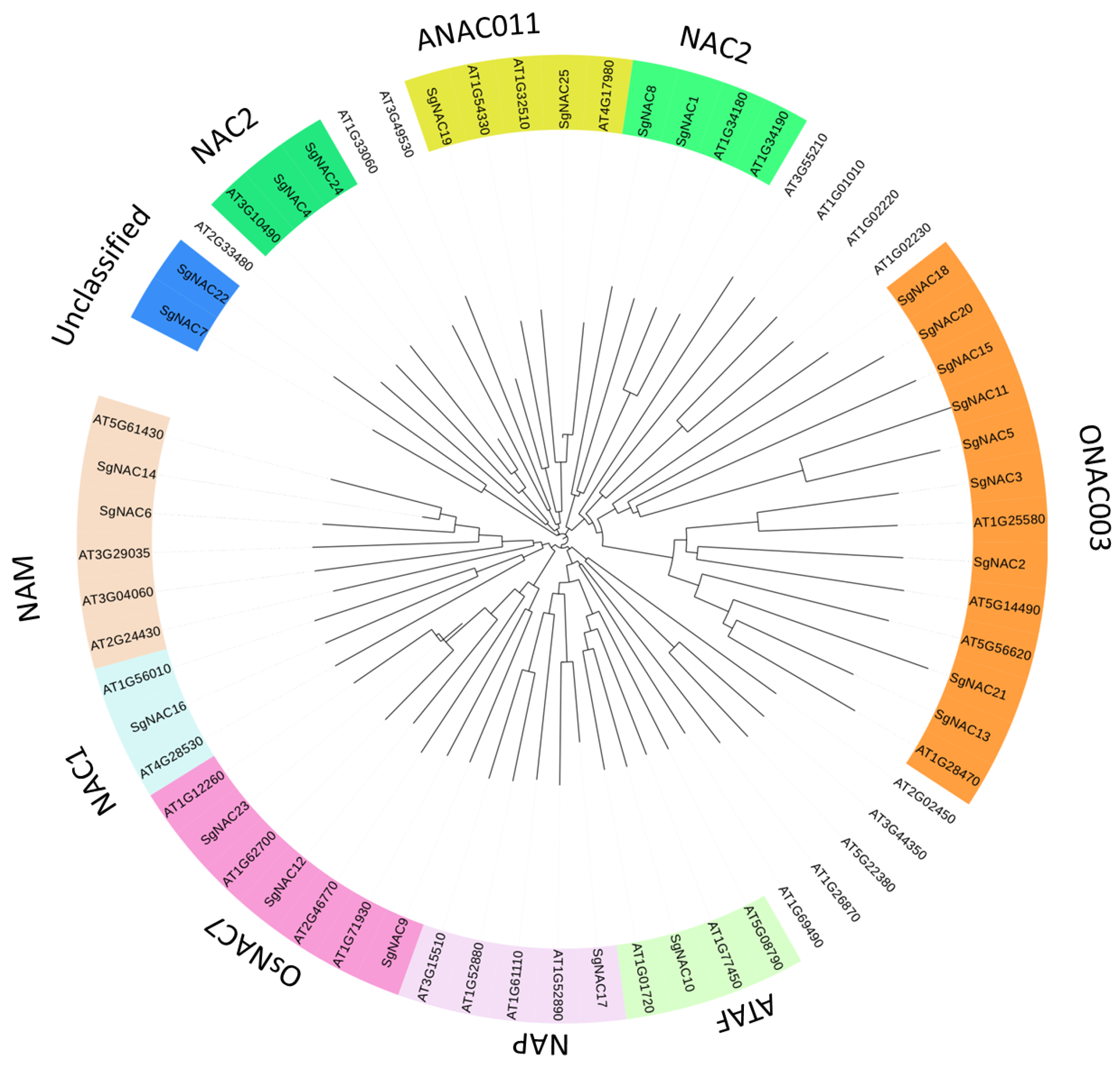
3.2. Evolutionary Analysis of NAC Families in S. glauca and A. thaliana
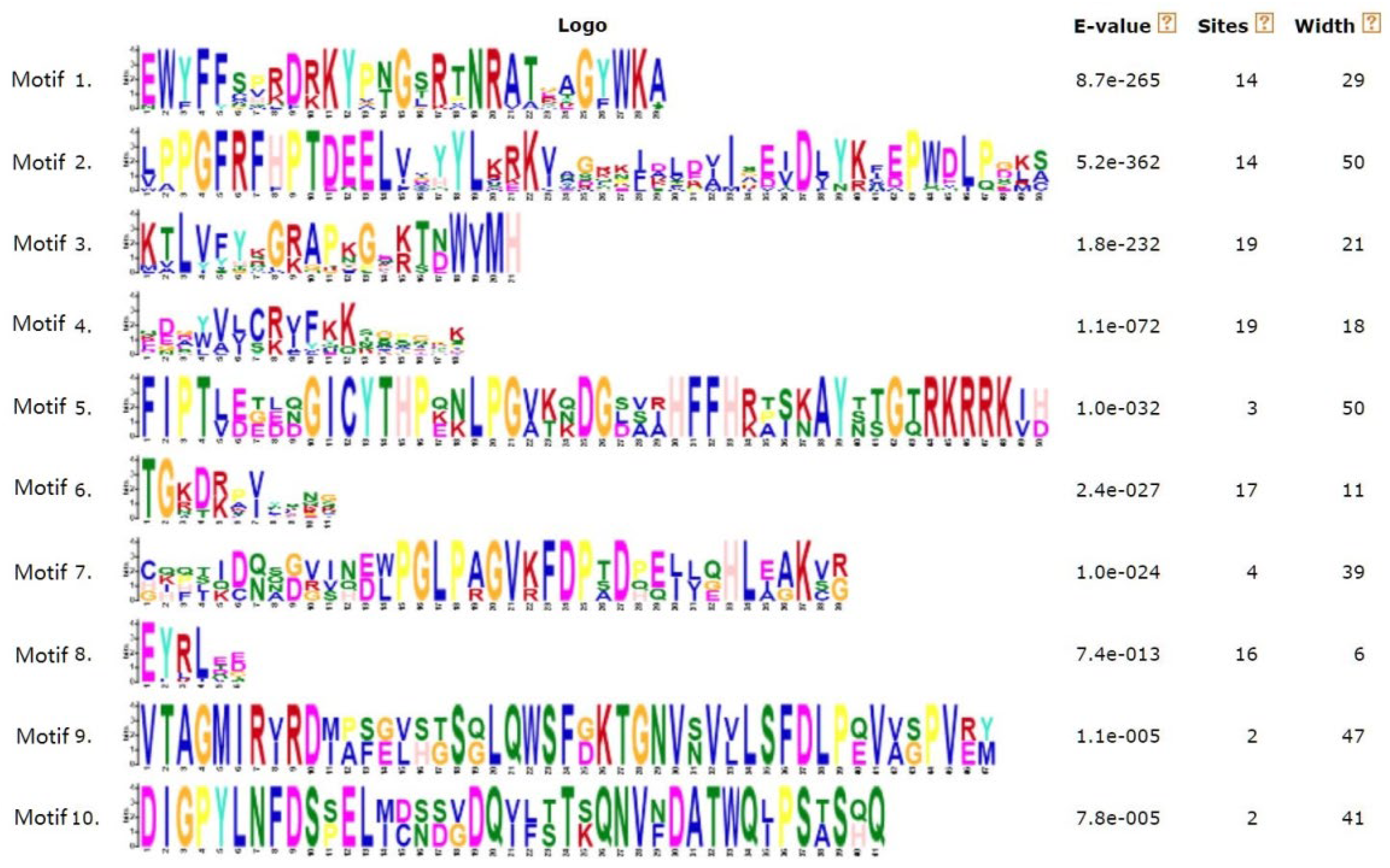
3.3. Conserved Motifs of NAC Family Transcription Factors in S. glauca
3.4. Expression Analysis of NAC Genes in S. glauca
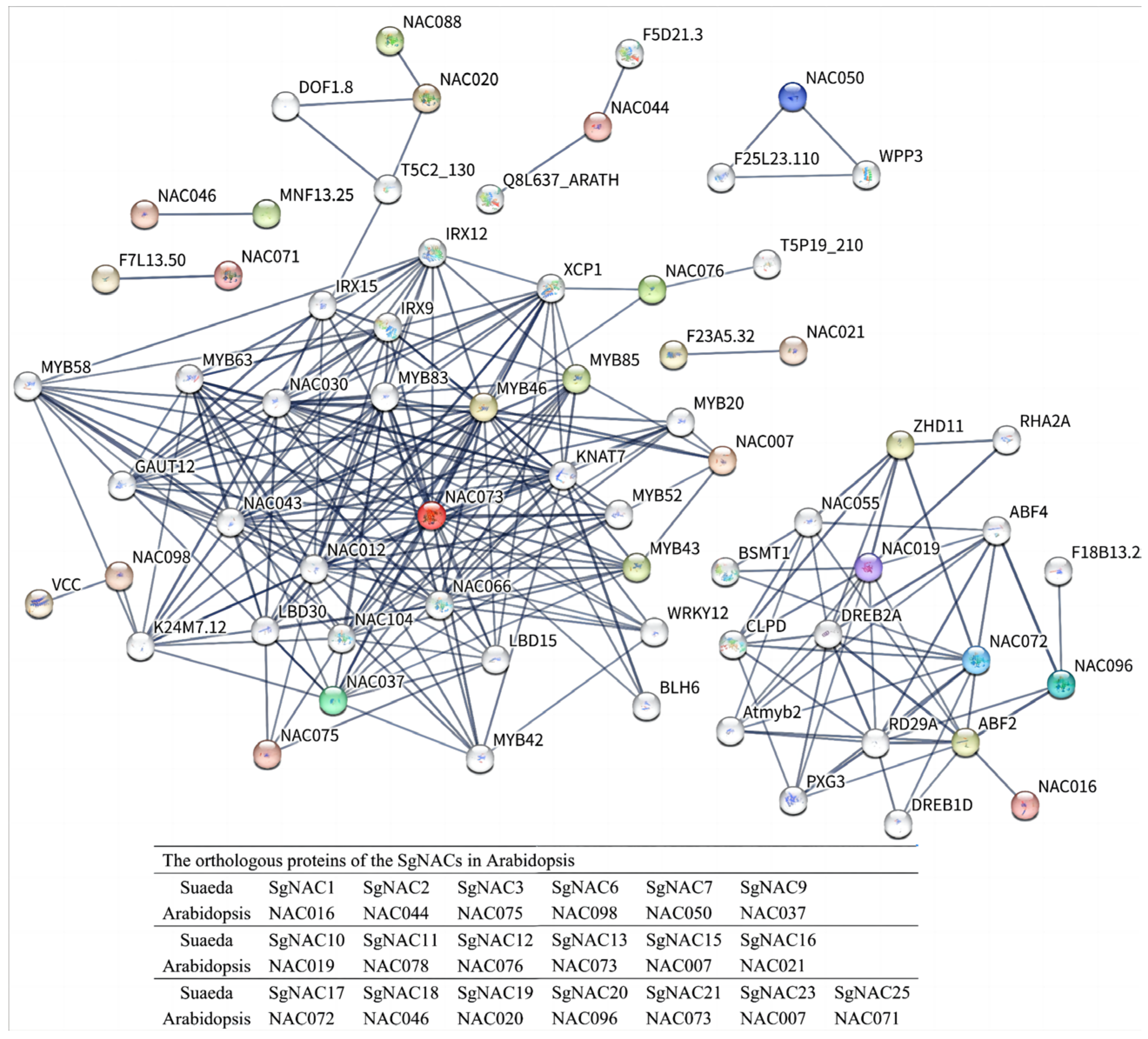
3.5. Analysis of NAC Protein–Protein Interaction Network
3.6. qRT-PCR Verification of Transcript Profiles
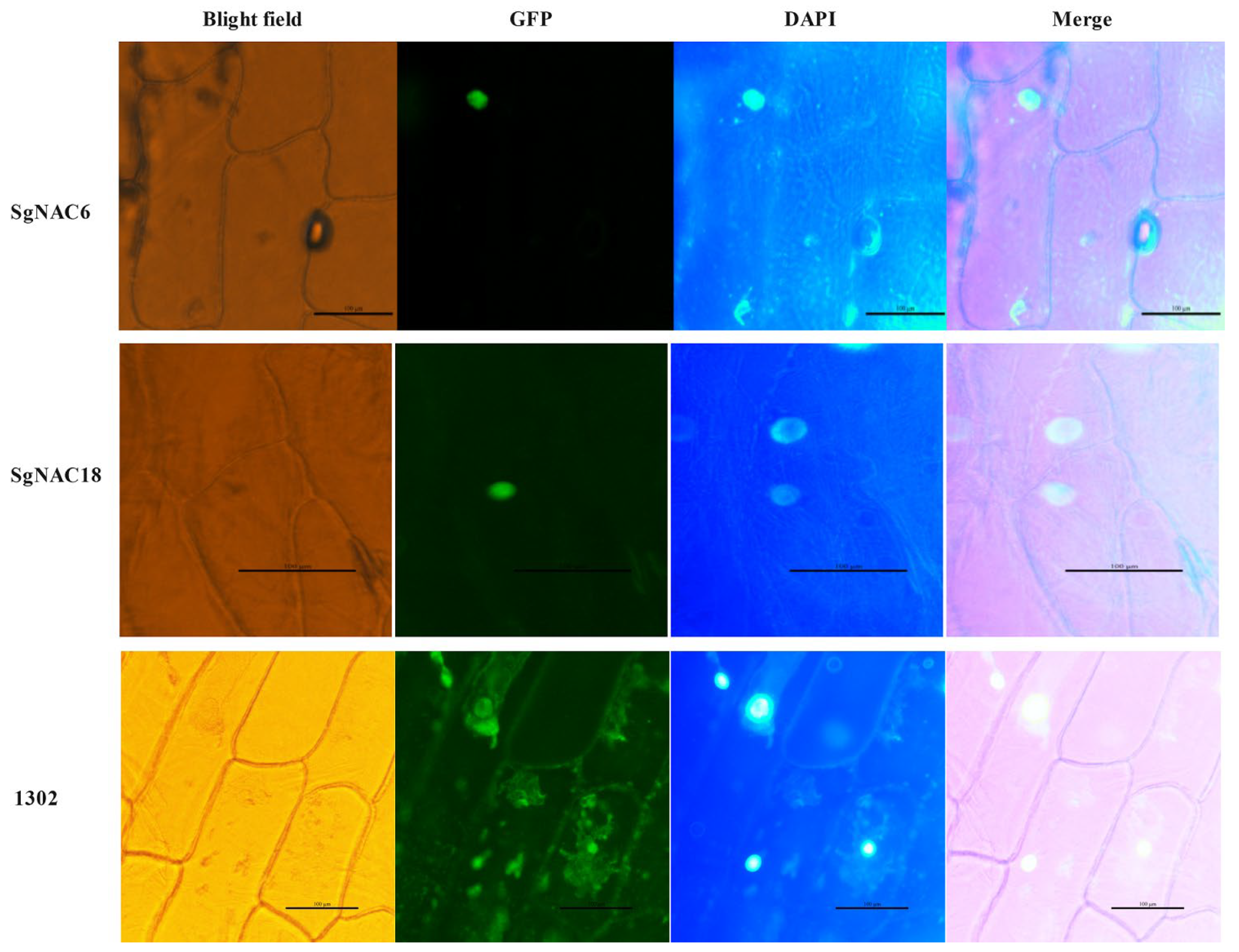
3.7. Subcellular Localization of SgNAC6 and SgNAC18 Proteins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bu, Q.; Jiang, H.; Li, C.B.; Zhai, Q.; Zhang, J.; Wu, X.; Sun, J.; Xie, Q.; Li, C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008, 18, 756–767. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, J.B.; Li, J.Q.; Huang, J.; Shen, R.F.; Zeng, D.L.; Zhu, X.F. A NAC transcription factor represses a module associated with xyloglucan content and regulates aluminum tolerance. Plant Physiol. 2024, kiae281. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, H.; Wei, Y.; Han, J.; Wang, Y.; Li, X.; Zhang, L.; Han, D. Overexpression of a Fragaria vesca NAM, ATAF, and CUC (NAC) Transcription Factor Gene (FvNAC29) Increases Salt and Cold Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2024, 25, 4088. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Mueller-Roeber, B.; Balazadeh, S. Arabidopsis NAC transcription factor JUNGBRUNNEN1 affects thermomemory-associated genes and enhances heat stress tolerance in primed and unprimed conditions. Plant Signal. Behav. 2012, 7, 1518–1521. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Zou, X.; Liang, Z.; Zhang, K.; Wu, D.; Jin, H.; Wang, H.; Shen, Q. GBF family member PfGBF3 and NAC family member PfNAC2 regulate rosmarinic acid biosynthesis under high light. Plant Physiol. 2024, 195, 1728–1744. [Google Scholar] [CrossRef] [PubMed]
- Nagahage, I.S.P.; Sakamoto, S.; Nagano, M.; Ishikawa, T.; Mitsuda, N.; Kawai-Yamada, M.; Yamaguchi, M. An Arabidopsis NAC domain transcription factor, ATAF2, promotes age-dependent and dark-induced leaf senescence. Physiol. Plant. 2020, 170, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Phung, J.; Zhai, Y.; Neff, M.M. Self-transcriptional repression of the Arabidopsis NAC transcription factor ATAF2 and its genetic interaction with phytochrome A in modulating seedling photomorphogenesis. Planta 2020, 252, 48. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Yu, Y.; Zhu, L.; Liu, H.; Chai, Q.; Guo, W. A cotton NAC transcription factor GhirNAC2 plays positive roles in drought tolerance via regulating ABA biosynthesis. Plant Sci. 2020, 296, 110498. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Zhang, H.; Qian, X.; Li, A.; Zhao, G.; Jing, R. TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 2012, 63, 2933–2946. [Google Scholar] [CrossRef] [PubMed]
- An, R.B.; Sohn, D.H.; Jeong, G.S.; Kim, Y.C. In vitro hepatoprotective compounds from Suaeda glauca. Arch. Pharmacal Res. 2008, 31, 594–597. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Dong, D.; Yang, Q.; Zhu, D. Salt-Responsive Transcriptome Profiling of Suaeda glauca via RNA Sequencing. PLoS ONE 2016, 11, e0150504. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Chen, Q.; Cui, X.; Abozeid, A.; Liu, Y.; Liu, J.; Tang, Z. Comparative metabolomics of two saline-alkali tolerant plants Suaeda glauca and Puccinellia tenuiflora based on GC-MS platform. Nat. Prod. Res. 2019, 35, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shi, D.; Wang, D. Comparative effects of salt and alkali stresses on growth, osmotic adjustment and ionic balance of an alkali-resistant halophyte Suaeda glauca (Bge.). Plant Growth Regul. 2008, 56, 179–190. [Google Scholar] [CrossRef]
- Zhao, N.; Yan, L.U.; Zuo, J.C.; Zhou-Min, L.U. Effect of Alkali Stress on Seed Germination of Suaeda glauca Bunge. North. Hortic. 2012, 1, 45–47. [Google Scholar]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ran, K.; Dong, Q.; Zhao, Q.; Shi, S. Cloning, sequencing, and expression analysis of 32 NAC transcription factors (MdNAC) in apple. PeerJ 2020, 8, e8249. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Ma, Z.; Sun, W.; Huang, L.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide analysis of the NAC transcription factor family in Tartary buckwheat (Fagopyrum tataricum). BMC Genom. 2019, 20, 113. [Google Scholar] [CrossRef]
- Jin, H.; Xu, G.; Meng, Q.; Huang, F.; Yu, D. GmNAC5, a NAC transcription factor, is a transient response regulator induced by abiotic stress in soybean. Sci. World J. 2013, 2013, 768972. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Woo, D.H.; Nguyen, L.V.; Tran, H.T.; Tarte, V.N.; Mehdi, S.M.M.; Lee, S.Y.; Moon, Y.H. Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 2017, 245, 329–341. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2016, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.G.; Chen, X.J. Identification a new NAC transcription factor OsNAC3 in rice. Acta Phytopathol. Sin. 2018, 48, 61–69. [Google Scholar]



| Gene | Primer Sequences |
|---|---|
| SgNAC2 | For-CCGAACTGCAAGTTTTCGAT |
| SgNAC2 | Rev-GGGTGTAGCAAATCCCTTGA |
| SgNAC14 | For-GCCACCATATCCCTCTTCAA |
| SgNAC14 | Rev-CAAATCCCATTTTCACCTTCA |
| SgNAC16 | For-TCGGATTGAAGGACCTTTTG |
| SgNAC16 | Rev-AGTCCATCAATGGCGGTAAG |
| SgNAC25 | For-CGCGATATGGAGTGGTTCTT |
| SgNAC25 | Rev-TCATGCATCACCCAATCAGT |
| Actin | For-CCGCAAAGATTACATACC |
| Actin | Rev-TCACCGAAAGTGCTTCTA |
| Name | Number of Amino Acids | Molecular Weight | Theoretical pI | Subcellular Location |
|---|---|---|---|---|
| SgNAC1 | 476 | 53,159.54 | 5.67 | Extracellular |
| SgNAC2 | 476 | 53,734.52 | 5.45 | Nuclear |
| SgNAC3 | 436 | 48,661.99 | 4.80 | Nuclear |
| SgNAC4 | 417 | 46,272.22 | 5.30 | Nuclear |
| SgNAC5 | 416 | 45,552.67 | 4.29 | Extracellular |
| SgNAC6 | 363 | 40,803.62 | 6.07 | Nuclear |
| SgNAC7 | 334 | 38,096.55 | 8.08 | Nuclear |
| SgNAC8 | 317 | 35,238.61 | 4.85 | Nuclear |
| SgNAC9 | 315 | 36,363.36 | 6.59 | Nuclear |
| SgNAC10 | 302 | 34,461.15 | 7.04 | Nuclear |
| SgNAC11 | 285 | 30,799.18 | 4.12 | Extracellular |
| SgNAC12 | 264 | 30,666.56 | 8.22 | Nuclear |
| SgNAC13 | 263 | 29,969.94 | 9.03 | Nuclear |
| SgNAC14 | 126 | 13,603.23 | 6.00 | Nuclear |
| SgNAC15 | 245 | 28,377.47 | 6.01 | Nuclear |
| SgNAC16 | 243 | 27,807.89 | 7.70 | Nuclear |
| SgNAC17 | 229 | 26,021.46 | 9.55 | Nuclear |
| SgNAC18 | 211 | 24,039.92 | 5.75 | Nuclear |
| SgNAC19 | 209 | 24,255.47 | 9.02 | Nuclear |
| SgNAC20 | 203 | 22,706.82 | 4.67 | Extracellular |
| SgNAC21 | 103 | 11,972.49 | 5.35 | Nuclear |
| SgNAC22 | 143 | 16,120.71 | 4.59 | Extracellular |
| SgNAC23 | 133 | 15,816.1 | 9.21 | Nuclear |
| SgNAC24 | 132 | 15,558.77 | 10.11 | Nuclear |
| SgNAC25 | 130 | 15,335.51 | 8.85 | Nuclear |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Zhu, L.; Yu, X.; Yang, Q.; Yuan, F.; Jin, H. Identification of NAC Transcription Factors in Suaeda glauca and Their Responses to Salt Stress. Curr. Issues Mol. Biol. 2024, 46, 8741-8751. https://doi.org/10.3390/cimb46080516
Fu X, Zhu L, Yu X, Yang Q, Yuan F, Jin H. Identification of NAC Transcription Factors in Suaeda glauca and Their Responses to Salt Stress. Current Issues in Molecular Biology. 2024; 46(8):8741-8751. https://doi.org/10.3390/cimb46080516
Chicago/Turabian StyleFu, Xujun, Longmin Zhu, Xiaomin Yu, Qinghua Yang, Fengjie Yuan, and Hangxia Jin. 2024. "Identification of NAC Transcription Factors in Suaeda glauca and Their Responses to Salt Stress" Current Issues in Molecular Biology 46, no. 8: 8741-8751. https://doi.org/10.3390/cimb46080516
APA StyleFu, X., Zhu, L., Yu, X., Yang, Q., Yuan, F., & Jin, H. (2024). Identification of NAC Transcription Factors in Suaeda glauca and Their Responses to Salt Stress. Current Issues in Molecular Biology, 46(8), 8741-8751. https://doi.org/10.3390/cimb46080516





