Novel Plant Breeding Techniques Shake Hands with Cereals to Increase Production
Abstract
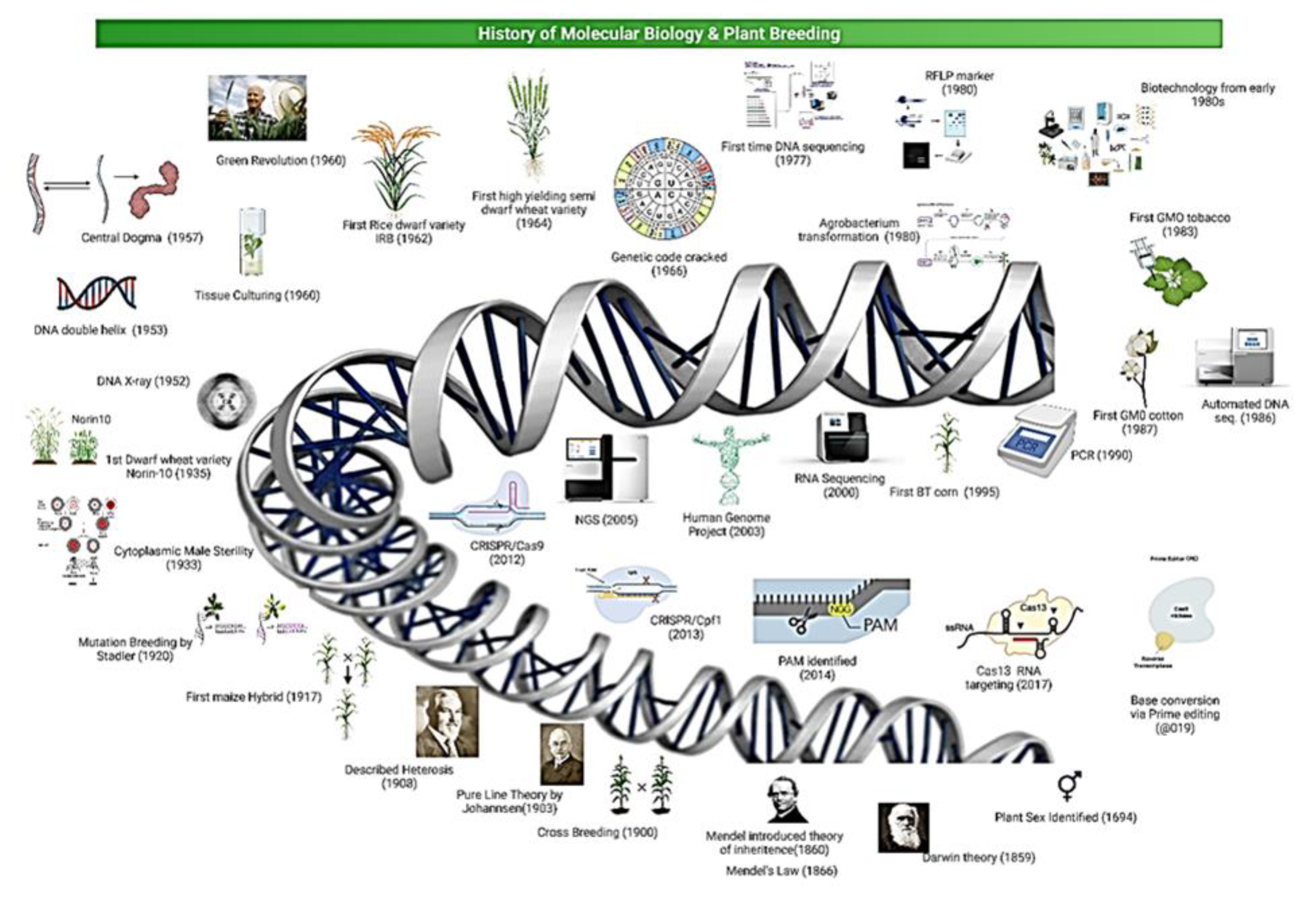
:1. Introduction
2. Development of Genomic Technologies and Role in Plant Breeding
3. Genome Editing with DSBs
3.1. CRISPR from Yogurt to Plant Breeding
3.2. CRISPR/Cas9 and Cpf1 in Genome Editing
3.3. Genome Editing (with DSBs) Role in Cereal’s Genome Improvement
3.4. Genome Editing without DSBs and Donor Template
3.4.1. Base Editing
3.4.2. Epigenetic Editing
3.4.3. Prime Editing
4. Genome Editing (without DSBs) Role in Cereals’ Improvement
5. Developing Genome-Edited Plants Free of Transgene
5.1. Isolate Segregants by Mendelian Segregation
5.2. Programmed Self-Elimination of Transgene Plants
5.3. Transient CRISPR/Cas9 Gene Expression by Protoplast
5.4. RNP-Mediated Genome Editing
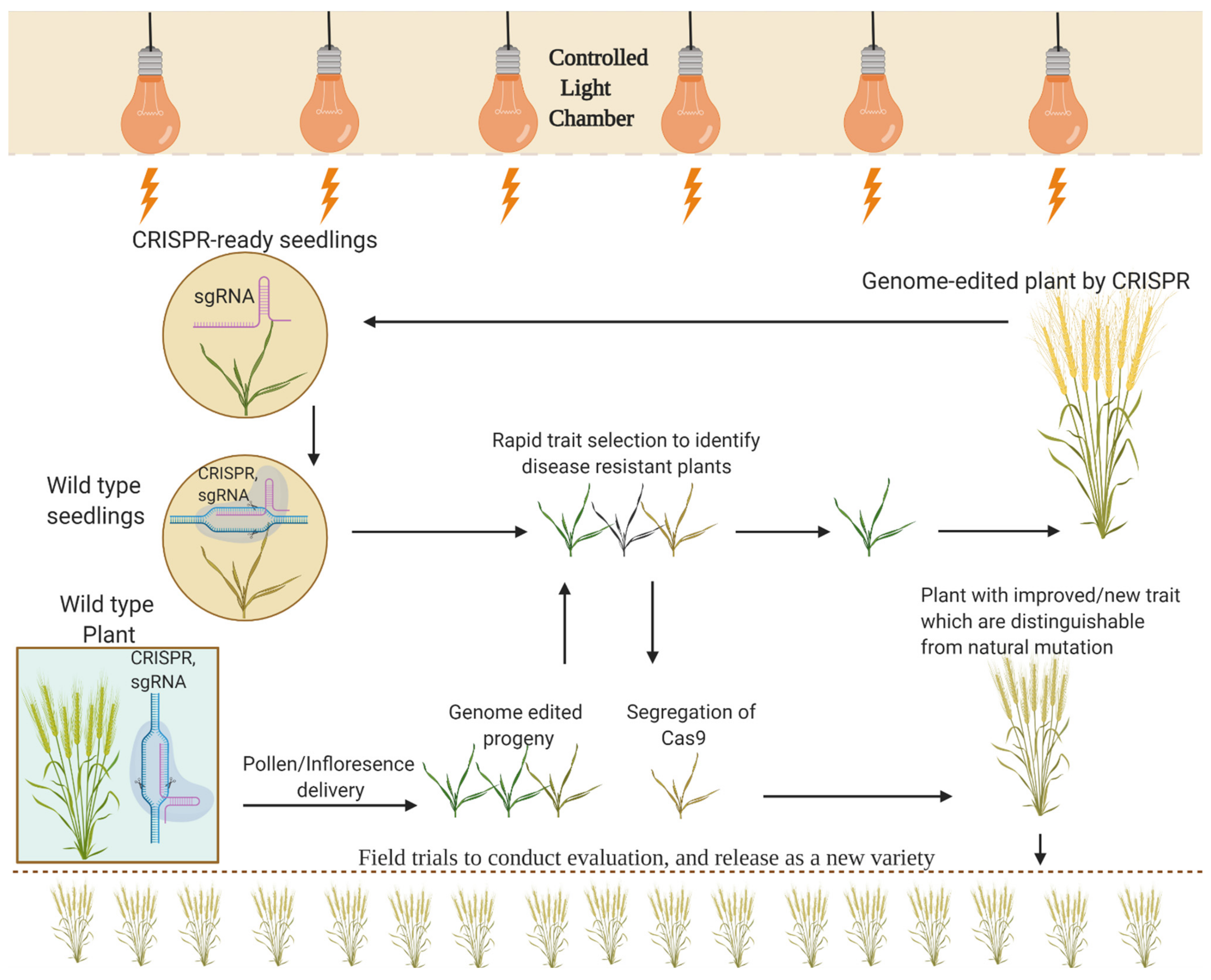
6. Speed Breeding
6.1. An Amalgamation of Speed Breeding with GETs
6.2. Achievements of Speed Breeding
7. Future Prospects
7.1. Solutions by Improved GETs
7.2. Increase in Regeneration Efficiency
7.3. Speed Breeding to Rule out GMOs and Shorten the Breeding Procedure
7.4. Transcriptional Factors
Author Contributions
Funding
Conflicts of Interest
References
- Haroon, M.; Idrees, F.; Naushahi, H.A.; Afzal, R.; Usman, M.; Qadir, T.; Rauf, H. Nitrogen Use Efficiency: Farming Practices and Sustainability. J. Exp. Agric. Int. 2019, 36, 1–11. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations (FAO). High Level Expert Forum—How to Feed the World in 2050. 2009, pp. 1–4. Available online: https://www.fao.org/wsfs/forum2050/wsfs-forum/en/ (accessed on 8 December 2021).
- Searchinger, T.; Hanson, C.; Ranganathan, J.; Lipinski, B.; Waite, R.; Winterbottom, R.; Dinshaw, A.; Heimlich, R.; Boval, M.; Chemineau, P. Creating a Sustainable Food Future. A Menu of Solutions to Sustainably Feed More Than 9 Billion People by 2050; World resources report 2013-14: Interim findings 2014; World Resources Institute: Washington, DC, USA, 2014. [Google Scholar]
- Ahmar, S.; Saeed, S.; Hafeez, M.; Khan, U.; Khan, S.U. A Revolution toward Gene-Editing Technology and Its Application to Crop A Revolution toward Gene-Editing Technology and Its Application to Crop Improvement. Int. J. Mol. Sci. 2020, 21, 5665. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Managing Land Carrying Capacity: Key to Achieving Sustainable Production Systems for Food Security. Land 2022, 11, 484. [Google Scholar] [CrossRef]
- Topp, C.N.; Jez, J.M. Introduction to emerging technologies in plant science. Emerg. Top. Life Sci. 2021, 5, 177–178. [Google Scholar] [CrossRef]
- Khush, G.S. Green revolution: The way forward. Nat. Rev. Genet. 2001, 2, 815–822. [Google Scholar] [CrossRef]
- Haroon, M.; Afzal, R.; Idrees, F.; Sunny, A.; Khan, A.S. Genome Editing and Speed Breeding; Game Changers to Boost the Crop Production. Int. J. Biol. Res. 2019, 2, 295–300. [Google Scholar]
- Pinstrup-Andersen, P.; Hazell, P.B.R. The impact of the green revolution and prospects for the future. Food Rev. Int. 1985, 1, 1–25. [Google Scholar] [CrossRef]
- Pearce, S. Towards the replacement of wheat ‘Green Revolution’genes. J. Exp. Bot. 2021, 72, 157–160. [Google Scholar] [CrossRef]
- Abdallah, N.A.; Prakash, C.S.; McHughen, A.G. Genome editing for crop improvement: Challenges and opportunities. GM Crops Food 2015, 6, 183–205. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Hua, K.; Lang, Z. Genome editing for horticultural crop improvement. Hortic. Res. 2019, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Osakabe, Y.; Osakabe, K. Genome editing with engineered nucleases in plants. Plant Cell Physiol. 2015, 56, 389–400. [Google Scholar] [CrossRef] [Green Version]
- Ahmar, S.; Gill, R.A.; Jung, K.H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Massel, K.; Godwin, I.D.; Gao, C. Applications and potential of genome editing in crop improvement. Genome Biol. 2018, 19, 210. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.-D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acquaah, G. Principles of Plant Genetics and Breeding; John Wiley & Sons: Hoboken, NJ, USA, 2009; ISBN 1444309013. [Google Scholar]
- Hickey, L.T.; Hafeez, A.N.; Robinson, H.; Jackson, S.A.; Leal-Bertioli, S.C.M.; Tester, M.; Gao, C.; Godwin, I.D.; Hayes, B.J.; Wulff, B.B.H. Breeding crops to feed 10 billion. Nat. Biotechnol. 2019, 37, 744–754. [Google Scholar] [CrossRef] [Green Version]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F.W. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol. J. 2016, 14, 496–502. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Jain, M. The CRISPR–Cas system for plant genome editing: Advances and opportunities. J. Exp. Bot. 2015, 66, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Billon, P.; Bryant, E.E.; Joseph, S.A.; Nambiar, T.S.; Hayward, S.B.; Rothstein, R.; Ciccia, A. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell 2017, 67, 1068–1079. [Google Scholar] [CrossRef] [Green Version]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Hua, K.; Tao, X.; Yuan, F.; Wang, D.; Zhu, J.-K. Precise A· T to G· C base editing in the rice genome. Mol. Plant 2018, 11, 627–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-guided genome editing for target gene mutations in wheat. G3 Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarts, D.C.; Jinek, M. Cas9 versus Cas12a/Cpf1: Structure–function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA 2018, 9, e1481. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Chen, B.; Hu, J.; Almeida, R.; Liu, H.; Balakrishnan, S.; Covill-Cooke, C.; Lim, W.A.; Huang, B. Expanding the CRISPR imaging toolset with Staphylococcus aureus Cas9 for simultaneous imaging of multiple genomic loci. Nucleic Acids Res. 2016, 44, e75. [Google Scholar] [CrossRef] [Green Version]
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 2016, 14, 483–495. [Google Scholar] [CrossRef]
- Endo, A.; Masafumi, M.; Kaya, H.; Toki, S. Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 2016, 6, 38169. [Google Scholar] [CrossRef] [Green Version]
- Safari, F.; Zare, K.; Negahdaripour, M.; Barekati-Mowahed, M.; Ghasemi, Y. CRISPR Cpf1 proteins: Structure, function and implications for genome editing. Cell Biosci. 2019, 9, 36. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, S.K. Molecular Genetics, Genomics, and Biotechnology in Crop Plant Breeding. Agronomy 2020, 10, 439. [Google Scholar] [CrossRef] [Green Version]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Zafar, M.M.; Ali, A.; Hafeez, A.; Sharif, F.; Guan, X.; Deng, X.; Pengtao, L.; Shi, Y.; Haroon, M.; et al. The Pivotal Role of Major Chromosomes of Sub-Genomes A and D in Fiber Quality Traits of Cotton. Front. Genet. 2022, 12, 642595. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, A.J.; Chao, S.; Asoro, F.G.; Heffner, E.L.; Hayashi, T.; Iwata, H.; Smith, K.P.; Sorrells, M.E.; Jannink, J.-L. Genomic selection in plant breeding: Knowledge and prospects. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2011; Volume 110, pp. 77–123. ISBN 0065-2113. [Google Scholar]
- Peleman, J.D.; van der Voort, J.R. Breeding by design. Trends Plant Sci. 2003, 8, 330–334. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of genomic tools in plant breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [Green Version]
- Visendi, P.; Batley, J.; Edwards, D. Next generation characterisation of cereal genomes for marker discovery. Biology 2013, 2, 1357–1377. [Google Scholar] [CrossRef] [Green Version]
- Muir, P.; Li, S.; Lou, S.; Wang, D.; Spakowicz, D.J.; Salichos, L.; Zhang, J.; Weinstock, G.M.; Isaacs, F.; Rozowsky, J.; et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016, 17, 53. [Google Scholar] [CrossRef] [Green Version]
- Sahu, P.K.; Sao, R.; Mondal, S.; Vishwakarma, G.; Gupta, S.K.; Kumar, V.; Singh, S.; Sharma, D.; Das, B.K. Next Generation Sequencing Based Forward Genetic Approaches for Identification and Mapping of Causal Mutations in Crop Plants: A Comprehensive Review. Plants 2020, 9, 1355. [Google Scholar] [CrossRef]
- Le Nguyen, K.; Grondin, A.; Courtois, B.; Gantet, P. Next-generation sequencing accelerates crop gene discovery. Trends Plant Sci. 2019, 24, 263–274. [Google Scholar] [CrossRef]
- Alkan, C.; Sajjadian, S.; Eichler, E.E. Limitations of next-generation genome sequence assembly. Nat. Methods 2011, 8, 61–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monson-Miller, J.; Sanchez-Mendez, D.C.; Fass, J.; Henry, I.M.; Tai, T.H.; Comai, L. Reference genome-independent assessment of mutation density using restriction enzyme-phased sequencing. BMC Genom. 2012, 13, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christina Szalinski Yogurt Shows the Way for a Revolution in Genome Editing. Available online: https://www.ascb.org/science-news/yogurt-shows-the-way-for-a-revolution-in-genome-editing/ (accessed on 8 December 2021).
- Jansen, R.; van Embden, J.D.A.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E. V Annotation and Classification of CRISPR-Cas Systems. Methods Mol. Biol. 2015, 1311, 47–75. [Google Scholar] [CrossRef] [Green Version]
- Haft, D.H.; Selengut, J.D.; Brinkac, L.M.; Zafar, N.; White, O. Genome Properties: A system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics 2005, 21, 293–306. [Google Scholar] [CrossRef] [Green Version]
- Hale, C.R.; Zhao, P.; Olson, S.; Duff, M.O.; Graveley, B.R.; Wells, L.; Terns, R.M.; Terns, M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009, 139, 945–956. [Google Scholar] [CrossRef] [Green Version]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Kazi, T.A.; Biswas, S.R. Chapter Four—CRISPR/dCas system as the modulator of gene expression. In Advances in CRISPR/Cas and Related Technologies; Academic Press: Cambridge, MA, USA, 2021; pp. 99–122. Volume 178, ISBN 1877-1173. [Google Scholar]
- Bortesi, L.; Zhu, C.; Zischewski, J.; Perez, L.; Bassié, L.; Nadi, R.; Forni, G.; Lade, S.B.; Soto, E.; Jin, X.; et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 2016, 14, 2203–2216. [Google Scholar] [CrossRef]
- Li, C.; Brant, E.; Budak, H.; Zhang, B. CRISPR/Cas: A Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J. Zhejiang Univ. Sci. B 2021, 22, 253–284. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, K.-Y.B.; Chan, Y.-H.; Abdullah, W.M.A.N.W.; Lim, S.-H.E.; Lai, K.-S. The CRISPR/Cas9 System for Crop Improvement: Progress and Prospects. Next Gener. Plant Breed. 2018, 129, 129–145. [Google Scholar] [CrossRef] [Green Version]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, L. Exploring class 1 CRISPR systems. Nat. Methods 2019, 16, 1079. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR–Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Makarova, K.S.; Zhang, F.; Koonin, E. V SnapShot: Class 1 CRISPR-Cas Systems. Cell 2017, 168, 946.e1. [Google Scholar] [CrossRef]
- Bin Moon, S.; Lee, J.M.; Kang, J.G.; Lee, N.-E.; Ha, D.-I.; Kim, D.Y.D.; Kim, S.H.; Yoo, K.; Kim, D.Y.D.; Ko, J.-H.; et al. Highly efficient genome editing by CRISPR-Cpf1 using CRISPR RNA with a uridinylate-rich 3’-overhang. Nat. Commun. 2018, 9, 3651. [Google Scholar] [CrossRef]
- Liu, Y.; Han, J.; Chen, Z.; Wu, H.; Dong, H.; Nie, G. Engineering cell signaling using tunable CRISPR–Cpf1-based transcription factors. Nat. Commun. 2017, 8, 2095. [Google Scholar] [CrossRef] [Green Version]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Alok, A.; Sandhya, D.; Jogam, P.; Rodrigues, V.; Bhati, K.K.; Sharma, H.; Kumar, J. The Rise of the CRISPR/Cpf1 System for Efficient Genome Editing in Plants. Front. Plant Sci. 2020, 11, 264. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Ming, M.; Ren, Q.; Pan, C.; He, Y.; Zhang, Y.; Liu, S.; Zhong, Z.; Wang, J.; Malzahn, A.A.; Wu, J.; et al. CRISPR–Cas12b enables efficient plant genome engineering. Nat. Plants 2020, 6, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Ueta, R.; Osakabe, Y.; Osakabe, K. Precision genome editing in plants: State-of-the-art in CRISPR/Cas9-based genome engineering. BMC Plant Biol. 2020, 20, 234. [Google Scholar] [CrossRef] [PubMed]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395–2410. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. = Yi chuan Xue Bao 2016, 43, 529. [Google Scholar] [CrossRef]
- Sánchez-León, S.; Gil-Humanes, J.; Ozuna, C.V.; Giménez, M.J.; Sousa, C.; Voytas, D.F.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Farhat, S.; Jain, N.; Singh, N.; Sreevathsa, R.; Dash, P.K.; Rai, R.; Yadav, S.; Kumar, P.; Sarkar, A.K.; Jain, A. CRISPR-Cas9 directed genome engineering for enhancing salt stress tolerance in rice. Semin. Cell Dev. Biol. 2019, 96, 91–99. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 2018, 60, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhou, W.; Ren, Y.; Tian, X.; Lv, T.; Wang, Z.; Fang, J.; Chu, C.; Yang, J.; Bu, Q. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. = Yi Chuan Xue Bao 2017, 44, 175. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Bo, W.; Zhaohui, Z.; Huanhuan, Z.; Xia, W.; Binglin, L.I.U.; Lijia, Y.; Xiangyan, H.A.N.; Deshui, Y.; Xuelian, Z.; Chunguo, W. Targeted mutagenesis of NAC transcription factor gene, OsNAC041, leading to salt sensitivity in rice. Rice Sci. 2019, 26, 98–108. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [Green Version]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.-J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, D.; Zhang, D.; Zhao, X.; Cao, X.; Dong, L.; Liu, J.; Chen, K.; Zhang, H.; Gao, C. Analysis of the functions of Ta GW 2 homoeologs in wheat grain weight and protein content traits. Plant J. 2018, 94, 857–866. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef] [PubMed]
- Smulders, M.J.M.; Jouanin, A.A.; Gilissen, L.J.W.J. Gene editing using CRISPR/Cas9 to modify or remove gliadins from wheat and produce coeliac disease epitope-free wheat. In Proceedings of the Proceedings of the 31st Meeting of the Working Group on Prolamin Analysis and Toxicity, Minden, Germany, 28–30 September 2018; pp. 63–68.
- Bhowmik, P.; Ellison, E.; Polley, B.; Bollina, V.; Kulkarni, M.; Ghanbarnia, K.; Song, H.; Gao, C.; Voytas, D.F.; Kagale, S. Targeted mutagenesis in wheat microspores using CRISPR/Cas9. Sci. Rep. 2018, 8, 6502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayani, S.; Sabzalian, M.R.; Mazaheri-Tirani, M. CRISPR/Cas9 Genome Editing in Bread Wheat (Triticum aestivum L.) Genetic Improvement. In Advances in Plant Breeding Strategies: Cereals; Springer: Berlin/Heidelberg, Germany, 2019; pp. 453–469. [Google Scholar]
- Liang, Z.; Chen, K.; Zhang, Y.; Liu, J.; Yin, K.; Qiu, J.-L.; Gao, C. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 2018, 13, 413. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. Cris. J. 2018, 1, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, M.; Kumar, M.; Albertsen, M.C.; Young, J.K.; Cigan, A.M. Concurrent modifications in the three homeologs of Ms45 gene with CRISPR-Cas9 lead to rapid generation of male sterile bread wheat (Triticum aestivum L.). Plant Mol. Biol. 2018, 97, 371–383. [Google Scholar] [CrossRef]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Li, J.; Zhang, H.; Si, X.; Tian, Y.; Chen, K.; Liu, J.; Chen, H.; Gao, C. Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genom. = Yi Chuan Xue Bao 2017, 44, 465. [Google Scholar] [CrossRef]
- Yao, L.; Zhang, Y.; Liu, C.; Liu, Y.; Wang, Y.; Liang, D.; Liu, J.; Sahoo, G.; Kelliher, T. OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants 2018, 4, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational Improvement of Rice Yield and Cold Tolerance by Editing the Three Genes OsPIN5b, GS3, and OsMYB30 With the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar] [CrossRef] [Green Version]
- Butt, H.; Eid, A.; Ali, Z.; Atia, M.A.M.; Mokhtar, M.M.; Hassan, N.; Lee, C.M.; Bao, G.; Mahfouz, M.M. Efficient CRISPR/Cas9-mediated genome editing using a chimeric single-guide RNA molecule. Front. Plant Sci. 2017, 8, 1441. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Shi, J.; Wang, H.; Wu, Y.; Hazebroek, J.; Meeley, R.B.; Ertl, D.S. The maize low-phytic acid mutant lpa2 is caused by mutation in an inositol phosphate kinase gene. Plant Physiol. 2003, 131, 507–515. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Anand, A.; Quick, P.; Bandyopadhyay, A. Editing a stomatal developmental gene in rice with CRISPR/Cpf1. In Plant Genome Editing with CRISPR Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 257–268. [Google Scholar]
- Malzahn, A.A.; Tang, X.; Lee, K.; Ren, Q.; Sretenovic, S.; Zhang, Y.; Chen, H.; Kang, M.; Bao, Y.; Zheng, X. Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol. 2019, 17, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhang, X.; Wang, W.; Guo, X.; Wu, Z.; Du, W.; Zhao, Y.; Xia, L. Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice. Mol. Plant 2018, 11, 995–998. [Google Scholar] [CrossRef] [Green Version]
- Xu, R.; Qin, R.; Li, H.; Li, D.; Li, L.; Wei, P.; Yang, J. Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 2017, 15, 713–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.; Mao, Y.; Lu, Y.; Tao, X.; Zhu, J.K. Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol. Plant 2017, 10, 1011–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Zhang, Y.; Kleinstiver, B.P.; Guo, J.A.; Aryee, M.J.; Miller, J.; Malzahn, A.; Zarecor, S.; Lawrence-Dill, C.J.; Joung, J.K. Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol. J. 2019, 17, 362–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, R.; Qin, R.; Li, H.; Li, J.; Yang, J.; Wei, P. Enhanced genome editing in rice using single transcript unit CRISPR-LbCpf1 systems. Plant Biotechnol. J. 2019, 17, 553. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Lowder, L.G.; Zhang, T.; Malzahn, A.A.; Zheng, X.; Voytas, D.F.; Zhong, Z.; Chen, Y.; Ren, Q.; Li, Q. A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants 2017, 3, 17018. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Zhang, Y.; You, Q.; Tang, X.; Ren, Q.; Liu, S.; Yang, L.; Wang, Y.; Liu, X.; Liu, B. Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol. Plant 2018, 11, 999–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390. [Google Scholar] [CrossRef]
- Char, S.N.; Unger-Wallace, E.; Frame, B.; Briggs, S.A.; Main, M.; Spalding, M.H.; Vollbrecht, E.; Wang, K.; Yang, B. Heritable site-specific mutagenesis using TALENs in maize. Plant Biotechnol. J. 2015, 13, 1002–1010. [Google Scholar] [CrossRef] [Green Version]
- Shan, Q.; Zhang, Y.; Chen, K.; Zhang, K.; Gao, C. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol. J. 2015, 13, 791–800. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Chen, K.; Liang, Z.; Li, J.; Zhang, Y.; Zhang, K.; Liu, J.; Voytas, D.F.; Zheng, X. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 2013, 6, 1365–1368. [Google Scholar] [CrossRef] [Green Version]
- Kelliher, T.; Starr, D.; Richbourg, L.; Chintamanani, S.; Delzer, B.; Nuccio, M.L.; Green, J.; Chen, Z.; McCuiston, J.; Wang, W.; et al. MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 2017, 542, 105–109. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; Meyer, A.; Römer, P.; Jordan, T.; Lahaye, T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006, 163, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Cantos, C.; Francisco, P.; Trijatmiko, K.R.; Slamet-Loedin, I.; Chadha-Mohanty, P.K. Identification of “safe harbor” loci in indica rice genome by harnessing the property of zinc-finger nucleases to induce DNA damage and repair. Front. Plant Sci. 2014, 5, 302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Ainley, W.M.; Sastry-Dent, L.; Welter, M.E.; Murray, M.G.; Zeitler, B.; Amora, R.; Corbin, D.R.; Miles, R.R.; Arnold, N.L.; Strange, T.L. Trait stacking via targeted genome editing. Plant Biotechnol. J. 2013, 11, 1126–1134. [Google Scholar] [CrossRef]
- Gao, H.; Smith, J.; Yang, M.; Jones, S.; Djukanovic, V.; Nicholson, M.G.; West, A.; Bidney, D.; Falco, S.C.; Jantz, D.; et al. Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J. 2010, 61, 176–187. [Google Scholar] [CrossRef]
- Djukanovic, V.; Smith, J.; Lowe, K.; Yang, M.; Gao, H.; Jones, S.; Nicholson, M.G.; West, A.; Lape, J.; Bidney, D. Male-sterile maize plants produced by targeted mutagenesis of the cytochrome P 450-like gene (MS 26) using a re-designed I–C reI homing endonuclease. Plant J. 2013, 76, 888–899. [Google Scholar] [CrossRef]
- Youssef, D.; Nihou, A.; Partier, A.; Tassy, C.; Paul, W.; Rogowsky, P.M.; Beckert, M.; Barret, P. Induction of targeted deletions in transgenic bread wheat (Triticum aestivum L.) using customized meganuclease. Plant Mol. Biol. Rep. 2018, 36, 71–81. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480–485. [Google Scholar] [CrossRef]
- Marzec, M.; Hensel, G. Prime Editing: Game Changer for Modifying Plant Genomes. Trends Plant Sci. 2020, 25, 722–724. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Joshi, R.K.; Zhao, K. Base editing in crops: Current advances, limitations and future implications. Plant Biotechnol. J. 2020, 18, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Li, J.; Liu, X.; Shan, T.; Qin, R.; Wei, P. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Commun. 2020, 1, 100043. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Yang, L.; Briggs, A.W.; Chew, W.L.; Mali, P.; Guell, M.; Aach, J.; Goodman, D.B.; Cox, D.; Kan, Y.; Lesha, E.; et al. Engineering and optimising deaminase fusions for genome editing. Nat. Commun. 2016, 7, 13330. [Google Scholar] [CrossRef] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Xie, N.; Zhou, Y.; Sun, Q.; Tang, B. Novel Epigenetic Techniques Provided by the CRISPR/Cas9 System. Stem Cells Int. 2018, 2018, 7834175. [Google Scholar] [CrossRef]
- Hauser, M.-T.; Aufsatz, W.; Jonak, C.; Luschnig, C. Transgenerational epigenetic inheritance in plants. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2011, 1809, 459–468. [Google Scholar] [CrossRef] [Green Version]
- Singroha, G.; Sharma, P. Epigenetic Modifications in Plants under Abiotic Stress. In Epigenetics; IntechOpen: London, UK, 2019. [Google Scholar]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Zheng, N.; Li, L.; Wang, X. Molecular mechanisms, off-target activities, and clinical potentials of genome editing systems. Clin. Transl. Med. 2020, 10, 412–426. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; He, S.; Huang, S.; Li, C.; Chen, Y.; Liu, Z.; Huang, X.; Wang, X. Efficient generation of mouse models with the prime editing system. Cell Discov. 2020, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Zong, Y.; Liu, Y.; Xue, C.; Li, B.; Li, X.; Wang, Y.; Li, J.; Liu, G.; Huang, X.; Cao, X.; et al. An engineered prime editor with enhanced editing efficiency in plants. Nat. Biotechnol. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhu, J.-K. Precise Editing of a Target Base in the Rice Genome Using a Modified CRISPR/Cas9 System. Mol. Plant 2017, 10, 523–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, A.; Ueguchi-Tanaka, M.; Sonoda, Y.; Kitano, H.; Koshioka, M.; Futsuhara, Y.; Matsuoka, M.; Yamaguchi, J. slender Rice, a Constitutive Gibberellin Response Mutant, Is Caused by a Null Mutation of the SLR1 Gene, an Ortholog of the Height-Regulating Gene GAI/RGA/RHT/D8. Plant Cell 2001, 13, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [Green Version]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y.; Qiu, J.-L.; Wang, D.; Gao, C. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438. [Google Scholar] [CrossRef]
- Gallego-Bartolomé, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.-C.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef] [Green Version]
- Stuitje, A.R.; Verbree, E.C.; Van Der Linden, K.H.; Mietkiewska, E.M.; Nap, J.; Kneppers, T.J.A. Seed-expressed fluorescent proteins as versatile tools for easy (co) transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol. J. 2003, 1, 301–309. [Google Scholar] [CrossRef]
- Durr, J.; Papareddy, R.; Nakajima, K.; Gutierrez-Marcos, J. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 2018, 8, 4443. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Y. Fluorescence marker-assisted isolation of Cas9-free and CRISPR-edited Arabidopsis plants. In Plant Genome Editing with CRISPR Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 147–154. [Google Scholar]
- Gao, X.; Chen, J.; Dai, X.; Zhang, D.; Zhao, Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016, 171, 1794–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, H.; Liu, S.; Xu, S.; Chen, W.; Zhou, X.; Tan, Y.; Huang, J.; Shu, Q. CRISPR-S: An active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 2017, 15, 1371. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, K.; Huang, W.; Liu, G.; Gao, Y.; Wang, J.; Huang, Q.; Ji, Y.; Qin, X.; Wan, L. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 2012, 24, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Zhu, M.; Wang, L.; Wu, J.; Wang, Q.; Wang, R.; Zhao, Y. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant 2018, 11, 1210–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.; Turesson, H.; Nicolia, A.; Fält, A.-S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.; Hsu, C.; Yang, L.; Lee, L.; Fu, J.; Cheng, Q.; Wu, F.; Hsiao, H.C.; Zhang, Y.; Zhang, R. Application of protoplast technology to CRISPR/Cas9 mutagenesis: From single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 2018, 16, 1295–1310. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Zhang, S.; Wang, W.; Xiong, X.; Meng, F.; Cui, X. Efficient targeted mutagenesis in potato by the CRISPR/Cas9 system. Plant Cell Rep. 2015, 34, 1473–1476. [Google Scholar] [CrossRef] [Green Version]
- Subburaj, S.; Chung, S.J.; Lee, C.; Ryu, S.-M.; Kim, D.H.; Kim, J.-S.; Bae, S.; Lee, G.-J. Site-directed mutagenesis in Petunia× hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016, 35, 1535–1544. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Murovec, J.; Guček, K.; Bohanec, B.; Avbelj, M.; Jerala, R. DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci. 2018, 9, 1594. [Google Scholar] [CrossRef]
- Chiurugwi, T.; Kemp, S.; Powell, W.; Hickey, L.T. Speed breeding orphan crops. Theor. Appl. Genet. 2019, 132, 607–616. [Google Scholar] [CrossRef]
- Razzaq, A.; Kaur, P.; Akhter, N.; Wani, S.H.; Saleem, F. Next-Generation Breeding Strategies for Climate-Ready Crops. Front. Plant Sci. 2021, 12, 620420. [Google Scholar] [CrossRef] [PubMed]
- Hickey, L.T.; Germán, S.E.; Pereyra, S.A.; Diaz, J.E.; Ziems, L.A.; Fowler, R.A.; Platz, G.J.; Franckowiak, J.D.; Dieters, M.J. Speed breeding for multiple disease resistance in barley. Euphytica 2017, 213, 64. [Google Scholar] [CrossRef]
- Alahmad, S.; Dinglasan, E.; Leung, K.M.; Riaz, A.; Derbal, N.; Voss-Fels, K.P.; Able, J.A.; Bassi, F.M.; Christopher, J.; Hickey, L.T. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods 2018, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.J.; Wright, G.C.; Dieters, M.J.; George, D.L.; Hunter, M.N.; Tatnell, J.R.; Fleischfresser, D.B. Development and application of speed breeding technologies in a commercial peanut breeding program. Peanut Sci. 2013, 40, 107–114. [Google Scholar] [CrossRef]
- Fiyaz, R.A.; Ajay, B.C.; Ramya, K.T.; Kumar, J.A.; Sundaram, R.M.; Rao, L.V.S. Speed Breeding: Methods and Applications. In Accelerated Plant Breeding, Volume 1; Springer: Berlin/Heidelberg, Germany, 2020; pp. 31–49. [Google Scholar]
- Ghosh, S.; Watson, A.; Gonzalez-Navarro, O.E.; Ramirez-Gonzalez, R.H.; Yanes, L.; Mendoza-Suárez, M.; Simmonds, J.; Wells, R.; Rayner, T.; Green, P.; et al. Speed breeding in growth chambers and glasshouses for crop breeding and model plant research. Nat. Protoc. 2018, 13, 2944–2963. [Google Scholar] [CrossRef] [Green Version]
- Samantara, K.; Bohra, A.; Mohapatra, S.R.; Prihatini, R.; Asibe, F.; Singh, L.; Reyes, V.P.; Tiwari, A.; Maurya, A.K.; Croser, J.S.; et al. Breeding More Crops in Less Time: A Perspective on Speed Breeding. Biology 2022, 11, 275. [Google Scholar] [CrossRef]
- Sysoeva, M.I.; Markovskaya, E.F.; Shibaeva, T.G. Plants under continuous light: A review. Plant Stress 2010, 4, 5–17. [Google Scholar]
- Jähne, F.; Hahn, V.; Würschum, T.; Leiser, W.L. Speed breeding short-day crops by LED-controlled light schemes. Theor. Appl. Genet. 2020, 133, 2335–2342. [Google Scholar] [CrossRef]
- Velez-Ramirez, A.I.; Van Ieperen, W.; Vreugdenhil, D.; Millenaar, F.F. Plants under continuous light. Trends Plant Sci. 2011, 16, 310–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, S.R.; Langton, F.A. Photoperiod and plant growth: A review. J. Hortic. Sci. Biotechnol. 2005, 80, 2–10. [Google Scholar] [CrossRef]
- Sharma, A. Speed Breeding: A Race against Time. Agrobios Newsletter, 1 May 2019; 73–74. [Google Scholar]
- Richardson, T.; Thistleton, J.; Higgins, T.J.; Howitt, C.; Ayliffe, M. Efficient Agrobacterium transformation of elite wheat germplasm without selection. Plant Cell, Tissue Organ Cult. 2014, 119, 647–659. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Linghu, Q.; Nagira, Y.; Miki, R.; Taoka, N.; Imai, R. An in planta biolistic method for stable wheat transformation. Sci. Rep. 2017, 7, 11443. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Lu, Y.; Botella, J.R.; Mao, Y.; Hua, K.; Zhu, J.-K. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Mol. Plant 2017, 10, 1007–1010. [Google Scholar] [CrossRef] [Green Version]
- Hen-Avivi, S.; Savin, O.; Racovita, R.C.; Lee, W.-S.; Adamski, N.M.; Malitsky, S.; Almekias-Siegl, E.; Levy, M.; Vautrin, S.; Bergès, H. A metabolic gene cluster in the wheat W1 and the barley Cer-cqu loci determines β-diketone biosynthesis and glaucousness. Plant Cell 2016, 28, 1440–1460. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wei, W.; Zhu, H.; Challa, G.S.; Bi, C.; Trick, H.N.; Li, W. W3 is a new wax locus that is essential for biosynthesis of β-diketone, development of glaucousness, and reduction of cuticle permeability in common wheat. PLoS ONE 2015, 10, e0140524. [Google Scholar] [CrossRef] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Rana, M.M.; Takamatsu, T.; Baslam, M.; Kaneko, K.; Itoh, K.; Harada, N.; Sugiyama, T.; Ohnishi, T.; Kinoshita, T.; Takagi, H. Salt tolerance improvement in rice through efficient SNP marker-assisted selection coupled with speed-breeding. Int. J. Mol. Sci. 2019, 20, 2585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shivakumar, M.; Nataraj, V.; Kumawat, G.; Rajesh, V.; Chandra, S.; Gupta, S.; Bhatia, V.S. Speed breeding for Indian Agriculture: A rapid method for development of new crop varieties. Curr Sci 2018, 115, 1241. [Google Scholar]
- Riaz, A. Unlocking New Sources of Adult Plant Resistance to Wheat Leaf Rust. Ph.D. Thesis, Qld Alliance for Agriculture and Food Innovation, The University of Queensland, St Lucia, QLD, Australia, 2018. [Google Scholar]
- Fund, S.A.D.; Ferrie, A.; Waterer, D. Development of Improved Spice Crops using Double Haploid Technology; Citeseer: Princeton, NJ, USA, 2009. [Google Scholar]
- Chaikam, V.; Molenaar, W.; Melchinger, A.E.; Boddupalli, P.M. Doubled haploid technology for line development in maize: Technical advances and prospects. Theor. Appl. Genet. 2019, 132, 3227–3243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, R.; Trethowan, R.; Ferrara, G.O.; Iwanaga, M.; Dodds, J.H.; Crouch, J.H.; Crossa, J.; Braun, H.-J. High yield potential, shuttle breeding, genetic diversity, and a new international wheat improvement strategy. Euphytica 2007, 157, 365–384. [Google Scholar] [CrossRef]
- Debernardi, J.M.; Tricoli, D.M.; Ercoli, M.F.; Hayta, S.; Ronald, P.; Palatnik, J.F.; Dubcovsky, J. A GRF–GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020, 38, 1274–1279. [Google Scholar] [CrossRef]
- Kong, J.; Martin-Ortigosa, S.; Finer, J.; Orchard, N.; Gunadi, A.; Batts, L.A.; Thakare, D.; Rush, B.; Schmitz, O.; Stuiver, M.; et al. Overexpression of the Transcription Factor GROWTH-REGULATING FACTOR5 Improves Transformation of Dicot and Monocot Species. Front. Plant Sci. 2020, 11, 572319. [Google Scholar] [CrossRef]
- Lee, S.-J.; Lee, B.H.; Jung, J.-H.; Park, S.K.; Song, J.T.; Kim, J.H. GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR Specify Meristematic Cells of Gynoecia and Anthers. Plant Physiol. 2018, 176, 717–729. [Google Scholar] [CrossRef] [Green Version]







| Functions | EMNs | ZFNs | TALENs | CRISPR/Cas9 | Base Editing | CRISPR/Cpf1 | References |
|---|---|---|---|---|---|---|---|
| Mode of action | In the target region direct conversion of information stand | In the target region double-strand breaks | In the targeted DNA region double-strand breaks | In the targetedDNA region double-strands or single-strand breaks | Single-Stranded Base changing | Double-stranded breaks | [19,20] |
| Target recognition | Good | Good | Good | Good | Very Good | Very Good | [20,21] |
| Mutation rate | Average | High | Average | Low | Low | High | [19,20,22,23] |
| Creation of large-scale libraries | Difficult to do | Impossible | Difficult to do | Possible | Possible | Possible | [20,24,25] |
| Multiplexing | Technically difficult | Hard to do | Hard to do | Possible | Possible | Possible | [19,20,26,27] |
| Components | Exogenous polynucleotide (chimeraplast) | Zn finger domains Nonspecific FokI nuclease domain | TALE DNA-binding domains Nonspecific FokI nuclease domain | Cas9 proteins, crRNA | CBEs, ABEs | Cpf1 proteins, crRNA | [20,21,28,29] |
| Structural protein | Dimeric protein | Dimeric protein | Dimeric protein | Monomeric Protein | Monomeric Protein | Monomeric Protein | [19,23] |
| Catalytic Domain | Absence of a catalytic domain | Restriction endonuclease FokI | Restriction endonuclease FokI | RuvC and HNH | RuvC and HNH | [20,28] | |
| Length of the target sequence (bp) | 68–88 | 24–36 | 24–59 | 20–22 | Point Mutation | 20–24 | [23,24,29,30] |
| Protein engineeringsteps | Not required | Required | Required | Not difficult to testgRNA | Not difficult to testgRNA | [20,25,31] | |
| Cloning | Unnecessary | Necessary | Necessary | Unnecessary | Unnecessary | [20,25,31] | |
| gRNA production | Not essential | Cannot apply | Cannot apply | Can produce easily | Can produce easily | [20,25,32] | |
| Target GETs | Not essential | ZFN Genome v2.0 ZifBASE Zinc-Finger Database (ZiFDB) Zinc-Finger Tool EENdb | TALE-NT 2.0 SPATA TALEN offer TALEN Library | CHOP CHOP CRISPRs web Server Crass: The CRISPRAssembler CRISPR Target | Cas nickase, Cpf1 adenosine deaminases, Cas13b | Breaking-Cas Cas-OFFinder CRISPORCCTOP | [29,31,33] |
| Off-target effects | Low off-target effect | Low off-target effect | Shows least off-target activities | Low off-target effect | Very Low | Low off-target effect | [20,22] |
| Cost of development | High | High | Higher | Low | Low | Low | [20,24,25] |
| Gene Editing Tool | Crop | Targeted Gene | Targeted Trait | Reference |
|---|---|---|---|---|
| CRISPR/Cas9 | Wheat | TaLOX2 | Development of grain | [72] |
| CRISPR/Cas9 | Maize | LIG1, Ms26. Ms45, ALS1, and ALS2 | Chlorsulfuron-resistant | [73] |
| CRISPR/Cas9 | Rice | GS3, GW2, GW5, TGW6, | Improved grain related parameters | [74] |
| CRISPR/Cas9 | Wheat | Gli-2 loci | Low-gluten foodstuff | [75] |
| CRISPR/Cas9 | Rice | OsPRX2 | Improved salt tolerance level | [76] |
| CRISPR/Cas9 | Wheat | TaInox, TaPds | Chlorophyll synthesis | [27] |
| CRISPR/Cas9 | Rice | Waxy | Enhanced glutinosity | [77] |
| CRISPR/Cas9 | Rice | Hd2, Hd4, Hd5 | Early heading | [78] |
| CRISPR/Cas9 | Maize | PPR, RPL | Reduced zein protein | [79] |
| CRISPR/Cas9 | Maize | ARGOS8 | Drought tolerance | [80] |
| CRISPR/Cas9 | Rice | OsNAC041 | Salt tolerant | [81] |
| CRISPR/Cas9 | Maize | ZmHKT1 | Salt tolerant | [82] |
| CRISPR/Cas9 | Rice | LAZY1 | Tiller spreading | [83] |
| CRISPR/Cas9 | Rice | Gn1a, GS3, DEP1 | Enhanced grain number, larger grain size, and dense erect panicles | [84] |
| CRISPR/Cas9 | Wheat | GW2 | Increased grain weight and protein content | [85] |
| CRISPR/Cas9 | Wheat | TaGASR7, TaGW2, TaDEP1, TdGASR7(durum wheat) | Grain development, kernel length, storability, and plant height and weight | [86] |
| CRISPR/Cas9 | Wheat | TaGW2, TaGASR7 | Grain and kernel length and weight | [87] |
| CRISPR/Cas9 | Wheat | α-gliadin, gamma-gliadins | Gliadins | [88] |
| CRISPR/Cas9 | Wheat | TaLOX2, TaUbil1 | Grain development | [89] |
| CRISPR/Cas9 | Wheat | TaDREB2,TaERF3 | Drought signaling | [90] |
| CRISPR/Cas9 | Wheat | TaCER9, TaLOX2,TaGW2 | Grain development | [91] |
| CRISPR/Cas9 | Wheat | TaGW2, TaLpx-1, TaMLO | Kernel width and weight; resistance to powdery mildew | [92] |
| CRISPR/Cas9 | Wheat | α-gliadin genes | Low-gluten wheat | [75] |
| CRISPR/Cas9 | Wheat | TaMs45 | Male fertility | [93] |
| CRISPR/Cas9 | Rice | OsSWEET13 | Bacterial blight resistance | [94] |
| CRISPR/Cas9 | Rice | SBEIIb | High amylose content | [95] |
| CRISPR/Cas9 | Wheat | EDR1 | Powdery mildew resistance | [96] |
| CRISPR/Cas9 | Rice | OsERF922 | Enhanced rice blast resistance | [97] |
| CRISPR/Cas9 | Rice | OsSWEET13 | Bacterial blight resistance | [97] |
| CRISPR/Cas9 | Maize | TMS5 | Thermosensitive male-sterile | [98] |
| CRISPR/Cas9 | Rice | OsMATL | Induction of haploid plants | [99] |
| CRISPR/Cas9 | Rice | OsPIN5b and GS3,OsMYB30 | High yielding and cold tolerance | [100] |
| CRISPR/Cas9 | Rice | ALS | Herbicide resistance | [72] |
| CRISPR/Cas9 | Rice | LAZY1 | Tiller spreading phenotype | [83] |
| CRISPR/Cas9 | Rice | Gn1a,DEP1, GS3 | Number of grains, erect panicles, specific for grain size | [84] |
| CRISPR/Cas9 | Rice | SBEIIb | High amylose rice | [95] |
| CRISPR/Cas9 | Rice | OsERF922 | Rice blast resistance | [94] |
| CRISPR/Cas9 | Rice | OsEPSPS | Glyphosate resistant | [101] |
| CRISPR/Cas9 | Rice | ALS | Herbicide resistance | [99] |
| CRISPR/Cas9 | Rice | ALS | Herbicide resistance | [102] |
| CRISPR/Cas9 | Rice | EPSPS | Herbicide resistance | [101] |
| CRISPR/Cas9 | Rice | ALS | Herbicide resistance | [103] |
| CRISPR/Cas9 | Maize | ALS | Herbicide resistance | [73] |
| CRISPR/Cas9 | Maize | ARGOS8 | Drought stress tolerance | [104] |
| CRISPR/Cas9 | Wheat | TaMLOA1, TaMLOB1,TaMLOD1 | Resistance to powderyMildew | [105] |
| CRISPR/Cas9 | Maize | PDS, IPK1A, IPK | Phytic acid content | [106] |
| CRISPR/Cpf1 | Rice | OsEPFL9 | To regulate the stomatal density in leaf | [107] |
| CRISPR/Cpf1 | Rice | OsROC5 and OsDEP1 | Editing efficiency was compared on varying temperature | [86,87] |
| CRISPR/Cpf1 | Maize | GL2 | Editing efficiency was compared on varying temperature | [108] |
| CRISPR/Cpf1 | Rice | DL, ALS, NCED1, AO1 | Drooping leaf phenotype | [32] |
| CRISPR/Cpf1 | Rice | OsPDS, OsBEL | Heritable mutations | [109,110] |
| CRISPR/Cpf1 | Rice | OsRLK, OsBEL | Albino phenotype | [111] |
| CRISPR/Cpf1 | Maize | glossy2 | Efficiency compared with CRISPR/Cas9 | [112] |
| CRISPR/Cpf1 | Rice | OsPDS, OsGS3 | Improved the editing efficiency | [113] |
| CRISPR/Cpf1 | Rice | OsDEP1, OsROC5, OsPDS | Tenfold reduction in miR159b transcription, transcriptional repression | [114] |
| CRISPR/Cpf1 | Rice | DEP1, PDS, EPFL9 | Efficient editing at all TTTV PAM sites | [115] |
| TALENs | Rice | OsSWEET14 | Bacterial blight resistance | [116] |
| TALENs | Wheat | TaMLO | Powdery mildew resistance | [105] |
| TALENs | Maize | ZmGL2 | Reduced epicuticular wax in leaves | [117] |
| TALENs | Rice | OsBADH2 | Fragrant rice | [118] |
| TALENs | Rice | DEP1, CKX2, BADH2, SD1 | Rapid and efficient gene modification in rice | [119] |
| TALENs | Maize | ZmMTL | Induction of haploid plants | [120] |
| TALENs | Maize | PDS, IPK1A, IPK and MRP4 | Reduce the phosphorous concentration | [121] |
| TALEN | Wheat | TaMLO | Powdery mildew resistance | [105] |
| ZFN | Maize | PAT | Herbicide resistance | [122] |
| ZFN | Rice | OsQQR | Detection of safe harbor loci herbicide | [123] |
| ZFNs | Maize | ZmIPK1 | Herbicide tolerant and phytate reduced maize | [124] |
| ZFNs | Maize | ZmTLP | Trait stacking | [125] |
| ZFNs | Rice | OsQQR | Trait stacking | [123] |
| MNs | Maize | lg1,ms26 | Targeted mutation | [126] |
| MNs | Maize | ms26 | Male sterility | [127] |
| MNs | Wheat | DsRed | Removed selectable markers | [128] |
| Species | Trait/Diseases Improved | No. of Generations per Year | References |
|---|---|---|---|
| Spring Wheat | Stripe rust, seminal root number and angel, and plant height. | 4–6 generations per year | [16] |
| Yellow spot | [169] | ||
| Black rust | [169] | ||
| High protein rate andtolerant to preharvest sprouting | [185] | ||
| Brown rust | [186] | ||
| Stem rust | |||
| Durum Wheat | Crown rust | Up to 6 generations per year | [166] |
| Canola | Pod shattering | 4 generations per year | [16] |
| Barley | Net form net blotch | 4–6 generations per year | [165] |
| Spot form net blotch | |||
| Spot blotch | |||
| Leaf rust | |||
| Glaucousness drought-tolerant trait | [16] | ||
| Rice | Salt tolerance | 4 generations per year | [184] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haroon, M.; Wang, X.; Afzal, R.; Zafar, M.M.; Idrees, F.; Batool, M.; Khan, A.S.; Imran, M. Novel Plant Breeding Techniques Shake Hands with Cereals to Increase Production. Plants 2022, 11, 1052. https://doi.org/10.3390/plants11081052
Haroon M, Wang X, Afzal R, Zafar MM, Idrees F, Batool M, Khan AS, Imran M. Novel Plant Breeding Techniques Shake Hands with Cereals to Increase Production. Plants. 2022; 11(8):1052. https://doi.org/10.3390/plants11081052
Chicago/Turabian StyleHaroon, Muhammad, Xiukang Wang, Rabail Afzal, Muhammad Mubashar Zafar, Fahad Idrees, Maria Batool, Abdul Saboor Khan, and Muhammad Imran. 2022. "Novel Plant Breeding Techniques Shake Hands with Cereals to Increase Production" Plants 11, no. 8: 1052. https://doi.org/10.3390/plants11081052
APA StyleHaroon, M., Wang, X., Afzal, R., Zafar, M. M., Idrees, F., Batool, M., Khan, A. S., & Imran, M. (2022). Novel Plant Breeding Techniques Shake Hands with Cereals to Increase Production. Plants, 11(8), 1052. https://doi.org/10.3390/plants11081052








