Abstract
The pine wood nematode (PWN) Bursaphelenchus xylophilus has caused disastrous losses of pine forests in many countries, and the success of PWN depends strongly on interactions with its insect vectors. Monochamus saltuarius is a newly recorded vector in Northeast China. Feeding (i.e., immature) and egg-laying (i.e., mature) Monochamus spp. target different host plants, and olfactory cues play important roles regarding host choice. Whether infestation with PWN affects olfactory mechanisms in M. saltuarius related to feeding and oviposition is of interest as this may affect the spread of nematodes to new healthy hosts. However, little is known about molecular mechanisms of the olfactory system of M. saltuarius. We identified chemosensory-related genes in adult M. saltuarius and examined the influence of B. xylophilus on the respective expression patterns. Fifty-three odorant-binding proteins (OBPs), 15 chemosensory proteins, 15 olfactory receptors (ORs), 10 gustatory receptors, 22 ionotropic receptors (IRs), and two sensory neuron membrane proteins were identified, and sex bias among non-infested beetles was mainly found with respect to expression of OBPs. Interestingly, OBPs and ORs were markedly down-regulated in male M. saltuarius infested with B. xylophilus, which may reduce olfactory sensitivity of male M. saltuarius and affect the spreading of B. xylophilus to new hosts. Our results will help understand the interactions between B. xylophilus and M. saltuarius, which may lead to the identification of new control targets in the olfactory system of M. saltuarius.
1. Introduction
Pine is the common name of Pinus species (family Pinaceae), and it is the largest genus of conifers, which is widely distributed throughout the world [1]. Pines are not only pioneer species in afforestation, but also have very high economic value owing to their by-products such as pine resin, rosin, and turpentine, which are important raw materials for producing medicinal compounds, organic solvents, and fuels. Currently, pine forests are threatened by pine wood nematodes (PWNs; Bursaphelenchus xylophilus) (Steiner and Buhrer, 1934) Nickle (1970) [2]. More than 60 pine species can be infected with B. xylophilus, and vast areas of pine forests in several countries have been devastated as a result of pine wilt disease. In China alone, an estimated 50 million trees were killed in an area of 80,000 ha between 1982 and 2000 [3].
In 1982, PWN was first found in Nanjing, China, and in only a few decades, it spread rapidly from Nanjing to vast areas of China (Li and Zhang, 2018). According to the announcement of the National Forestry and Grassland Administration in 2021, there were 721 PWN-epidemic areas at county level, which were widely distributed in 16 provinces, thus seriously threatening pine forests in northern China.
The spread of B. xylophilus relies on insects of the genus Monochamus [4,5,6,7]. B. xylophilus is attracted by newly emerged Monochamus adults, and when infested beetles feed on shoots of healthy trees, the nematode can disperse to these trees [8,9]. With the spread of PWN from South Central China to Northeast China, its main vector insects changed from Monochamus alternatus (Coleoptera: Cerambycidae) to Monochamus saltuarius [10]. The interactions among trees, vector insects, and B. xylophilus are complex, considering previous studies on M. alternatus [8,11]; moreover, elucidating such interactions may be of importance for controlling the spread of PWN. M. saltuarius has been identified as a vector of B. xylophilus in Japan and Korea [12]; however, only limited information is available regarding its biology, pheromones, and potential control measures of M. saltuarius [12,13,14,15]. Few studies so far focused on the interactions between nematodes and M. saltuarius [16].
Sexual maturity of Monochamus spp. lasts for almost one month, and these insects undergo an immature (nutrition) period and mature reproduction period, which in females includes oviposition [17]. Nematodes in the spiracles of Monochamus spp. depart to invade new healthy trees through feeding wounds inflicted to the trees [9] or through oviposition incisions [18]. Monochamus spp. preferably feed on healthy trees and lay eggs in weak or dying trees [17]. Volatile organic compounds of plants [19,20,21] and insect olfactory systems [22] play important roles regarding insect host choice for feeding and oviposition; for example, terpene volatiles (such as α-pinene) are the main attractants for M. alternatus to feed [23,24], and ethanol exuded from weak or dying trees can attract female longicorn beetles to lay eggs [25,26,27]. However, it is unknown whether the olfactory recognition sensitivity of insect vectors may be affected by infestation with nematodes, which may then also influence the spreading of nematodes. For example, the spread of nematodes to new healthy hosts mainly depends on the feeding behavior of Monochamus spp.; if the olfactory sensitivity of vectors to healthy trees is affected by infestation with nematodes, the spreading efficiency of PWN may also be affected by such mechanisms.
The olfactory system is critical for insect feeding and reproduction (including mate selection and oviposition site searching) [28]. Several classes of genes are associated with insect olfactory performance, including two classes of binding proteins (odorant-binding proteins, [OBPs] and chemosensory proteins [CSPs]) [29], three receptor gene classes (olfactory receptors [ORs], ionotropic receptors [IRs], and gustatory receptors [GRs]), and sensory neuron membrane proteins (SNMPs) [30]. The binding proteins can preliminarily screen and transport organic hydrophobic volatile molecules from the environment (such as host volatiles and pheromones) to the olfactory neurons through hydrophilic sensillum lymph [31,32]. These receptors can detect and discriminate olfactory signals and translate them into bioelectric signals, and different receptor types frequently respond to different signals. For example, ORs mainly respond to olfactory signals [33], and GRs respond to taste signals such as sugars, bitter compounds, and other contact stimuli [34].
Identification and expression analysis of olfactory-related genes are the basis for further exploration of olfactory mechanisms in insects. However, information on olfactory-related genes in Coleoptera is scarce. Before 2013, members of the major chemosensory gene families had been identified only in T. castaneum [35] and in two bark beetles, Ips typographus and Dendroctonus ponderosae [36]. Chemosensory genes of M. alternatus were identified in 2014, and this is also the first longicorn beetle species whose olfactory recognition was examined [3]. Molecular olfactory reception mechanisms have received more attention [37,38], however, very little is known about the molecular mechanisms of olfactory recognition by M. saltuarius, which is a new and important vector of B. xylophilus in northeast China. This limits our general understanding of the olfactory cues underlying the spread of this pest.
Here, we aim to identify the chemosensory genes of M. saltuarius, and detect the influence of B. xylophilus infestation on chemosensory gene expression in M. saltuarius. This work may provide a basis for further research on the chemosensory mechanisms of M. saltuarius and help us to further understand the interactions between B. xylophilus and M. saltuarius further.
2. Materials and Methods
2.1. Insects
M. saltuarius at the mating stage were collected on the Dahuofang Experimental Forest Farm, Fushun City, Shenyang Province, China, in May 2019. Samples (n = 40) were collected in the forest. We searched M. saltuarius carefully in the area where some of the host trees were weak, and once mating couples were found (a male and a female M. saltuarius were mating, usually located on the trunk), they would be put it in a breathable plastic tube and taken back to the laboratory. Antennae were collected as follows: both antennae of each M. saltuarius were cut off at the base, placed in a marked centrifuge tube, and were immediately frozen in liquid nitrogen. Then, the respective donor individual was examined for nematodes using the Baermann funnel method. Four categories were used, i.e., males and females without nematodes (here termed ‘healthy’) and with nematodes (‘infested’), respectively. Five biological replicates were used per category, and each of the five biological replicates consisted of antennae coming from one beetle.
2.2. RNA-Seq Library Preparation and Sequencing
Total RNA was isolated from M. saltuarius antennae using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) [39,40], and genomic DNA was removed using DNase I (TaKara, Dalian, Liaoning, China). Integrity and purity of the extracted RNA were determined using a 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA), and RNA was quantified using a ND-2000 device (NanoDrop Thermo Scientific, Wilmington, DE, USA). Only high-quality RNA samples (OD260/280 = approximately 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 8.0, 28S:18S ≥ 1.0) were used to prepare sequencing libraries. One microgram total RNA was used to synthesize cDNA [41,42]. Sequencing libraries were prepared using the Illumina TruSeq™ RNA Sample Preparation Kit (Illumina, San Diego, CA, USA). Libraries were selected for cDNA target fragments of 200–300 bp, followed by PCR amplification using Phusion DNA polymerase (New England Biolabs, Boston, MA, USA) with 15 amplification cycles. After quantification, the sequencing of 2 × 150 bp paired-end reads was performed on an Illumina HiseqXTen sequencer (Illumina).
2.3. De-Novo Sequence Assembly
Clean data were obtained from raw sequencing data by trimming and quality control using SeqPrep (https://github.com/jstjohn/SeqPrep, accessed on 11 November 2019) and Sickle (https://github.com/najoshi/sickle, accessed on 6 January 2022) with default parameters. Clean data were then used to perform de novo assembly with Trinity software (http://trinityrnaseq.sourceforge.net/, accessed on 20 November 2019) [43], and redundancies were removed using TGICL [44].
2.4. Annotation
All assembled transcripts were searched against six public databases (NR, Swiss-Prot, Pfam, COG, GO, and KEGG) using BLAST to identify proteins with the highest sequence similarity with the produced transcripts to produce functional annotations (cut-off e-value < 1−5). BLAST2GO (http://www.blast2go.com/b2ghome, accessed on 25 November 2019) [45,46]. Software was used to produce gene ontology (GO) annotations of unique assembled transcripts to describe biological processes, molecular functions, and cellular components [47,48]. Metabolic pathway analysis was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/, accessed on 26 November 2019).
Chemosensory genes were identified in two ways: first, contig tBLASTx searches were performed using an M. saltuarius assembled unigene database with query sequences as chemosensory genes from other insects, such as Drosophila melanogaster, Bombyx mori, and other long-horned beetles. Second, annotation information of the unigenes was used. Open reading frames of the candidate genes from the two ways were identified and used to as confirmation through BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 6 January 2022).
2.5. Gene Expression Quantification
The expression level of each transcript was calculated according to the fragments per kilobase of exon per million mapped reads (FRKM) method [49]. The R software package EdgeR vision 2.12 (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html, accessed on 10 October 2019) was used for differential expression analysis [50]. A compatible-hits-norm model was used to normalize unigene expression levels and DEGs differentially expressed genes [51].
2.6. Phylogenetic Analyses
IQ-TREE web service was used to construct phylogenetic trees of the predicted protein sequences and orthologous genes from other insects [52]. The sequences were first aligned using BioEdit software with the ClustalW multiple alignment tool. Bootstrap phylogenetic trees were generated using 1000 replicates, and the best models were selected using the model fit result of IQ-TREE. iTOL was used to annotate the phylogenetic trees [53].
3. Results
3.1. M. saltuarius Antennae Transcriptomes
More than 7 Gbp clean sequence data were produced from most samples (except for one sample from an infested male, which produced 6.98 Gb data), and the Q30 value was > 94%. De-novo transcriptome assembly resulted in 38,198 contigs with an N50 of 1798 bp, and the largest unigene was 32,017 bp long. The length distribution of the transcriptome assembly is shown in Figure S1. The mapping rates of clean data were >78.16% for each sample. Gene annotation based on the NR, Swiss-Prot, Pfam, COG, GO, and KEGG databases yielded annotations for 20,931transcripts.
3.2. M. saltuarius Chemosensory Genes
We identified 53 OBPs, 15 CSPs, 15 ORs, 10 GRs, 22 IRs, and 2 SNMPs (Table S1). The identified 117 chemosensory genes were submitted to NCBI under the accession numbers MT008336–MT008452.
3.3. OBPs of M. saltuarius with and without B. xylophilus
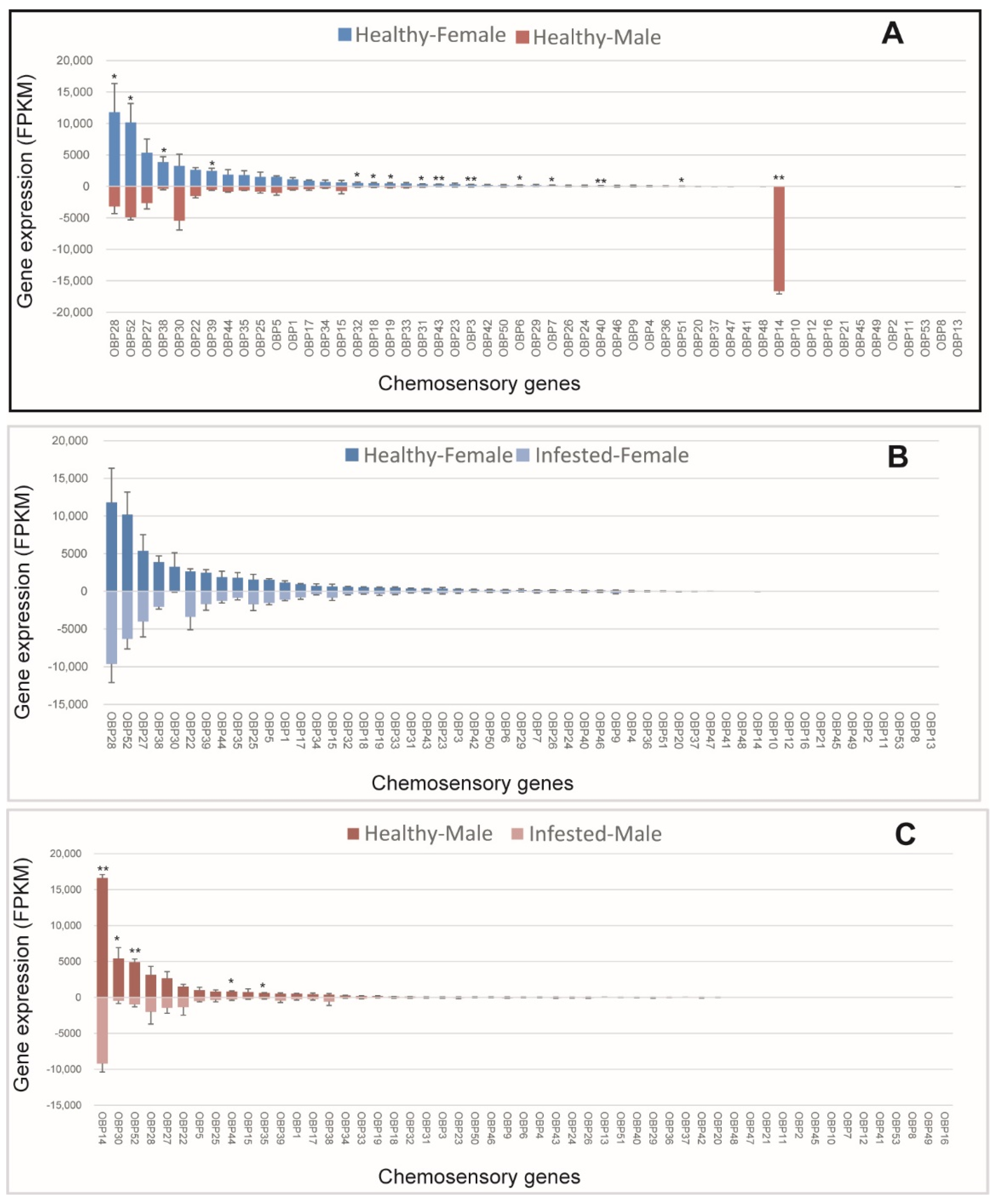
We analyzed sex-specific expression patterns of OBPs in non-B. xylophilus-infested M. saltuarius, and 15 OBPs were expressed differentially between sexes. Most differentially expressed OBPs (14/15) showed higher expression in females (Figure 1A). Among OBPs with female-biased expression in healthy beetles, OBP28, OBP52, OBP38, and OBP39 had high expression levels, and the other female-biased OBPs (OBP32, OBP18, OBP19, OBP31, OBP43, OBP3, OBP6, OBP7, OBP40, and OBP51) showed low expression. OBP14 expression was male-biased among healthy beetles, and, the expression of OBP14 was the highest in males and very low in females (Figure 1A). Thus, OBP14 may be related to sex pheromone recognition in male M. saltuarius. Further studies are needed to understand the reason of its differential expression between males and females. Effects of B. xylophilus on the expression patterns of OBPs in male and female beetles was examined, and none of the OBPs showed altered expression levels in females after one month of infestation with B. xylophilus (Figure 1B). However, expressions of five OBPs (including OBP14, OBP30, OBP52, OBP44, and OBP35) were significantly lower in infested male beetles than in non-infested ones (Figure 1C). Thus, OBP expression in male M. saltuarius was affected by infestation with B. xylophilus.
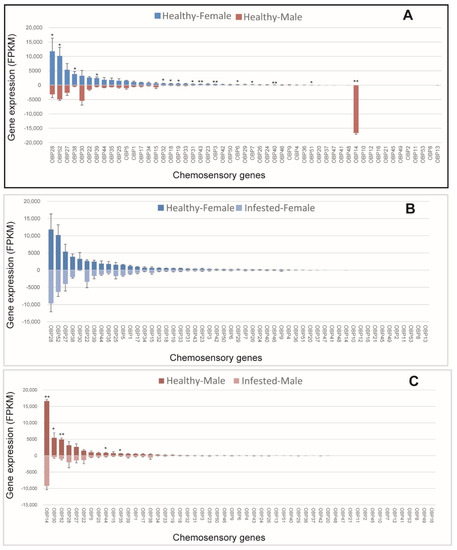
Figure 1.
Expression patterns of odorant-binding protein in healthy and Bursaphelenchus xylophilus-infested Monochamus saltuarius. (A) Differences between sexes of healthy M. saltuarius; (B) Differences between healthy and infested female M. saltuarius; (C) Differences between healthy and infested male M. saltuarius. Transcript levels are expressed as reads per kilobase of exon per million mapped reads (RPKM), and the expression data were shown as mean ±SE. The stars above each bar of figure indicate significant differences (p < 0.05 * and p < 0.05 **).
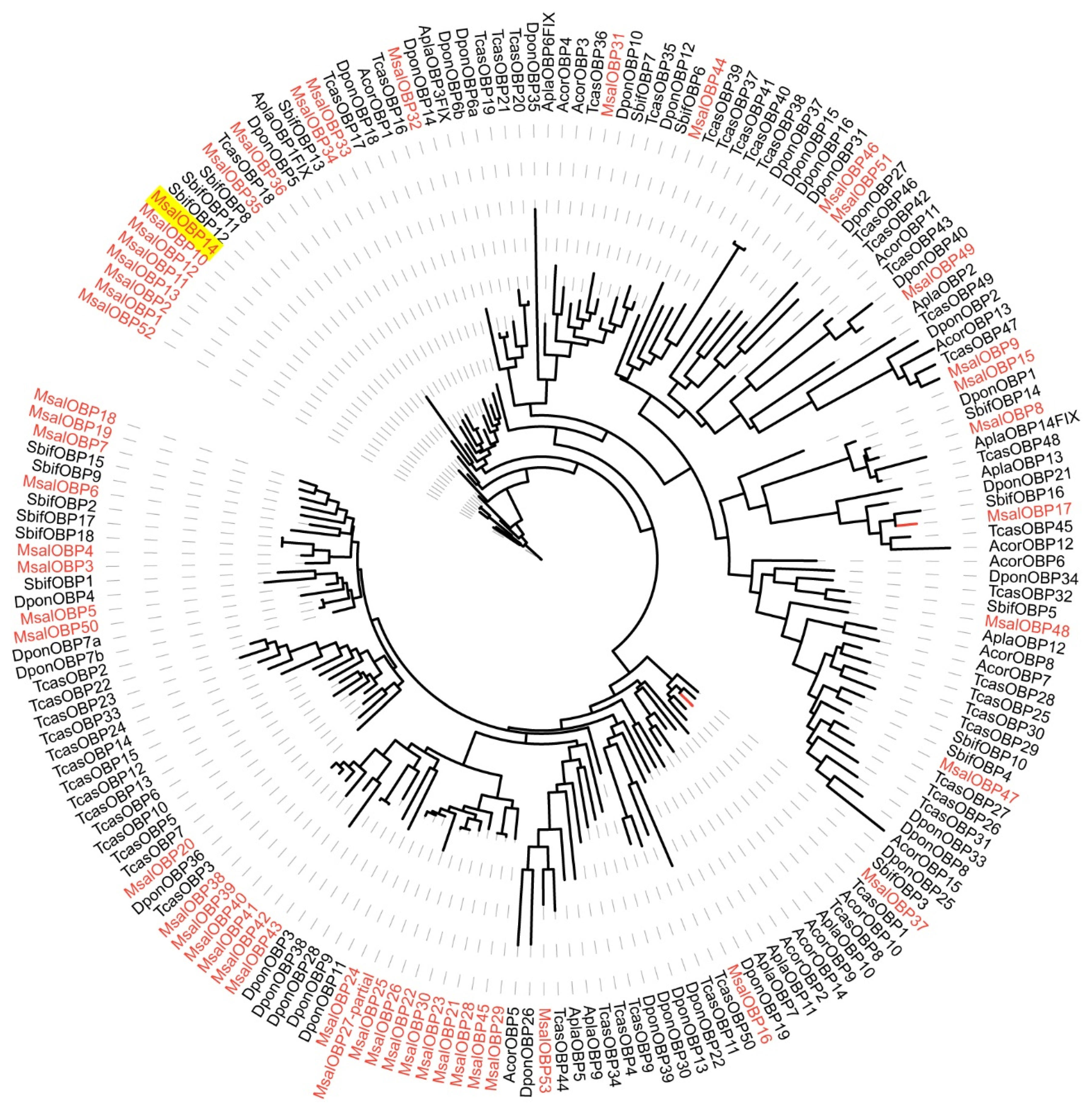
A phylogenetic tree was constructed to analyze the characteristics of OBPs from M. saltuarius, based on orthologs to OBP sequences from several other Coleoptera insects Semanotus bifasciatus [54], Tribolium castaneum [55], Anomala corpulenta [56], Agrilus planipennis, and Dendroctonus ponderosae [57] (Figure 2). OBPs from M. saltuarius seems to be homologous to OBPs of Coleopteran insects whose genomic information are available, and indicates the relative complete identification of OBPs from M. saltuarius. MsalOBP14, which expressed at an especially high level in male adults, seems not to be a typical PBPs. We did not get a special PBP clade in Figure 2 (MsalOBP14 was clustered together with other M. saltuarius OBPs) and MsalOBP14 was not crusted in PBP clade in another phylogenetic tree containing more model insect species (Figure S2). However, Pfam search hits PBP/GOBP family (PF01395.24, E-value is 4.8 × 10 –18) (http://pfam.xfam.org, accessed on 2 February 2019). The function of MsalOBP14 deserves further study.
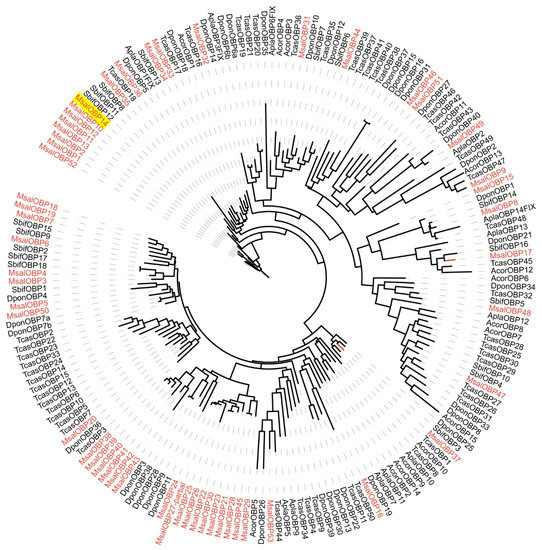
Figure 2.
Maximum likelihood dendrogram based on protein sequences of candidate odorant-binding proteins (OBP) in M. saltuarius and orthologs from other Coleopteran insects.JTT + F + R6 model based on the best fit result of IQ-tree model finding, and bootstrap consensus tree inferred from 1000 replicates were used. Monochamus saltuarius (Msal) (red), Semanotus bifasciatus (Sbif), Tribolium castaneum (Tcas), Anomala corpulenta (Acor), Agrilus planipennis (Apla), and Dendroctonus ponderosae (Dpon). MsalOBP14 expressed at an especially high level in male antennae of M. saltuarius was marked with yellow background.
3.4. CSPs of M. saltuarius with and without B. xylophilus
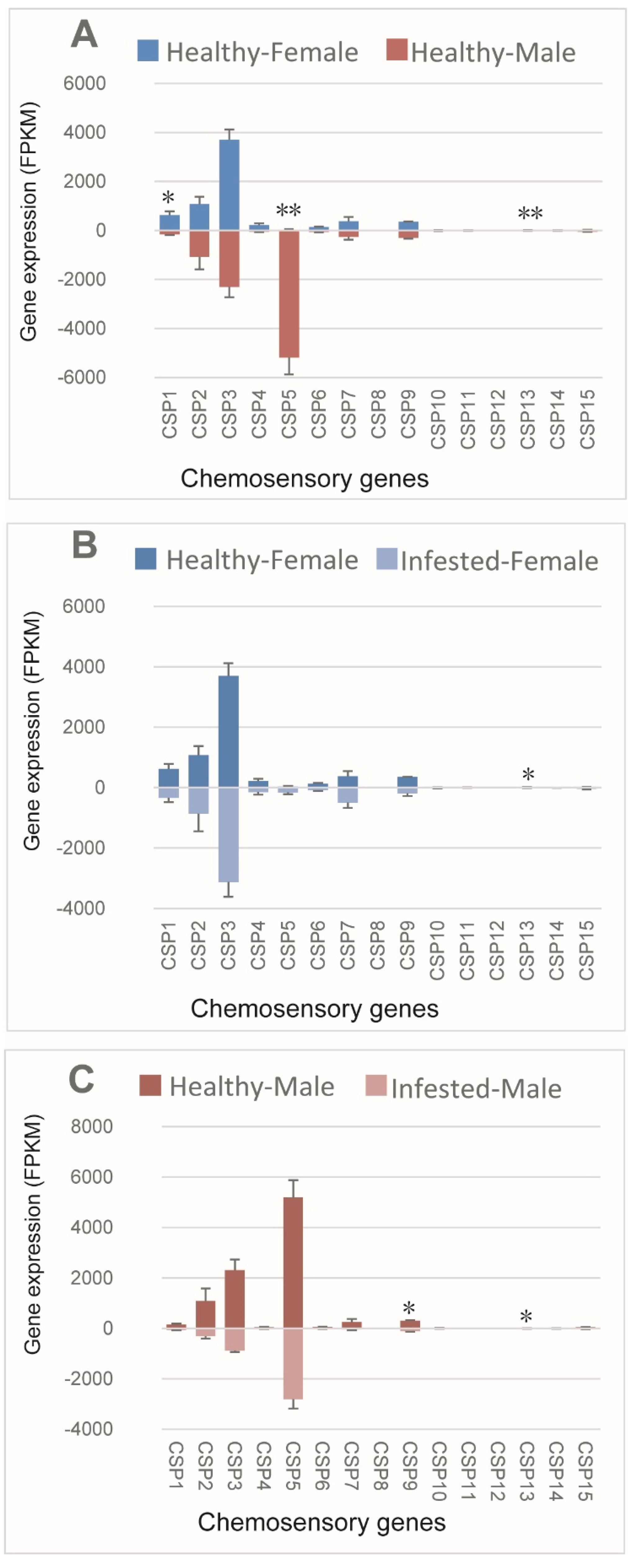
Sex-specific expression of CSPs was assessed in healthy M. saltuarius (Figure 3A), and three CPSs were expressed differently between sexes; two of these (CSP1 and CSP13) showed higher expression in females, and one (CSP5) showed extremely high expression in males. Infestation with B. xylophilus slightly affected CSP expression, only CSP13 in female (Figure 3B), and CSP9 and CSP13 male (Figure 3C) were down-regulated in the infested beetles (please see supplementary Table S2).
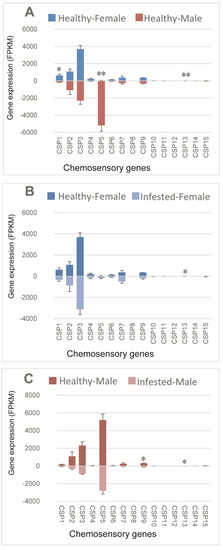
Figure 3.
Expression patterns of chemosensory genes in healthy and B. xylophilus-infested M. saltuarius. (A) Differences between sexes of healthy M. saltuarius; (B) Differences between healthy and infested female M. saltuarius; (C) Differences between healthy and infested male M. saltuarius. Transcript levels are expressed as reads per kilobase of exon per million mapped reads (RPKM), and the expression data were shown as mean±SE. The stars above each bar of figure indicate significant differences (p < 0.05 * and p < 0.05 **).
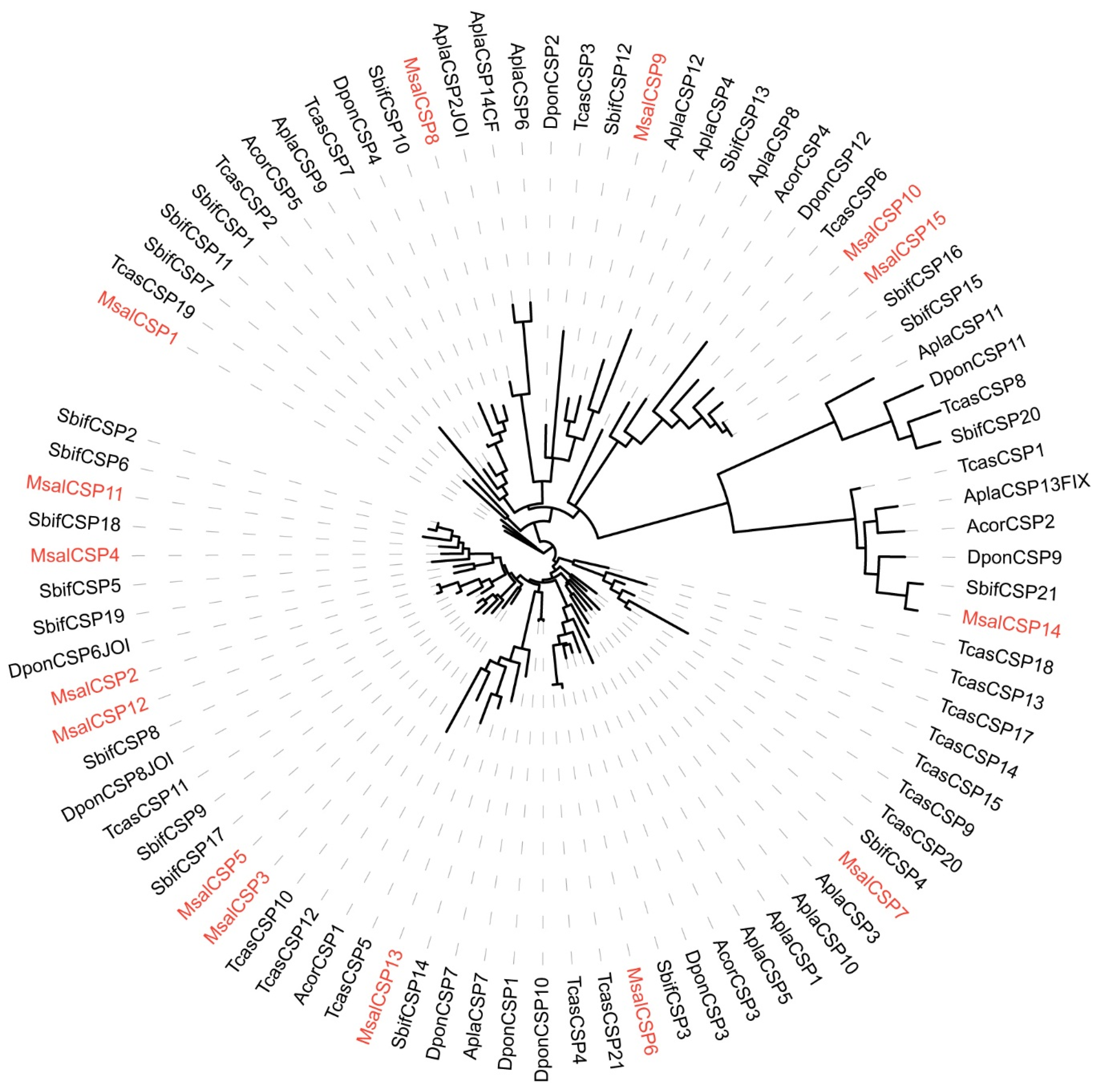
A phylogenetic tree of CSPs was constructed based on orthologs to CPS sequences from M. saltuarius, S. bifasciatus, T. castaneum, A. corpulenta, A. planipennis, and D. ponderosae (Figure 4). The identified CSPs from M. saltuarius were homologous to CSPs of Coleopteran insects whose genomic information were available, indicating relative complete identification of CSPs from M. saltuarius.
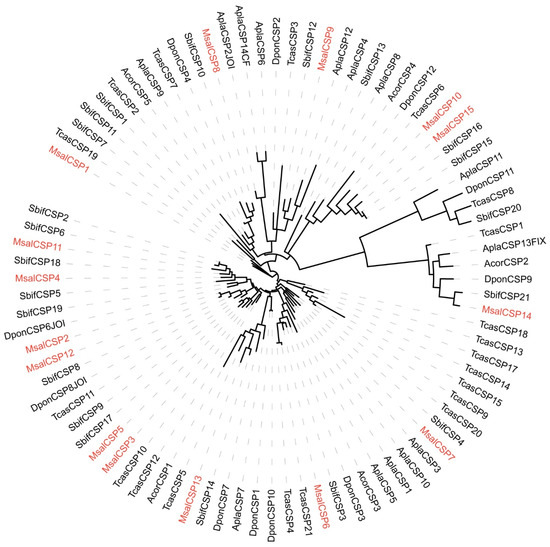
Figure 4.
Maximum likelihood dendrogram based on protein sequences of candidate chemosensory genes (CSP) in M. saltuarius and orthologs from other Coleopteran insects. LG + I + G4 model based on the best fit result of IQ-tree model finding, and bootstrap consensus tree inferred from 1000 replicates were used. Monochamus saltuarius (Msal) (red), Semanotus bifasciatus (Sbif), Tribolium castaneum (Tcas), Anomala corpulenta (Acor), Agrilus planipennis (Apla), and Dendroctonus ponderosae (Dpon).
3.5. ORs of M. saltuarius with and without B. xylophilus
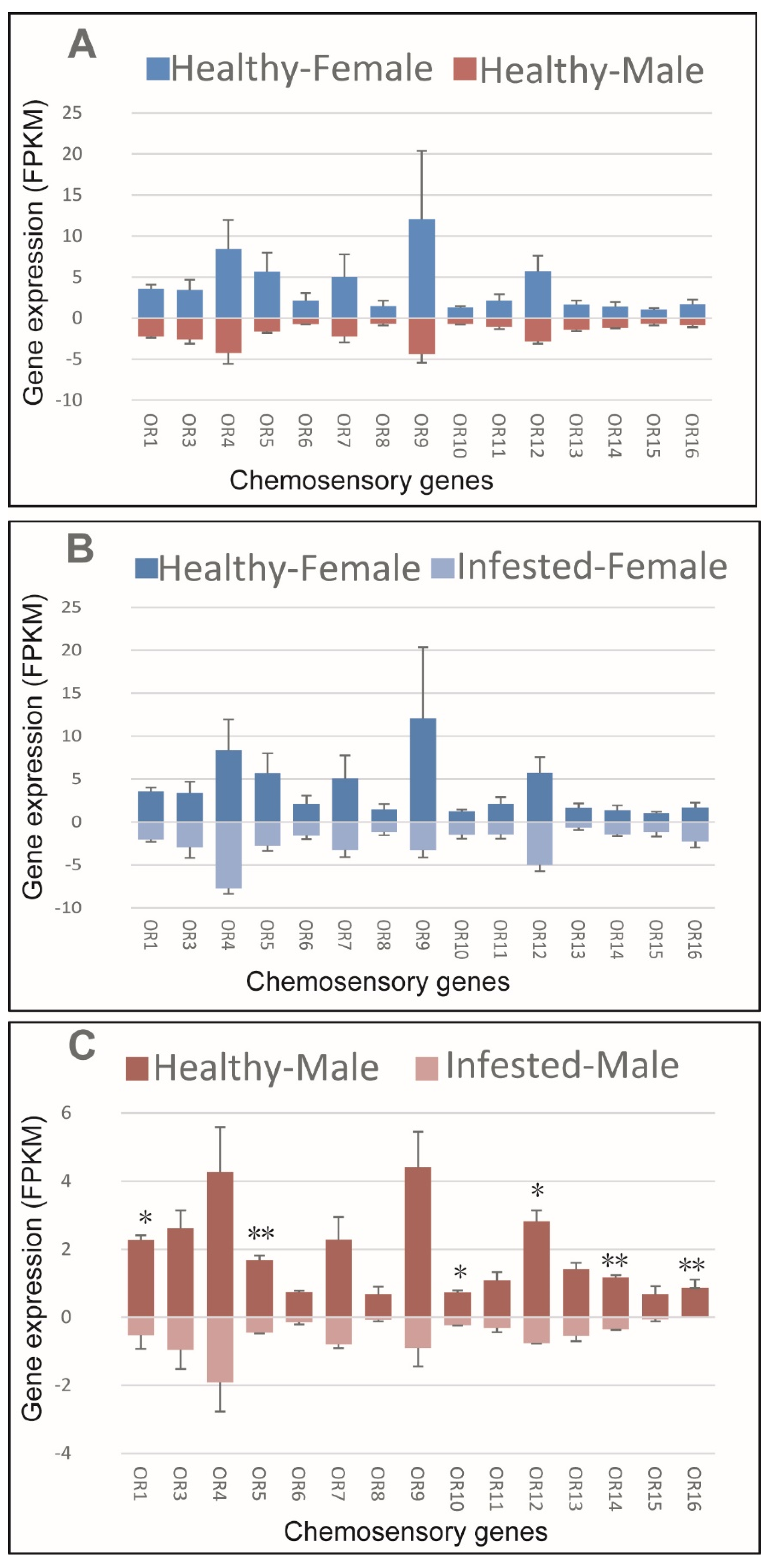
No sex bias was observed regarding OR expression in healthy M. saltuarius (Figure 5A). Infestation with B. xylophilus did not affect OR expression in female M. saltuarius (Figure 5B). Marked changes in OR expression were detected in male M. saltuarius carrying B. xylophilus compared with non-infested individuals (Figure 5C). OR1, OR5, OR10, OR12, OR14, and OR16 showed higher expression in healthy M. saltuarius than those that were infested. Thus, B. xylophilus can reduce olfactory sensitivity of male M. saltuarius.
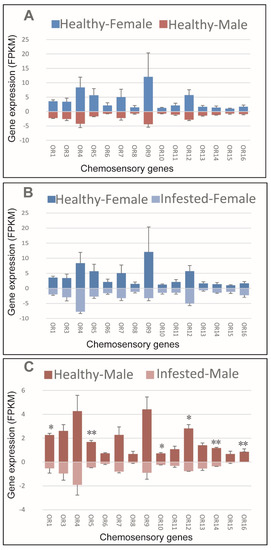
Figure 5.
Expression patterns of olfactory receptors in healthy and B. xylophilus-infested M. saltuarius. (A) Differences between sexes of healthy M. saltuarius; (B) Differences between healthy and infested female M. saltuarius; (C) Differences between healthy and infested male M. saltuarius. Transcript levels are expressed as reads per kilobase of exon per million mapped reads (RPKM), and the expression data were shown as mean±SE. The stars above each bar of figure indicate significant differences (p < 0.05 * and p < 0.05 **).
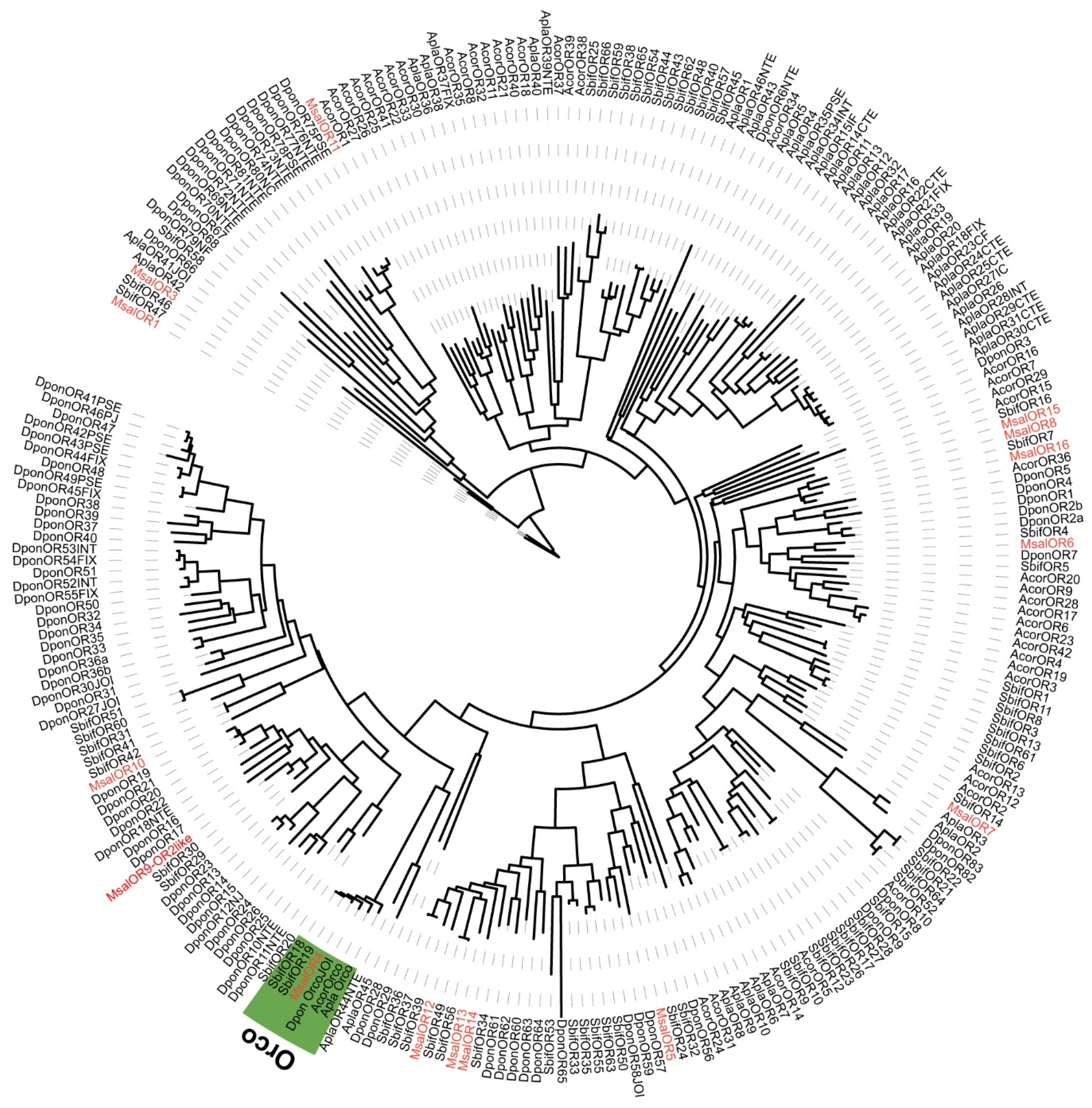
A phylogenetic analysis of ORs was performed based on the OR sequence orthologs from M. saltuarius, S. bifasciatus, A. corpulenta, A. planipennis, and D. ponderosae (Figure 6). Generally, almost all M. saltuarius ORs found orthology to other Coleoptera ORs, including the insect OR obligatory co-receptor, Orco. However, the total number of M. saltuarius ORs may have not been recovered due to its integrity in this transcriptomic study. This is obvious after comparison with genomic data from A. planipennis and D. ponderosae.
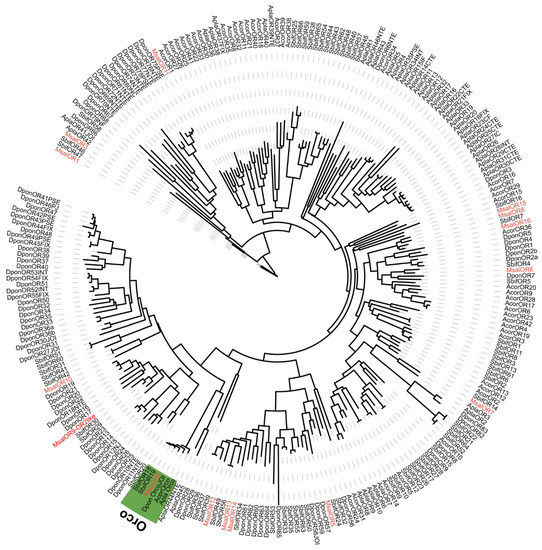
Figure 6.
Maximum likelihood dendrogram based on protein sequences of candidate olfactory receptors (OR) in M. saltuarius and orthologs from other Coleopteran insects. JTT + F + R6 model based on the best fit result of IQ-tree model finding, and bootstrap consensus tree inferred from 1000 replicates were used. Monochamus saltuarius (Msal) (red), S. bifasciatus (Sbif), A. corpulenta (Acor), A. planipennis (Apla), and D. ponderosae (Dpon). Orco clade marked with green background.
3.6. IRs, GRs, and SNMPs in Healthy and Infested M. saltuarius
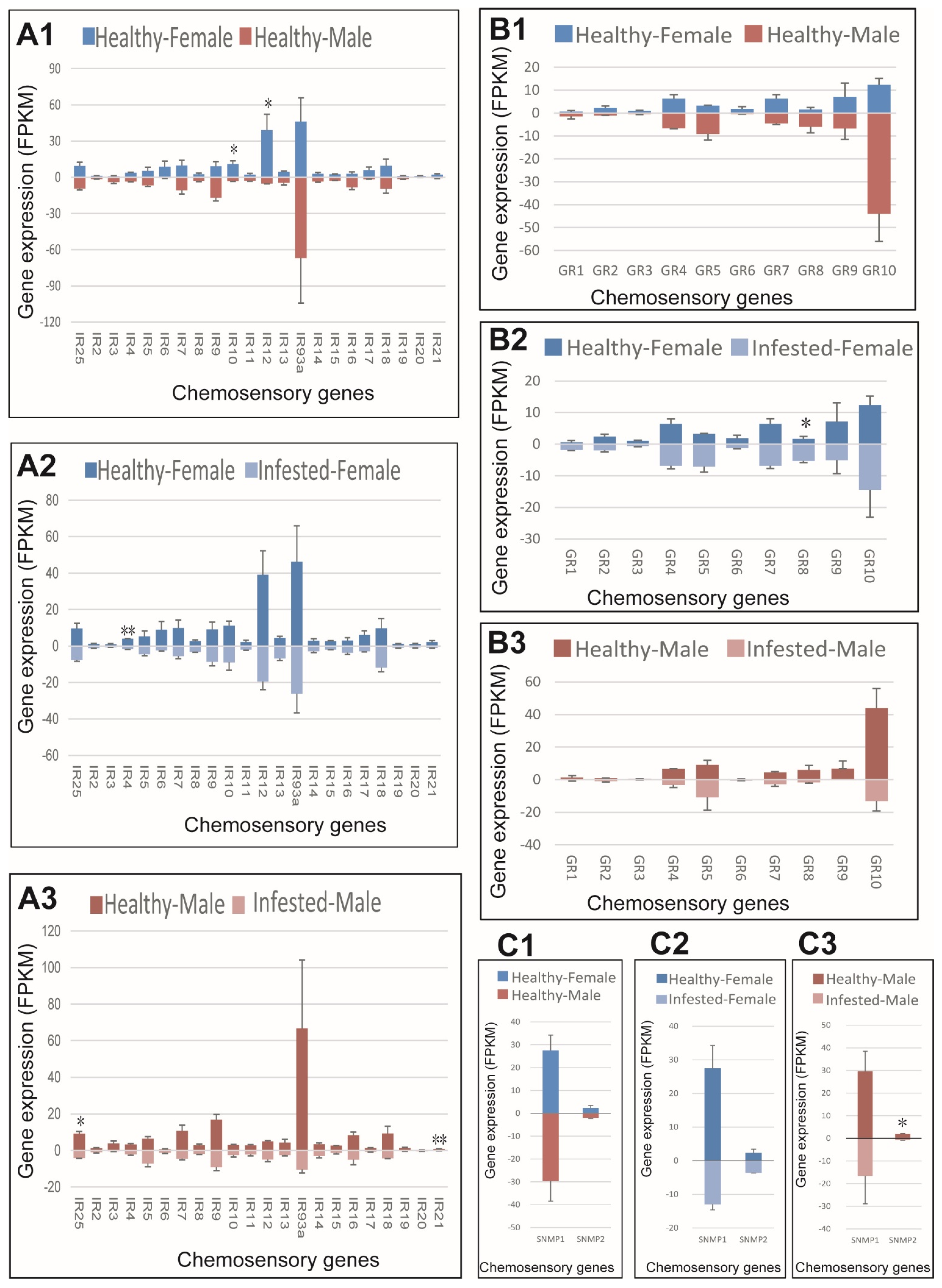
Regarding IRs, IR10 and IR12 were expressed higher in healthy females compared to healthy males, IR4 expressed higher in healthy than infested females, while IR25 and IR21 were expressed higher in healthy than infested males (Figure 7(A1–A3)). Regarding GRs, no sex bias was observed in healthy adults, and only GR8 showed higher expression in female M. saltuarius infested with B. xylophilus (Figure 7(B1–B3)). GR8 was the only up-regulated chemosensory gene in M. saltuarius infested with B. xylophilus. Regarding SNMP, no sex bias was observed in healthy M. saltuarius and SNMP2 showed lower expression in infested males than in healthy ones (Figure 7(C1–C3)).
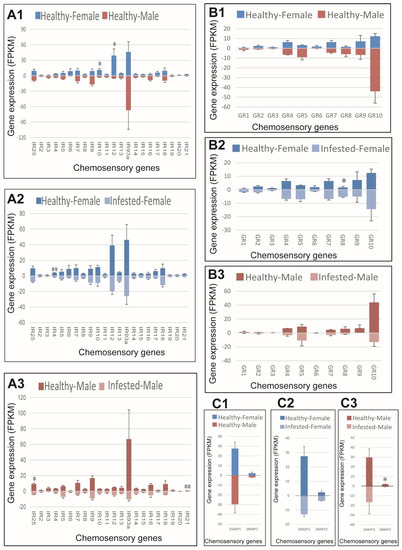
Figure 7.
Expression patterns of ionotropic receptors (IRs), gustatory receptors (GRs), and sensory neuron membrane proteins (SNMPs) in healthy and B. xylophilus-infested M. saltuarius. (A1), IRs expression differences between sexes of healthy M. saltuarius; (A2), IRs expression differences between healthy and infested female M. saltuarius; (A3), IRs expression differences between healthy and infested male M. saltuarius. (B1), GRs expression differences between sexes of healthy M. saltuarius; (B2), GRs expression differences between healthy and infested female M. saltuarius; (B3), GRs expression differences between healthy and infested male M. saltuarius. (C1), SNMPs expression differences between sexes of healthy M. saltuarius; (C2), SNMPs expression differences between healthy and infested female M. saltuarius; (C3), SNMPs expression differences between healthy and infested male M. saltuarius. Transcript levels are expressed as reads per kilobase of exon per million mapped reads (RPKM), and the expression data were shown as mean±SE. The stars above each bar of figure indicate significant differences (p < 0.05 * and p < 0.05 **).
4. Discussion
Olfactory cues play important roles in the host choice of immature and mature M. saltuarius. The spread of nematodes to new healthy hosts mainly relies on the feeding activity of immature M. saltuarius. If infestation with B. xylophilus for approximately one month can affect olfactory recognition in longicorn beetles it could also affect the nematode’s spreading interval duration. In the present study, we identified 53 OBPs, 15CSPs, 15 ORs, 10 GRs, 22 IRs, and 2 SNMPs in M. saltuarius, a new vector of B. xylophilus in northern China. Sex-specific expression patterns in healthy beetles occurred mainly regarding OBPs. Interestingly, we found that OBPs and ORs were down-regulated in male M. saltuarius infested with B. xylophilus, which may reduce olfactory sensitivity and delay their maturation; consequently, this may prolong the time period, during which B. xylophilus can be transmitted through infested beetles.
The numbers of OBPs and CSPs in M. saltuarius are consistent with other Coleopteran insects whose genomic information are available [55,57], indicating relative complete identification of these two chemosensory families in M. saltuarius. However, the number of OR genes in M. saltuarius was lower than the number of other Coleopteran insects, this may be due to some missing genes down-regulated in the adults, especially in sexually mature adults. Another reason maybe that these insects have mated and some of the chemosensory genes were down-regulated. However, S. bifasciatus ORs were also identified from antennae transcriptome data, and its numbers were much higher than M. saltuarius [54]. To elucidate the underlying causation, we reviewed olfactory gene identifications of other longicorn beetles. To date, olfactory-related gene identification and respective studies have only been performed on a few longicorn beetles. Fifty-two OBPs and 132 ORs were identified in the genome of Anoplophora glabripennis [58], and 57 ORs were identified in Megacyllene caryae [59]. However, 31 OBPs and only nine ORs were identified in the antenna transcriptome of M. alternatus [3]. Both Monochamus species possess considerably fewer ORs than other longicorn beetles, which may be a particular characteristic of this genus. However, further studies on the olfactory system of Monochamus spp. based on genome data are needed to confirm this prediction.
The present study is the first to confirm that infestation with B. xylophilus can influence olfactory gene expression in the vector M. saltuarius. Interactions between B. xylophilus and their vector insects are complex throughout all stages of development. Before accumulation in its vector, B. xylophilus can secrete ascarosides to promote beetle pupation. After developing into the adult stage, the beetle secretes ascarosides to attract the B. xylophilus 4th instar larvae [8]. B. xylophilus 4th instar larvae development relies on C16 and C18 fatty acid ethyl esters, which are abundantly produced by beetle pupae at emergence and newly emerged adults [60]. Thus, B. xylophilus can promote and use signals from vector beetles to facilitate its accumulation on its vector before the newly emerged beetles leave their old host plants. When the beetles disperse for foraging, the time span for B. xylophilus to spread on healthy trees is critical; a longer foraging time increases the probability that B. xylophilus will spread to healthy trees. Our results indicate that B. xylophilus can reduce the olfactory sensitivity of male M. saltuarius, which may delay sexual maturity and further prolong the beetle’s feeding activity, thus increasing the likelihood of spreading PWNs to new healthy hosts. Conversely, the olfactory system of female M. saltuarius was not affected by longtime B. xylophilus infestation, which may be a compromise for B. xylophilus as it requires female M. saltuarius to lay eggs, thereby initiating a new spreading cycle. However, further research is required to test whether GR8 upregulation in female M. saltuarius infested with B. xylophilus (Figure 7(B1–B3)) can facilitate feeding activity in female beetles and therefore enhance egg maturity and efficient oviposition. Further studies should be undertaken to test whether GR up regulation in infested mated females may help discriminate adequate hosts for oviposition and thereby facilitate the spread of nematode to damaged trees.
On the other hand, the blocking of chemosensation in infested M. saltuarius may be for the self-preservation of the insect. Sexually mature males need to find dead logs and trees where females are waiting for mating. Chemosensory gene down-regulation in males would reduce their sensitivity and influence mating success, and thus transfer less nematodes to its host trees. Anyway, whether these hypotheses are correct requires further physiological and gene function researches, and clarification of the complex relationships between nematodes-vectors-trees will imagine new pest control strategies.
Genes affected by B. xylophilus infestation may be used as new molecular targets for M. saltuarius control in the future. However, the functions of the regulated ORs and OBPs genes in male M. saltuarius are unclear, and further research is needed to elucidate the respective interactions between nematodes and their beetle vectors. This work extends our understanding of the complex relationship between nematodes-vectors-trees; this is critical for us to grasp new control targets and develop new control methods to protect large areas of pine forest.
Conclusion: firstly, we identified 53 OBPs, 15 CSPs, 15 ORs, 10 GRs, 22 IRs, and two SNMPs in M. saltuarius. Secondly, sex expression bias of the chemosensory genes was analyzed. Finally, we found that B. xylophilus infestation mainly down-regulated the OBPs and ORs expression levels in male M. saltuarius at the mating stage.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13020258/s1. Figure S1. Sequence length distribution of the Monochamus saltuarius antennal transcriptomes assembly. Figure S2. Maximum likelihood dendrogram based on protein sequences of candidate odorant binding protein (OBP) in M. saltuarius and orthologs from other insects. IQ-tree best fit model (VT+R6) was used and bootstrap consensus tree inferred from 1000 replicates. Monochamus saltuarius (Msal) (red), S. bifasciatus (Sbif), T. castaneum (Tcas), A. corpulenta (Acor), A. planipennis (Apla), D. ponderosae (Dpon), Drosophila melanogaster (Dmel), and Bombyx mori (Bmor). Orco clade was marked with green background. Table S1. Candidate chemosensory genes of Monochamus saltuarius. Table S2. FPKM and p values of the differently expressed chemosensory genes of Monochamus saltuarius.
Author Contributions
S.Z., X.W. and Y.Z. (Yanlong Zhang) designed the experiments; S.Z., X.W., Y.Z. (Yanlong Zhang) and Y.Z. (Yanan Zheng) collected samples, S.Z., X.W., Z.F. and R.Z. examined the samples; S.Z. analyzed the data and drafted the manuscript. Y.Z. (Yanlong Zhang), Y.Z. (Yanlong Zhang) and X.W. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Fundamental Research Funds for the Central Non-profit Research Institution of CAF: CAFYBB2018SZ006, CAFYBB2020QC001.
Data Availability Statement
The identified chemosensory genes were submitted to NCBI under the accession numbers MT008336–MT008452.
Acknowledgments
This research was funded by the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (CAFYBB2018SZ006, CAFYBB2020QC001).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shen, H.; Wang, F. The most plants in Pinus. Plants 1991, 6, 40. [Google Scholar]
- Nickle, W.R. A taxonomic review of the genera of the Aphelenchoidea (Fuchs, 1937) Thorne, 1949 (Nematoda: Tylenchida). J. Nematol. 1970, 2, 375–392. [Google Scholar]
- Wang, J.; Li, D.Z.; Min, S.F.; Mi, F.; Zhou, S.S.; Wang, M.Q. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp. Biochem. Physiol. Part D Genom. Proteom. 2014, 11, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Iwasaki, A. Role of Monochamus alternatus (Coleoptera: Cerambycidae) as a vector of Bursaphelenchus lignicolus (Nematoda: Aphelenchoididae). Nihon Ringakkai Shi J. Jpn. For. Soc. 1972, 54, 177–183. [Google Scholar]
- Zhang, J.; Zhang, R.; Chen, J. Species and their dispersal ability of Monochamus as vectors to transmit Bursaphelenchus xylophilus. J. Zhejiang For. Coll. 2007, 24, 350–356. [Google Scholar]
- Linit, M.J.; Kondo, E.; Smith, M.T. Insects associated with the pinewood nematode, Bursaphelenchus xylophilus (Nematoda: Aphelenchoididae), in missouri. Environ. Entomol. 1983, 12, 467–470. [Google Scholar] [CrossRef]
- Kobayashi, F.; Yamane, A.; Ikeda, T. The japanese pine sawyer beetle as the vector of pine wilt disease. Annu. Rev. Entomol. 1984, 29, 115–135. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, X.; Wei, Y.; Zhou, J.; Zhang, W.; Qin, P.; Chinta, S.; Kong, X.; Liu, Y.; Yu, H.; et al. Ascarosides coordinate the dispersal of a plant-parasitic nematode with the metamorphosis of its vector beetle. Nat. Commun. 2016, 7, 12341. [Google Scholar] [CrossRef] [Green Version]
- Enda, N.; Mamiya, Y. Transmission of Bursaphelenchus lignicolus (Nematoda: Aphelenchoididae) by Monochamus alternatus (Coleoptera: Cerambycidae). Nematologica 1972, 18, 159–162. [Google Scholar]
- Meng, Y. Monitoring, identification and control of pine wood nematode in Dandong City, Liaoning Province. Agric. Technol. 2018, 38, 62–63. [Google Scholar]
- Wu, Y.; Wickham, J.D.; Zhao, L.; Sun, J. Co2 drives the pine wood nematode off its insect vector. Curr. Biol. 2019, 29, R619–R620. [Google Scholar] [CrossRef]
- Cho, W.S.; Koo, H.N.; Yun, S.H.; Lee, J.S.; Kim, G.H. Electron beam-induced sterility and inhibition of ovarian development in the sakhalin pine longicorn, Monochamus saltuarius (Coleoptera: Cerambycidae). J. Econ. Entomol. 2018, 2, 725–731. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, X. Analysis on the trend of invasion and expansion of Bursaphelenchus xylophilus. For. Pest Dis. 2018, 37, 1–4. [Google Scholar]
- Jung, C.S.; Koh, S.-H.; Nam, Y.; Ahn, J.J.; Lee, C.Y.; Choi, W.I. A model for predicting spring emergence of Monochamus saltuarius (Coleoptera: Cerambycidae) from Korean white pine, pinus koraiensis. J. Econ. Entomol. 2015, 108, 1830–1836. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-R.; Lee, S.-C.; Lee, D.H.; Choi, W.-S.; Jung, C.-S.; Jeon, J.-H.; Kim, J.-E.; Park, I.-K. Identification of the aggregation-sex pheromone produced by male Monochamus saltuarius, a major insect vector of the pine wood nematode. J. Chem. Ecol. 2017, 43, 670–678. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, H.; Sheng, R.-C.; Sun, H.; Sun, S.-H.; Chen, F.-M. The first record of Monochamus saltuarius (Coleoptera; Cerambycidae) as vector of Bursaphelenchus xylophilus and its new potential hosts in china. Insects 2020, 11, 636. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.L. The southern pine sawyer. USDA Bur. Entomol. Bull. 1909, 58, 41–56. [Google Scholar]
- Wingfield, M.J. Transmission of pine wood nematode to cut timber and girdled trees. Plant Dis. 1983, 67, 35–37. [Google Scholar] [CrossRef]
- Skoczek, A.; Piesik, D.; Wenda-Piesik, A.; Buszewski, B.; Bocianowski, J.; Wawrzyniak, M. Volatile organic compounds released by maize following herbivory or insect extract application and communication between plants. J. Appl. Entomol. 2017, 141, 630–643. [Google Scholar] [CrossRef]
- Piesik, D.; Rochat, D.; Delaney, K.J.; Marion-Poll, F. Orientation of European Corn Borer first instar larvae to synthetic green leaf volatiles. J. Appl. Entomol. 2013, 137, 234–240. [Google Scholar] [CrossRef] [Green Version]
- Piesik, D.; Rochat, D.; van der Pers, J.; Marion-Poll, F. Pulsed odors from maize or spinach elicit orientation in European Corn Borer neonate larvae. J. Chem. Ecol. 2009, 35, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- De Bruyne, M.; Baker, T.C. Odor detection in insects: Volatile codes. J. Chem. Ecol. 2008, 34, 882–897. [Google Scholar] [CrossRef]
- Toshiya, I.; Nobuo, E.; Akiomi, Y.; Katsuo, O.; Takaaki, T. Attractants for the Japanese pine sawyer, Monochamus alternatus hope (Coleoptera: Cerambycidae). Nihon Ringakkai Shi J. Jpn. For. Soc. 1980, 15, 358–361. [Google Scholar]
- Fan, J.; Zhang, D.; Zhang, Z.; Meng, J.; Wang, Y. Feeding behavior of Monochamus alternatus and its relationship with the host volatiles. J. Zhejiang AF Univ. 2014, 31, 78–82. [Google Scholar]
- Hao, D.; Fenglin, M.A.; Wang, Y.; Zhang, Y.; Dai, H. Electroantennogram and behavioral responses of Monochamus alternatus to the volatiles from pinus thunbergii with different physiological status. Chin. J. Appl. Ecol. 2006, 17, 1070. [Google Scholar]
- Fettköther, R.; Reddy, G.V.P.; Noldt, U.; Dettner, K. Effect of host and larval frass volatiles on behavioural response of the old house borer, Hylotrupes bajulus (L.) (Coleoptera: Cerambycidae), in a wind tunnel bioassay. Chemoecology 2000, 10, 1–10. [Google Scholar] [CrossRef]
- Phillips, T.W.; Wilkening, A.J.; Atkinson, T.H.; Nation, J.L.; Wilkinson, R.C.; Foltz, J.L. Synergism of turpentine and ethanol as attractants for certain pine-infesting beetles (Coleoptera). Environ. Entomol. 1988, 17, 456–462. [Google Scholar] [CrossRef]
- Wynand, V.D.G.V.N.; Carlson, J.R. Insects as chemosensors of humans and crops. Nature 2006, 444, 302–307. [Google Scholar]
- Pelosi, P.; Zhou, J.-J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef]
- Pelosi, P.; Calvello, M.; Ban, L. Diversity of odorant-binding proteins and chemosensory proteins in insects. Chem. Senses 2005, 30, i291–i292. [Google Scholar] [CrossRef] [Green Version]
- Rützler, M.; Zwiebel, L. Molecular biology of insect olfaction:Recent progress and conceptual models. J. Comp. Physiol. A 2005, 191, 777–790. [Google Scholar] [CrossRef]
- Su, C.Y.; Menuz, K.; Carlson, J.R. Olfactory perception: Receptors, cells, and circuits. Cell 2009, 139, 45–59. [Google Scholar] [CrossRef] [Green Version]
- Vosshall, L.B.; Stocker, R.F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007, 30, 505–533. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Latief, M. A family of chemoreceptors in Tribolium castaneum (Tenebrionidae: Coleoptera). PLoS ONE 2007, 2, e1319. [Google Scholar] [CrossRef]
- Andersson, M.N.; Grosse-Wilde, E.; Keeling, C.I.; Bengtsson, J.M.; Yuen, M.M.S.; Li, M.; Hillbur, Y.; Bohlmann, J.; Hansson, B.S.; Schlyter, F. Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genom. 2013, 14, 198. [Google Scholar] [CrossRef] [Green Version]
- Ali, S.; Ahmed, M.Z.; Li, N.; Ali, S.A.I.; Wang, M.Q. Functional characteristics of chemosensory proteins in the sawyer beetle Monochamus alternatus Hope. Bull. Entomol. Res. 2018, 109, 34–42. [Google Scholar] [CrossRef]
- Hu, P.; Wang, J.; Cui, M.; Tao, J.; Luo, Y. Antennal transcriptome analysis of the asian longhorned beetle Anoplophora glabripennis. Sci. Rep. 2016, 6, 26652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.; Zhang, Z.; Wang, H.; Kong, X. Antennal transcriptome analysis and comparison of olfactory genes in two sympatric defoliators, Dendrolimus houi and Dendrolimus kikuchii (lepidoptera: Lasiocampidae). Insect Biochem. Mol. Biol. 2014, 52, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-F.; Liu, H.-H.; Kong, X.-B.; Wang, H.-B.; Liu, F.; Zhang, Z. Identification and expression profiling of chemosensory genes in Dendrolimus punctatus walker. Front. Physiol. 2017, 8, 471. [Google Scholar] [CrossRef] [PubMed]
- Bogdanova, E.A.; Shagin, D.A.; Lukyanov, S.A. Normalization of full-length enriched cdna. Mol. BioSystems 2008, 4, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Zhulidov, P.A.; Bogdanova, E.A.; Shcheglov, A.S.; Vagner, L.L.; Khaspekov, G.L.; Kozhemyako, V.B.; Matz, M.V.; Meleshkevitch, E.; Moroz, L.L.; Lukyanov, S.A.; et al. Simple cDNA normalization using Kamchatka Crab duplex-specific nuclease. Nucleic Acids Res. 2004, 32, e37. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from rna-seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Krieger, J.; Grosse-Wilde, E.; Gohl, T.; Dewer, Y.M.E.; Raming, K.; Breer, H. Genes encoding candidate pheromone receptors in a moth (Heliothis virescens). Proc. Natl. Acad. Sci. USA 2004, 101, 11845–11850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level—The DESeq Package; European Molecular Biology Laboratory: Heidelberg, Germany, 2012. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shen, S.; Zhang, S.; Wang, H.; Kong, X.; Liu, F.; Zhang, Z. Chemosensory characteristics of two semanotus bifasciatus populations. Forests 2019, 10, 655. [Google Scholar] [CrossRef] [Green Version]
- Vieira, F.G.; Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the arthropoda: Origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 2011, 3, 476–490. [Google Scholar] [CrossRef]
- Xiao, L.; Qian, J.; Jie, W.; Fei, L.; Jiang, X.; Hu, J.; Qu, M. Chemosensory gene families in adult antennae of Anomala corpulenta motschulsky (Coleoptera: Scarabaeidae: Rutelinae). PLoS ONE 2015, 10, e0144214. [Google Scholar]
- Andersson, M.N.; Keeling, C.I.; Mitchell, R.F. Genomic content of chemosensory genes correlates with host range in wood-boring beetles (Dendroctonus ponderosae, Agrilus planipennis, and Anoplophora glabripennis). BMC Genom. 2019, 20, 690. [Google Scholar] [CrossRef] [Green Version]
- McKenna, D.D.; Scully, E.D.; Pauchet, Y.; Hoover, K.; Kirsch, R.; Geib, S.M.; Mitchell, R.F.; Waterhouse, R.M.; Ahn, S.-J.; Arsala, D.; et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016, 17, 227. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, R.F.; Hughes, D.T.; Luetje, C.W.; Millar, J.G.; Soriano-Agatón, F.; Hanks, L.M.; Robertson, H.M. Sequencing and characterizing odorant receptors of the cerambycid beetle Megacyllene caryae. Insect Biochem. Mol. 2012, 42, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Zhang, S.; Wei, W.; Hao, H.; Zhang, B.; Butcher, R.; Sun, J. Chemical signals synchronize the life cycles of a plant-parasitic nematode and its vector beetle. Curr. Biol. 2013, 23, 2038. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).