Insights into the Binding of Dietary Phenolic Compounds to Human Serum Albumin and Food-Drug Interactions
Abstract
:1. Introduction
2. Plasma Proteins
3. Phenolic Compounds and Plasma Protein Binding
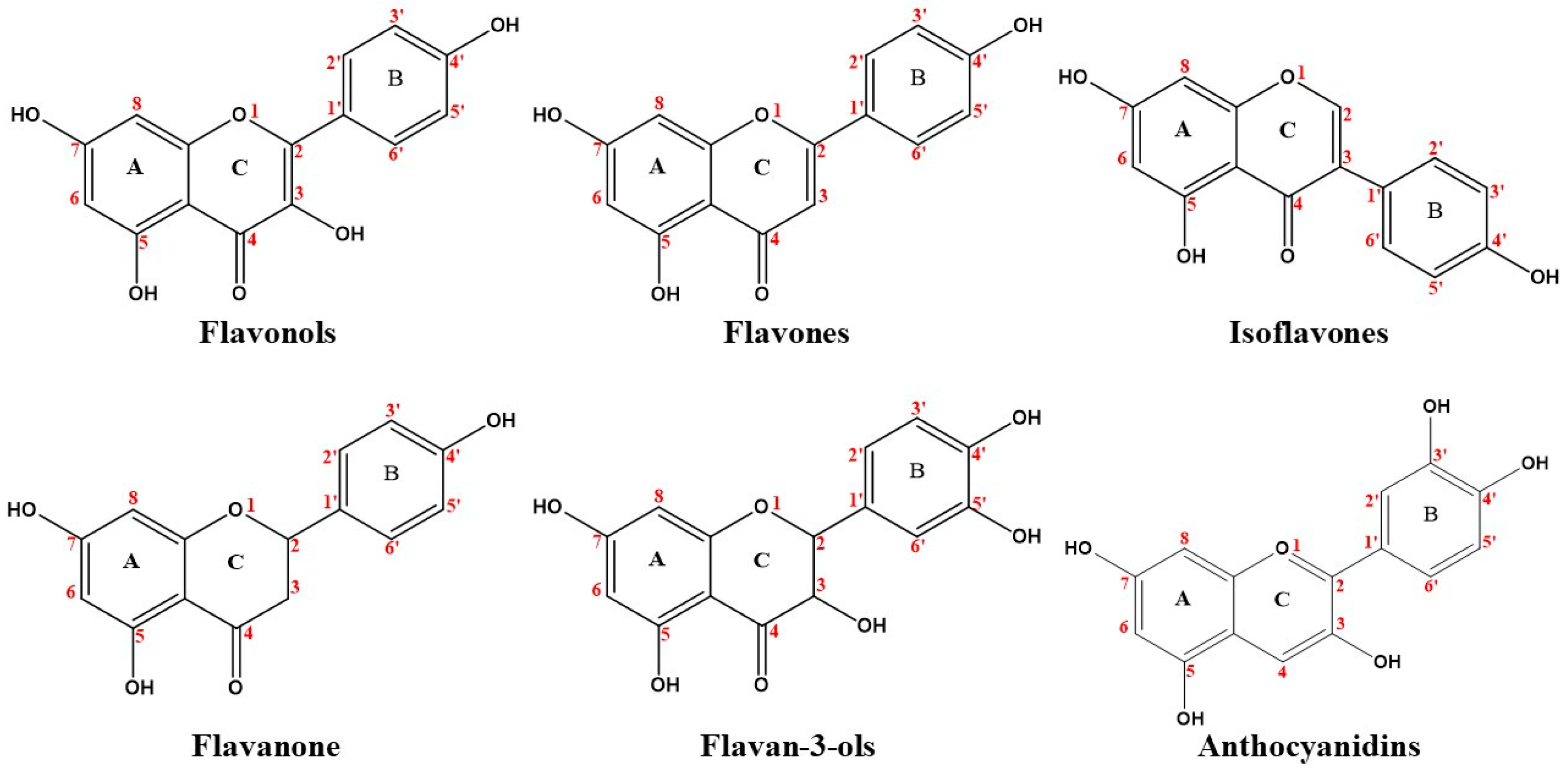
3.1. Flavonoids
3.1.1. Flavonols
3.1.2. Flavones
3.1.3. Isoflavones
3.1.4. Flavanones
3.1.5. Flavan-3-Ols
3.1.6. Anthocyanidins
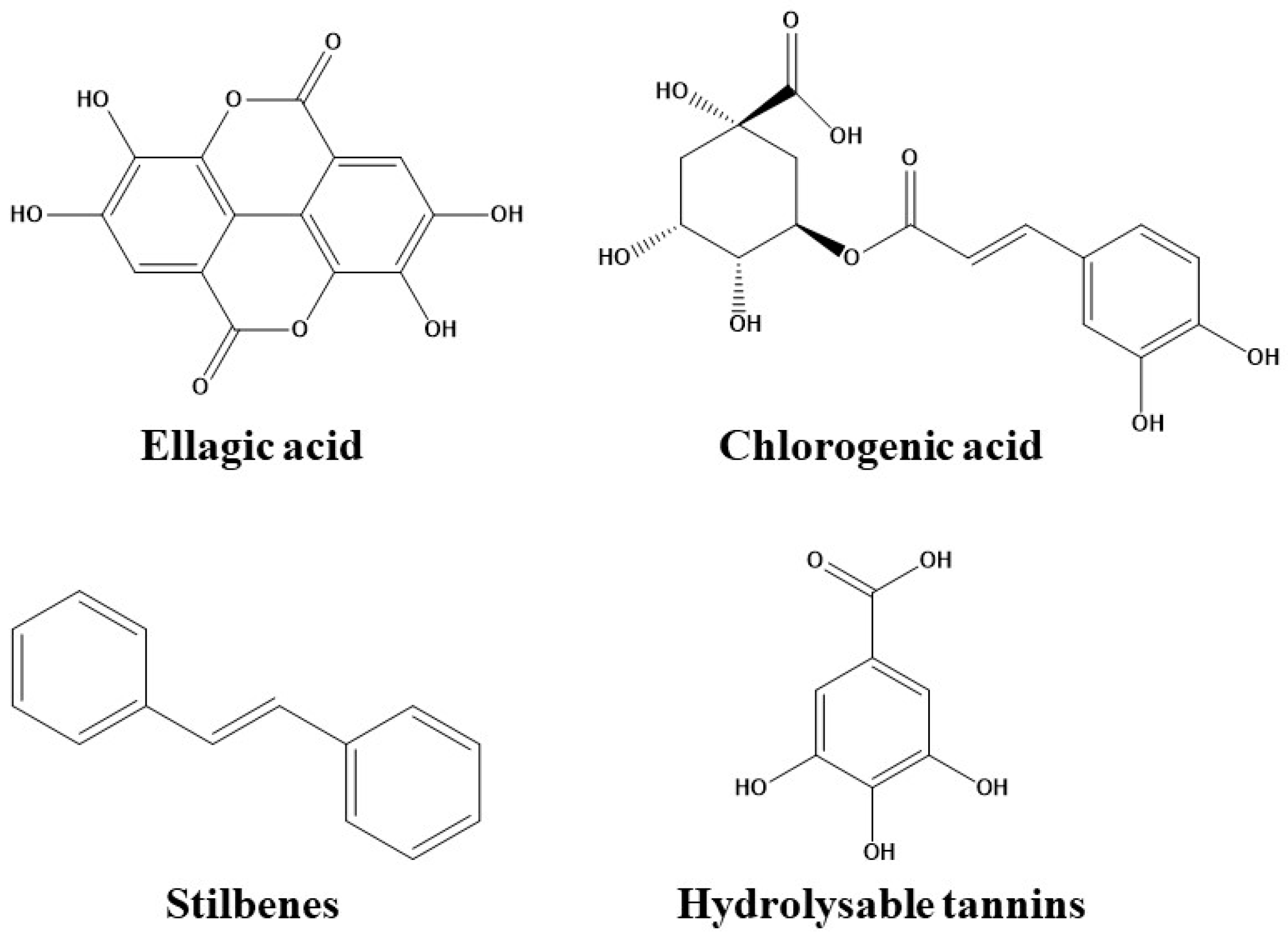
3.2. Phenolic Acids
3.2.1. Hydroxybenzoic Acids
3.2.2. Hydroxycinnamic Acids
3.3. Stilbenes
3.4. Hydrolysable Tannins
4. Interactions between Phenolic Compounds and Plasma Proteins
4.1. Non-Covalent Bonds
4.2. Hydrolysable Tannins
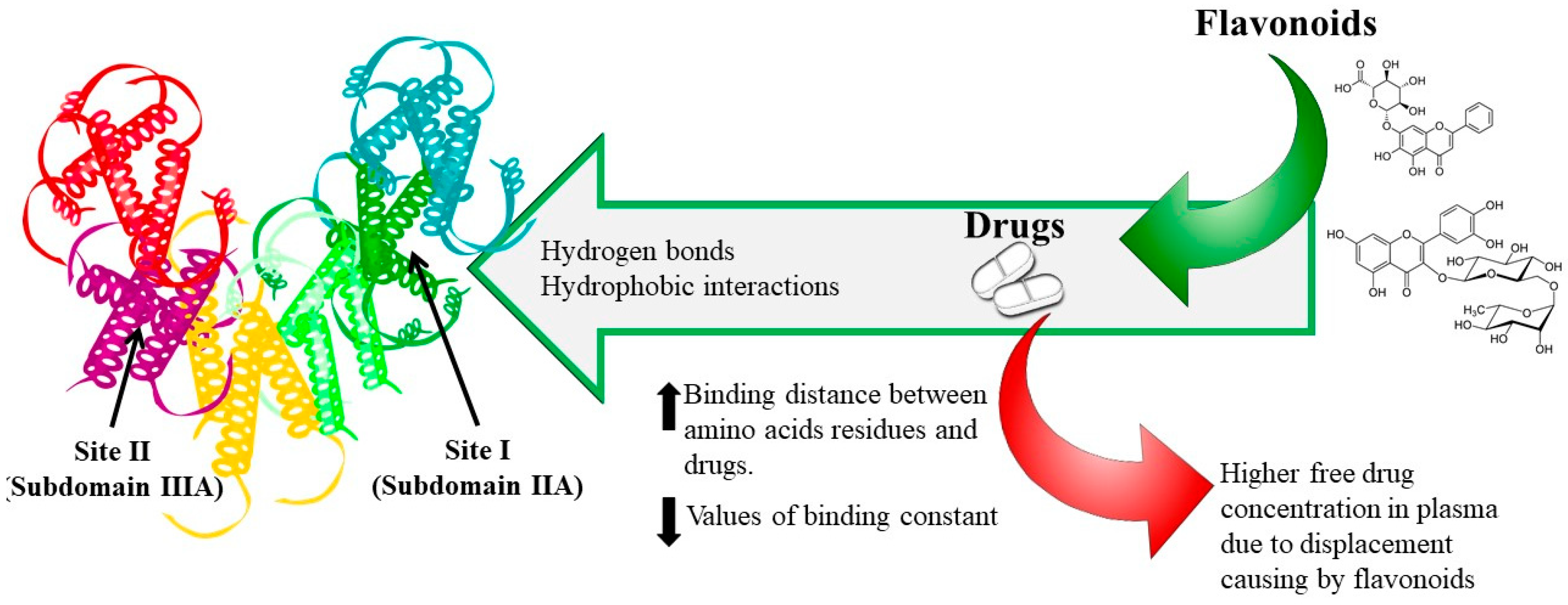
5. Binding Sites
6. Phenolic Compounds–Drug Interaction
7. Conclusions
Funding
Conflicts of Interest
References
- Xiao, J.; Kai, G. A review of dietary polyphenol-plasma protein interactions: Characterization, influence on the bioactivity, and structure-affinity relationship. Crit. Rev. Food Sci. Nutr. 2012, 52, 85–101. [Google Scholar] [CrossRef] [PubMed]
- Ribas-Agustí, A.; Martín-Belloso, O.; Soliva-Fortuny, R.; Elez-Martínez, P. Food processing strategies to enhance phenolic compounds bioaccessibility and bioavailability in plant-based foods. Crit. Rev. Food Sci. Nutr. 2018, 58, 2531–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poloni, D.M.; Dangles, O.; Vinson, J.A. Binding of plant polyphenols to serum albumin and LDL: Healthy implications for heart disease. J. Agric. Food Chem. 2019, 67, 9139–9147. [Google Scholar] [CrossRef] [PubMed]
- FDA, G. Bioavailability and Bioequivalence Studies for Orally Administered Drug Products—General; Food and Drug Administration: Rockville, MD, USA, 2002. [Google Scholar]
- Neilson, A.P.; Goodrich, K.M.; Ferruzzi, M.G. Bioavailability and Metabolism of Bioactive Compounds from Foods. In Nutrition in the Prevention and Treatment of Disease; Elsevier: Amsterdam, The Netherlands, 2017; pp. 301–319. ISBN 978-0-12-802928-2. [Google Scholar]
- Luca, S.V.; Macovei, I.; Bujor, A.; Miron, A.; Skalicka-Woźniak, K.; Aprotosoaie, A.C.; Trifan, A. Bioactivity of dietary polyphenols: The role of metabolites. Crit. Rev. Food Sci. Nutr. 2020, 60, 626–659. [Google Scholar] [CrossRef]
- Serreli, G.; Deiana, M. Biological relevance of extra virgin olive oil polyphenols metabolites. Antioxidants 2018, 7, 170. [Google Scholar] [CrossRef] [Green Version]
- Caldwell, J.; Gardner, I.; Swales, N. An introduction to drug disposition: The basic principles of absorption, distribution, metabolism, and excretion. Toxicol. Pathol. 1995, 23, 102–114. [Google Scholar] [CrossRef]
- Talevi, A.; Quiroga, P.A.M. (Eds.) ADME Processes in Pharmaceutical Sciences: Dosage, Design, and Pharmacotherapy Success; Springer: Cham, Switzerland, 2018; ISBN 978-3-319-99592-2. [Google Scholar]
- Gupta, A. Plasma Proteins. In Comprehensive Biochemistry for Dentistry: Textbook for Dental Students; Gupta, A., Ed.; Springer: Singapore, 2019; pp. 67–75. ISBN 978-981-13-1035-5. [Google Scholar]
- Otagiri, M. A molecular functional study on the interactions of drugs with plasma proteins. Drug Metab. Pharmacokinet. 2005, 20, 309–323. [Google Scholar] [CrossRef] [Green Version]
- Kamble, S.; Loadman, P.; Abraham, M.H.; Liu, X. Structural properties governing drug-plasma protein binding determined by high-performance liquid chromatography method. J. Pharm. Biomed. Anal. 2018, 149, 16–21. [Google Scholar] [CrossRef] [Green Version]
- Dasgupta, A. Monitoring Free Drug Concentration. In Clinical Challenges in Therapeutic Drug Monitoring; Elsevier: Amsterdam, The Netherlands, 2016; pp. 71–100. ISBN 978-0-12-802025-8. [Google Scholar]
- Roberts, J.A.; Pea, F.; Lipman, J. The clinical relevance of plasma protein binding changes. Clin. Pharmacokinet. 2013, 52, 1–8. [Google Scholar] [CrossRef]
- Vallner, J.J. Binding of Drugs by Albumin Plasma Protein. J. Pharm. Sci. 1977, 66, 447–465. [Google Scholar] [CrossRef]
- Smith, D.A.; Di, L.; Kerns, E.H. The effect of plasma protein binding on in vivo efficacy: Misconceptions in drug discovery. Nat. Rev. Drug Discov. 2010, 9, 929–939. [Google Scholar] [CrossRef] [PubMed]
- Beasley, V. Absorption, Distribution, Metabolism, and Elimination: Differences among Species. In Veterinary Toxicology; International Veterinary Information Service: Ithaca, NY, USA, 1999; pp. 1–19. [Google Scholar]
- Hartmut, D.; Schmidt, S. Clinical Pharmacokinetics and Pharmacodynamics: Concepts and Applications, 5th ed.; Wolters Kluwer Health/Lippincott William & Wilkins: Philadelphia, PA, USA, 2020. [Google Scholar]
- Rowland, M.; Tozer, T.N. Clinical Pharmacokinetics and Pharmacodynamics: Concepts and Applications, 4th ed.; Wolters Kluwer Health/Lippincott William & Wilkins: Philadelphia, PA, USA, 2011; ISBN 978-0-7817-5009-7. [Google Scholar]
- Kratochwil, N.A.; Huber, W.; Müller, F.; Kansy, M.; Gerber, P.R. Predicting plasma protein binding of drugs: A new approach. Biochem. Pharmacol. 2002, 64, 1355–1374. [Google Scholar] [CrossRef]
- Nation, R.L.; Theuretzbacher, U.; Tsuji, B.T. Concentration-dependent plasma protein binding: Expect the unexpected. Eur. J. Pharm. Sci. 2018, 122, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Merlot, A.M.; Kalinowski, D.S.; Richardson, D.R. Unraveling the mysteries of serum albumin—More than just a serum protein. Front. Physiol. 2014, 5, 299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poulin, P.; Haddad, S. Albumin and Uptake of Drugs in Cells: Additional Validation Exercises of a Recently Published Equation that Quantifies the Albumin-Facilitated Uptake Mechanism(s) in Physiologically Based Pharmacokinetic and Pharmacodynamic Modeling Research. J. Pharm. Sci. 2015, 104, 4448–4458. [Google Scholar] [CrossRef]
- Vuignier, K.; Schappler, J.; Veuthey, J.-L.; Carrupt, P.-A.; Martel, S. Drug–protein binding: A critical review of analytical tools. Anal. Bioanal. Chem. 2010, 398, 53–66. [Google Scholar] [CrossRef]
- Schmidt, S.; Gonzalez, D.; Derendorf, H. Significance of Protein Binding in Pharmacokinetics and Pharmacodynamics. J. Pharm. Sci. 2010, 99, 1107–1122. [Google Scholar] [CrossRef]
- Howard, M.; Hill, J.; Galluppi, G.; McLean, M. Plasma Protein Binding in Drug Discovery and Development. Comb. Chem. High Throughput Screen. 2010, 13, 170–187. [Google Scholar] [CrossRef]
- Trainor, G.L. The importance of plasma protein binding in drug discovery. Expert Opin. Drug Discov. 2007, 2, 51–64. [Google Scholar] [CrossRef]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef]
- Sheng, F.; Wang, Y.; Zhao, X.; Tian, N.; Hu, H.; Li, P. Separation and Identification of Anthocyanin Extracted from Mulberry Fruit and the Pigment Binding Properties toward Human Serum Albumin. J. Agric. Food Chem. 2014, 62, 6813–6819. [Google Scholar] [CrossRef] [PubMed]
- Varshney, A.; Sen, P.; Ahmad, E.; Rehan, M.; Subbarao, N.; Khan, R.H. Ligand binding strategies of human serum albumin: How can the cargo be utilized? Chirality 2010, 22, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A. Comprehensive Biochemistry for Dentistry: Textbook for Dental Students; Springer: Singapore, 2018; ISBN 9789811310348. [Google Scholar]
- Yeggoni, D.P.; Rachamallu, A.; Subramanyam, R. A comparative binding mechanism between human serum albumin and α-1-acid glycoprotein with corilagin: Biophysical and computational approach. RSC Adv. 2016, 6, 40225–40237. [Google Scholar] [CrossRef]
- Xiao, J.; Zhao, Y.; Wang, H.; Yuan, Y.; Yang, F.; Zhang, C.; Yamamoto, K. Noncovalent Interaction of Dietary Polyphenols with Common Human Plasma Proteins. J. Agric. Food Chem. 2011, 59, 10747–10754. [Google Scholar] [CrossRef]
- Das, P.; Chaudhari, S.K.; Das, A.; Kundu, S.; Saha, C. Interaction of flavonols with human serum albumin: A biophysical study showing structure-activity relationship and enhancement when coated on silver nanoparticles. J. Biomol. Struct. Dyn. 2019, 37, 1414–1426. [Google Scholar] [CrossRef]
- Cao, H.; Liu, X.; Ulrih, N.P.; Sengupta, P.K.; Xiao, J. Plasma protein binding of dietary polyphenols to human serum albumin: A high performance affinity chromatography approach. Food Chem. 2019, 270, 257–263. [Google Scholar] [CrossRef]
- Poór, M.; Boda, G.; Needs, P.W.; Kroon, P.A.; Lemli, B.; Bencsik, T. Interaction of quercetin and its metabolites with warfarin: Displacement of warfarin from serum albumin and inhibition of CYP2C9 enzyme. Biomed. Pharmacother. 2017, 88, 574–581. [Google Scholar] [CrossRef]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Probing the binding of the flavonoid, quercetin to human serum albumin by circular dichroism, electronic absorption spectroscopy and molecular modelling methods. Biochem. Pharmacol. 2003, 65, 447–456. [Google Scholar] [CrossRef]
- Dufour, C.; Dangles, O. Flavonoid–serum albumin complexation: Determination of binding constants and binding sites by fluorescence spectroscopy. Biochim. Biophys. Acta Gen. Subj. 2005, 1721, 164–173. [Google Scholar] [CrossRef]
- Rimac, H.; Debeljak, Ž.; Šakić, D.; Weitner, T.; Gabričević, M.; Vrček, V.; Zorc, B.; Bojić, M. Structural and electronic determinants of flavonoid binding to human serum albumin: An extensive ligand-based study. RSC Adv. 2016, 6, 75014–75022. [Google Scholar] [CrossRef] [Green Version]
- Cao, H.; Chen, L.; Xiao, J. Binding Citrus flavanones to human serum albumin: Effect of structure on affinity. Mol. Biol. Rep. 2011, 38, 2257–2262. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.K.; Rakotomanomana, N.; Dufour, C.; Dangles, O. Binding of citrus flavanones and their glucuronides and chalcones to human serum albumin. Food Funct. 2011, 2, 617. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S. Study on the interaction of (+)-catechin with human serum albumin using isothermal titration calorimetry and spectroscopic techniques. New J. Chem. 2015, 39, 386–395. [Google Scholar] [CrossRef]
- Ishii, T.; Ichikawa, T.; Minoda, K.; Kusaka, K.; Ito, S.; Suzuki, Y.; Akagawa, M.; Mochizuki, K.; Goda, T.; Nakayama, T. Human Serum Albumin as an Antioxidant in the Oxidation of (−)-Epigallocatechin Gallate: Participation of Reversible Covalent Binding for Interaction and Stabilization. Biosci. Biotechnol. Biochem. 2011, 75, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Minoda, K.; Ichikawa, T.; Katsumata, T.; Onobori, K.; Mori, T.; Suzuki, Y.; Ishii, T.; Nakayama, T. Influence of the Galloyl Moiety in Tea Catechins on Binding Affinity for Human Serum Albumin. J. Nutr. Sci. Vitaminol. 2010, 56, 331–334. [Google Scholar] [CrossRef] [Green Version]
- Tang, L.; Zuo, H.; Shu, L. Comparison of the interaction between three anthocyanins and human serum albumins by spectroscopy. J. Lumin. 2014, 153, 54–63. [Google Scholar] [CrossRef]
- Zhang, J.; Zuo, B.; Ulrih, N.P.; Sengupta, P.K.; Zheng, X.; Xiao, J. Structure-affinity relationship of dietary anthocyanin–HSA interaction. J. Berry Res. 2018, 8, 1–9. [Google Scholar] [CrossRef]
- Cahyana, Y.; Gordon, M.H. Interaction of anthocyanins with human serum albumin: Influence of pH and chemical structure on binding. Food Chem. 2013, 141, 2278–2285. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, S.; Qin, Y.; Liu, J.; Liu, J.; Wang, Q.; Ren, F.; Zhang, H. Interaction of phenolic acids and their derivatives with human serum albumin: Structure–affinity relationships and effects on antioxidant activity. Food Chem. 2018, 240, 1072–1080. [Google Scholar] [CrossRef]
- Peng, X.; Wang, X.; Qi, W.; Su, R.; He, Z. Affinity of rosmarinic acid to human serum albumin and its effect on protein conformation stability. Food Chem. 2016, 192, 178–187. [Google Scholar] [CrossRef]
- Kang, J.; Liu, Y.; Xie, M.; Li, S.; Jiang, M.; Wang, Y. Interactions of human serum albumin with chlorogenic acid and ferulic acid. Biochim. Biophys. Acta Gen. Subj. 2004, 1674, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Huang, Y.; Ma, X.; Liao, X.; Wang, Q.; Xiong, X.; Li, H. Multispectroscopic and docking studies on the binding of chlorogenic acid isomers to human serum albumin: Effects of esteryl position on affinity. Food Chem. 2016, 212, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Min, J.; Meng-Xia, X.; Dong, Z.; Yuan, L.; Xiao-Yu, L.; Xing, C. Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J. Mol. Struct. 2004, 692, 71–80. [Google Scholar] [CrossRef]
- Cao, H.; Jia, X.; Shi, J.; Xiao, J.; Chen, X. Non-covalent interaction between dietary stilbenoids and human serum albumin: Structure–affinity relationship, and its influence on the stability, free radical scavenging activity and cell uptake of stilbenoids. Food Chem. 2016, 202, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.S. Spectroscopic study on the interaction of resveratrol and pterostilbene with human serum albumin. J. Photochem. Photobiol. B Biol. 2015, 149, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Sekowski, S.; Bitiucki, M.; Ionov, M.; Zdeb, M.; Abdulladjanova, N.; Rakhimov, R.; Mavlyanov, S.; Bryszewska, M.; Zamaraeva, M. Influence of valoneoyl groups on the interactions between Euphorbia tannins and human serum albumin. J. Lumin. 2018, 194, 170–178. [Google Scholar] [CrossRef]
- Sekowski, S.; Ionov, M.; Kaszuba, M.; Mavlyanov, S.; Bryszewska, M.; Zamaraeva, M. Biophysical studies of interaction between hydrolysable tannins isolated from Oenothera gigas and Geranium sanguineum with human serum albumin. Colloids Surf. B Biointerfaces 2014, 123, 623–628. [Google Scholar] [CrossRef]
- Wang, B.; Qin, Q.; Chang, M.; Li, S.; Shi, X.; Xu, G. Molecular interaction study of flavonoids with human serum albumin using native mass spectrometry and molecular modeling. Anal. Bioanal. Chem. 2018, 410, 827–837. [Google Scholar] [CrossRef]
- Boulton, D.W.; Walle, U.K.; Walle, T. Extensive Binding of the Bioflavonoid Quercetin to Human Plasma Proteins. J. Pharm. Pharmacol. 1998, 50, 243–249. [Google Scholar] [CrossRef]
- Dangles, O.; Dufour, C.; Bret, S. Flavonol–serum albumin complexation. Two-electron oxidation of flavonols and their complexes with serum albumin. J. Chem. Soc. Perkin Trans. 1999, 2, 737–744. [Google Scholar] [CrossRef]
- Xiao, J.; Cao, H.; Wang, Y.; Yamamoto, K.; Wei, X. Structure-affinity relationship of flavones on binding to serum albumins: Effect of hydroxyl groups on ring A. Mol. Nutr. Food Res. 2010, 54, S253–S260. [Google Scholar] [CrossRef] [PubMed]
- Mahesha, H.G.; Singh, S.A.; Srinivasan, N.; Rao, A.G.A. A spectroscopic study of the interaction of isoflavones with human serum albumin. FEBS J. 2006, 273, 451–467. [Google Scholar] [CrossRef]
- Mandeville, J.-S.; Froehlich, E.; Tajmir-Riahi, H.A. Study of curcumin and genistein interactions with human serum albumin. J. Pharm. Biomed. Anal. 2009, 49, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Trnková, L.; Boušová, I.; Staňková, V.; Dršata, J. Study on the interaction of catechins with human serum albumin using spectroscopic and electrophoretic techniques. J. Mol. Struct. 2011, 985, 243–250. [Google Scholar] [CrossRef]
- Roy, D.; Dutta, S.; Maity, S.S.; Ghosh, S.; Roy, A.S.; Ghosh, K.S.; Dasgupta, S. Spectroscopic and docking studies of the binding of two stereoisomeric antioxidant catechins to serum albumins. J. Lumin. 2012, 132, 1364–1375. [Google Scholar] [CrossRef]
- Pattanayak, R.; Basak, P.; Sen, S.; Bhattacharyya, M. An insight to the binding of ellagic acid with human serum albumin using spectroscopic and isothermal calorimetry studies. Biochem. Biophys. Rep. 2017, 10, 88–93. [Google Scholar] [CrossRef]
- Tang, J.H.; Liang, G.B.; Zheng, C.Z.; Lian, N. Investigation on the Binding Behavior of Ellagic Acid to Human Serum Albumin in Aqueous Solution. J. Solut. Chem. 2013, 42, 226–238. [Google Scholar] [CrossRef]
- Nanda, R.K.; Sarkar, N.; Banerjee, R. Probing the interaction of ellagic acid with human serum albumin: A fluorescence spectroscopic study. J. Photochem. Photobiol. Chem. 2007, 192, 152–158. [Google Scholar] [CrossRef]
- He, J.; Wang, Q.; Zhang, L.; Lin, X.; Li, H. Docking simulations and spectroscopy of the interactions of ellagic acid and oleuropein with human serum albumin. J. Lumin. 2014, 154, 578–583. [Google Scholar] [CrossRef]
- Muralidhara, B.K.; Prakash, V. Interaction of 3′-O-caffeoyl d-quinic acid with human serum albumin. Int. J. Pept. Protein Res. 2009, 46, 1–8. [Google Scholar] [CrossRef]
- Sinisi, V.; Forzato, C.; Cefarin, N.; Navarini, L.; Berti, F. Interaction of chlorogenic acids and quinides from coffee with human serum albumin. Food Chem. 2015, 168, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukpoé-Kossi, C.N.N.; St-Louis, C.; Beauregard, M.; Subirade, M.; Carpentier, R.; Hotchandani, S.; Tajmir-Riahi, H.A. Resveratrol Binding to Human Serum Albumin. J. Biomol. Struct. Dyn. 2006, 24, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Diniz, A.; Escuder-Gilabert, L.; Lopes, N.P.; Villanueva-Camañas, R.M.; Sagrado, S.; Medina-Hernández, M.J. Characterization of interactions between polyphenolic compounds and human serum proteins by capillary electrophoresis. Anal. Bioanal. Chem. 2008, 391, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Rohn, S. Possibilities and limitations in the analysis of covalent interactions between phenolic compounds and proteins. Food Res. Int. 2014, 65, 13–19. [Google Scholar] [CrossRef]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef]
- Appel, H.M. Phenolics in ecological interactions: The importance of oxidation. J. Chem. Ecol. 1993, 19, 1521–1552. [Google Scholar] [CrossRef]
- Bian, Q.; Liu, J.; Tian, J.; Hu, Z. Binding of genistein to human serum albumin demonstrated using tryptophan fluorescence quenching. Int. J. Biol. Macromol. 2004, 34, 275–279. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Dong, L.; Li, J.; He, W.; Chen, X.; Hu, Z. Investigation of the interaction between naringin and human serum albumin. J. Mol. Struct. 2008, 875, 1–8. [Google Scholar] [CrossRef]
- Ding, F.; Diao, J.-X.; Sun, Y.; Sun, Y. Bioevaluation of Human Serum Albumin–Hesperidin Bioconjugate: Insight into Protein Vector Function and Conformation. J. Agric. Food Chem. 2012, 60, 7218–7228. [Google Scholar] [CrossRef]
- Xie, M.-X.; Xu, X.-Y.; Wang, Y.-D. Interaction between hesperetin and human serum albumin revealed by spectroscopic methods. Biochim. Biophys. Acta Gen. Subj. 2005, 1724, 215–224. [Google Scholar] [CrossRef]
- Feroz, S.R.; Mohamad, S.B.; Bakri, Z.S.D.; Malek, S.N.A.; Tayyab, S. Probing the Interaction of a Therapeutic Flavonoid, Pinostrobin with Human Serum Albumin: Multiple Spectroscopic and Molecular Modeling Investigations. PLoS ONE 2013, 8, e76067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, B.; Wang, Y.; Mi, R.; Ouyang, Y.; Hu, Y.-J. Evaluation of the interaction between naringenin and human serum albumin: Insights from fluorescence spectroscopy, electrochemical measurement and molecular docking. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 149, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Chanphai, P.; Tajmir-Riahi, H.A. Tea polyphenols bind serum albumins: A potential application for polyphenol delivery. Food Hydrocoll. 2019, 89, 461–467. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, L.; Li, J.; Chen, X. Studies on the interaction of gallic acid with human serum albumin in membrane mimetic environments. Talanta 2008, 76, 246–253. [Google Scholar] [CrossRef]
- Kaldas, M.I.; Walle, U.K.; van der Woude, H.; McMillan, J.M.; Walle, T. Covalent Binding of the Flavonoid Quercetin to Human Serum Albumin. J. Agric. Food Chem. 2005, 53, 4194–4197. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Modifications of human serum albumin and their binding effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef] [Green Version]
- Matei, I.; Hillebrand, M. Interaction of kaempferol with human serum albumin: A fluorescence and circular dichroism study. J. Pharm. Biomed. Anal. 2010, 51, 768–773. [Google Scholar] [CrossRef]
- Yuan, J.-L.; Lv, Z.; Liu, Z.-G.; Hu, Z.; Zou, G.-L. Study on interaction between apigenin and human serum albumin by spectroscopy and molecular modeling. J. Photochem. Photobiol. Chem. 2007, 191, 104–113. [Google Scholar] [CrossRef]
- Liu, H.; Bao, W.; Ding, H.; Jang, J.; Zou, G. Binding Modes of Flavones to Human Serum Albumin: Insights from Experimental and Computational Studies. J. Phys. Chem. B 2010, 114, 12938–12947. [Google Scholar] [CrossRef]
- Gokara, M.; Sudhamalla, B.; Amooru, D.G.; Subramanyam, R. Molecular Interaction Studies of Trimethoxy Flavone with Human Serum Albumin. PLoS ONE 2010, 5, e8834. [Google Scholar] [CrossRef] [Green Version]
- Nozaki, A.; Kimura, T.; Ito, H.; Hatano, T. Interaction of Polyphenolic Metabolites with Human Serum Albumin: A Circular Dichroism Study. Chem. Pharm. Bull. 2009, 57, 1019–1023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Guo, X.-Y.; Xu, L.; Liu, B.; Zhou, L.-L.; Wang, X.-F.; Wang, D.; Sun, T. Studies on the competitive binding of cleviprex and flavonoids to plasma protein by multi-spectroscopic methods: A prediction of food-drug interaction. J. Photochem. Photobiol. B Biol. 2017, 175, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, L.-L.; Liu, B.; Xu, L.; Wang, X.-F.; Sun, T. Decrease of the affinity of theophylline bind to serum proteins induced by flavonoids and their synergies on protein conformation. Int. J. Biol. Macromol. 2018, 107, 1066–1073. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y.; He, L.-L.; Liu, B.; Zhang, S.-Y.; Ye, X.; Jing, J.-J.; Zhang, J.-F.; Gao, M. Spectroscopic investigation on the food components–drug interaction: The influence of flavonoids on the affinity of nifedipine to human serum albumin. Food Chem. Toxicol. 2015, 78, 42–51. [Google Scholar] [CrossRef] [PubMed]
- He, L.-L.; Wang, Z.-X.; Wang, Y.-X.; Liu, X.-P.; Yang, Y.-J.; Gao, Y.-P.; Wang, X.; Liu, B. Studies on the interaction between promethazine and human serum albumin in the presence of flavonoids by spectroscopic and molecular modeling techniques. Colloids Surf. B Biointerfaces 2016, 145, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.-M.; Zhang, J.; Bai, C.-L.; Wang, X.; Qiu, X.-Z.; Wang, X.-L.; Ji, H.; Liu, B. Spectroscopic study on flavonoid–drug interactions: Competitive binding for human serum albumin between three flavonoid compounds and ticagrelor, a new antiplatelet drug. J. Lumin. 2015, 168, 69–76. [Google Scholar] [CrossRef]
- Mohseni-Shahri, F.S.; Housaindokht, M.R.; Bozorgmehr, M.R.; Moosavi-Movahedi, A.A. The influence of the flavonoid quercetin on the interaction of propranolol with human serum albumin: Experimental and theoretical approaches. J. Lumin. 2014, 154, 229–240. [Google Scholar] [CrossRef]
- Maciążek-Jurczyk, M.; Maliszewska, M.; Pożycka, J.; Równicka-Zubik, J.; Góra, A.; Sułkowska, A. Tamoxifen and curcumin binding to serum albumin. Spectroscopic study. J. Mol. Struct. 2013, 1044, 194–200. [Google Scholar] [CrossRef]
- Rimac, H.; Dufour, C.; Debeljak, Ž.; Zorc, B.; Bojić, M. Warfarin and Flavonoids Do Not Share the Same Binding Region in Binding to the IIA Subdomain of Human Serum Albumin. Molecules 2017, 22, 1153. [Google Scholar] [CrossRef]
| Class | Subclass | Reaction | Effect | References |
|---|---|---|---|---|
| Flavonoids | Flavonols | Hydroxylation | The binding affinity of flavonols for HSA * is structure-dependent, increasing with the number of OH groups in the A, B ring. | [33,34,35] |
| Glucuronidation | Decrease the binding constants. | [36,37] | ||
| Sulfation | Decrease the binding constants. | [38] | ||
| Flavones | Hydroxylation | The HSA affinity of flavones was influenced by the location and number of the OH group. | [33,35,39,40] | |
| Hydrogenation | Hydrogenation of the unsaturated C2=C3 double bond can reduce binding affinities for CHPP **. | |||
| Methoxylation | This reaction enhanced hydrophobicity and hydrophobic interactions increasing affinity for HSA. | [33,35,40] | ||
| Isoflavones | Hydroxylation | The hydroxylation at positions 5 and 7 on the A ring increased HSA binding affinity rates. A weakening of isoflavones binding affinities for CHPP after of hydroxylation in C-5 (A ring) and C-3’ (B ring). | [33,35] | |
| Flavanones | Hydrogenation | The C2=C3 double bond conjugated with the oxo group at C-4 plays an important role in flavanone affinity for plasma proteins. | [40] | |
| Hydroxylation | Affinity increased by the addition of a hydroxyl group on the A ring (C-5 and C-7) and the B ring (C-2’). | [35] | ||
| Methoxylation | Slightly increased the protein binding rate. | [40] | ||
| Glucuronidation | Glucuronidation in the B-ring weakly destabilizes the flavanone-HSA complex. | [41] | ||
| Flavan-3-ols | Hydroxylation | The number of hydroxyl groups on the B ring and the presence of a galloyl (3,4,5-trihydroxybenzoyl) moiety increase binding affinities for HSA. | [42,43,44] | |
| Anthocyanidins | Hydroxylation | The binding affinities increase with the number of hydroxyl groups on the B ring. | [45,46,47] | |
| Methoxylation | The methoxylation could either strengthen or reduce the anthocyanin affinity for HSA. | [46,47] | ||
| Phenolic Acids | Hydroxybenzoic acids | Hydroxylation | In the case of benzoic acid, the introduction of (1) an OH group at C-2 on the benzene ring exerted a positive effect and (2) a hydroxy substituent at C-4 had a negative influence. | [48] |
| Methoxylation | Both methylation of the hydroxy groups and substituting the hydroxy groups with methyl groups at C-3 and C-4 on the benzene ring resulted in an increase of binding affinity. | |||
| Hydroxycinnamic acids | Minimal modifications of the chemical structure led to significant changes in binding. | [49,50,51,52] | ||
| Stilbenes | Hydroxylation | The stilbenoid–HSA affinity was increased. | [35,53] | |
| Methylation | The stilbenoid–HSA affinity was reduced. | [54] | ||
| Methoxylation | [53] | |||
| Hydrolysable Tannins | Hydroxylation | The intensity of the interaction depends not only on the number of OH groups, but also on the bulk, flexibility and hydrophobicity of the chemical structure. | [55,56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Yerena, A.; Perez, M.; Vallverdú-Queralt, A.; Escribano-Ferrer, E. Insights into the Binding of Dietary Phenolic Compounds to Human Serum Albumin and Food-Drug Interactions. Pharmaceutics 2020, 12, 1123. https://doi.org/10.3390/pharmaceutics12111123
López-Yerena A, Perez M, Vallverdú-Queralt A, Escribano-Ferrer E. Insights into the Binding of Dietary Phenolic Compounds to Human Serum Albumin and Food-Drug Interactions. Pharmaceutics. 2020; 12(11):1123. https://doi.org/10.3390/pharmaceutics12111123
Chicago/Turabian StyleLópez-Yerena, Anallely, Maria Perez, Anna Vallverdú-Queralt, and Elvira Escribano-Ferrer. 2020. "Insights into the Binding of Dietary Phenolic Compounds to Human Serum Albumin and Food-Drug Interactions" Pharmaceutics 12, no. 11: 1123. https://doi.org/10.3390/pharmaceutics12111123
APA StyleLópez-Yerena, A., Perez, M., Vallverdú-Queralt, A., & Escribano-Ferrer, E. (2020). Insights into the Binding of Dietary Phenolic Compounds to Human Serum Albumin and Food-Drug Interactions. Pharmaceutics, 12(11), 1123. https://doi.org/10.3390/pharmaceutics12111123







