Immunological Analysis of Nodavirus Capsid Displaying the Domain III of Japanese Encephalitis Virus Envelope Protein
Abstract
1. Introduction
2. Materials and Methods
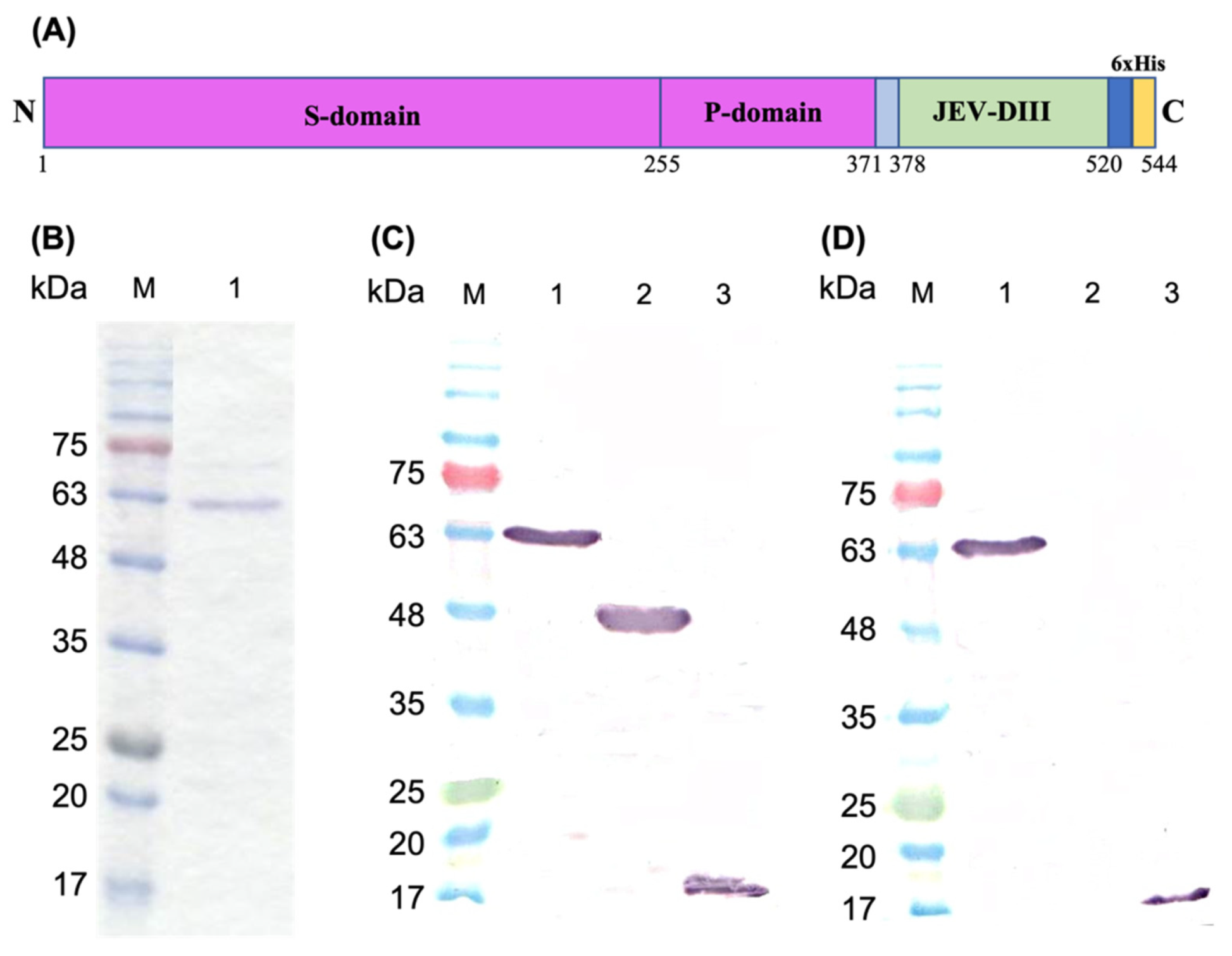
2.1. Construction of Recombinant Plasmids
2.2. Protein Expression and Purification
2.3. SDS-Polyacrylamide Gel Electrophoresis and Western Blotting
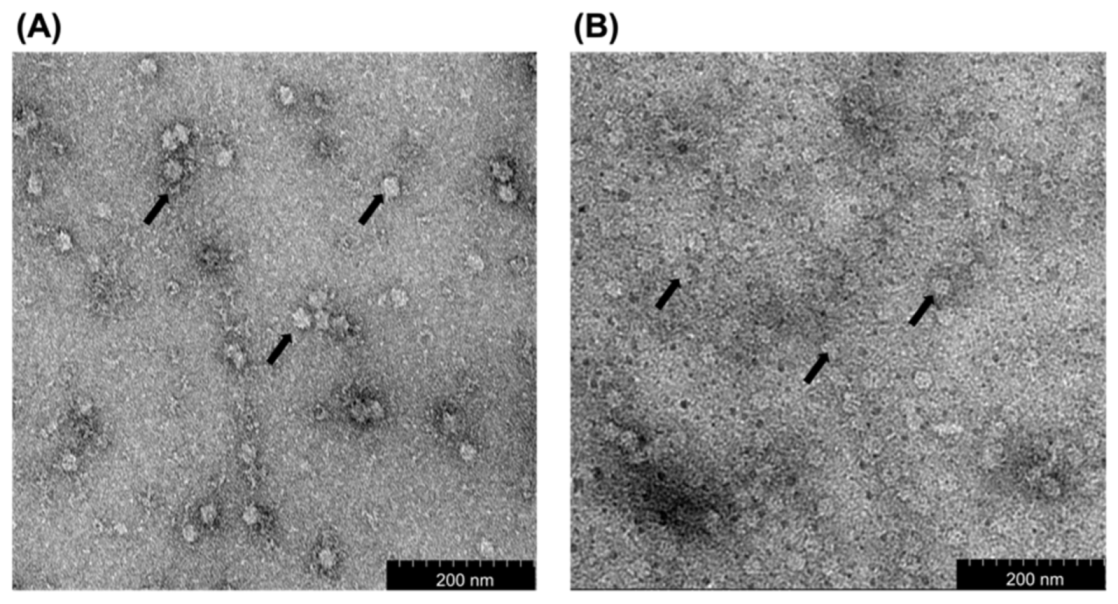
2.4. Transmission Electron Microscopy (TEM)
2.5. Dynamic Light Scattering (DLS)
2.6. Antigenicity Assay
2.7. Immunization of Mice
2.8. Immunogenicity Analysis
2.9. Splenocyte Isolation and Flow Cytometry
2.10. Cytokine Quantification through Sandwich Enzyme-Linked Immunosorbent Assay
2.11. Statistical Analysis
3. Results
3.1. Construction of Recombinant Plasmid, Protein Expression, and Purification
3.2. Transmission Electron Microscopy
3.3. Dynamic Light Scattering (DLS) Analysis
3.4. Analysis of the Antigenicity of the Fusion Protein by Enzyme-Linked Immunosorbent Assay (ELISA)
3.5. Immunogenicity of the Fusion Protein
3.6. Immunophenotyping of Mouse Splenocytes
3.7. Measurement of Cytokine Concentrations in Mouse Serum Samples
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, K.; Arshad, S.S.; Selvarajah, G.T.; Abu, J.; Toung, O.P.; Abba, Y.; Yasmin, A.R.; Bande, F.; Sharma, R.; Ong, B.L. Japanese encephalitis in Malaysia: An overview and timeline. Acta. Trop. 2018, 185, 219–229. [Google Scholar] [CrossRef]
- Kumar, K.; Arshad, S.S.; Toung, O.P.; Abba, Y.; Selvarajah, G.T.; Abu, J.; A, R.Y.; Ong, B.L.; Bande, F. The distribution of important sero-complexes of flaviviruses in Malaysia. Trop. Anim. Health Prod. 2019, 51, 495–506. [Google Scholar] [CrossRef]
- Platonov, A.; Rossi, G.; Karan, L.; Mironov, K.; Busani, L.; Rezza, G. Does the Japanese encephalitis virus (JEV) represent a threat for human health in Europe? Detection of JEV RNA sequences in birds collected in Italy. Euro. Surveill. 2012, 17, 20241. [Google Scholar] [CrossRef]
- Ravanini, P.; Huhtamo, E.; Ilaria, V.; Crobu, M.G.; Nicosia, A.M.; Servino, L.; Rivasi, F.; Allegrini, S.; Miglio, U.; Magri, A.; et al. Japanese encephalitis virus RNA detected in Culex pipiens mosquitoes in Italy. Euro. Surveill. 2012, 17, 20221. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Faye, O.; Prot, M.; Casademont, I.; Fall, G.; Fernandez-Garcia, M.D.; Diagne, M.M.; Kipela, J.M.; Fall, I.S.; Holmes, E.C.; et al. Autochthonous Japanese encephalitis with yellow fever coinfection in Africa. N. Engl. J. Med. 2017, 376, 1483–1485. [Google Scholar] [CrossRef]
- Ghosh, D.; Basu, A. Japanese encephalitis-a pathological and clinical perspective. PLoS Negl. Trop. Dis. 2009, 3, e437. [Google Scholar] [CrossRef] [PubMed]
- Campbell, G.L.; Hills, S.L.; Fischer, M.; Jacobson, J.A.; Hoke, C.H.; Hombach, J.M.; Marfin, A.A.; Solomon, T.; Tsai, T.F.; Tsu, V.D.; et al. Estimated global incidence of Japanese encephalitis: A systematic review. Bull. World Health Organ. 2011, 89, 766–774, 774A–774E. [Google Scholar] [CrossRef] [PubMed]
- Solomon, T.; Dung, N.M.; Kneen, R.; Thao le, T.T.; Gainsborough, M.; Nisalak, A.; Day, N.P.; Kirkham, F.J.; Vaughn, D.W.; Smith, S.; et al. Seizures and raised intracranial pressure in Vietnamese patients with Japanese encephalitis. Brain 2002, 125, 1084–1093. [Google Scholar] [CrossRef]
- Liu, W.; Fu, S.; Ma, X.; Chen, X.; Wu, D.; Zhou, L.; Yin, Q.; Li, F.; He, Y.; Lei, W.; et al. An outbreak of Japanese encephalitis caused by genotype Ib Japanese encephalitis virus in China, 2018: A laboratory and field investigation. PLoS Negl. Trop. Dis. 2020, 14, e0008312. [Google Scholar] [CrossRef] [PubMed]
- Sumiyoshi, H.; Mori, C.; Fuke, I.; Morita, K.; Kuhara, S.; Kondou, J.; Kikuchi, Y.; Nagamatu, H.; Igarashi, A. Complete nucleotide sequence of the Japanese encephalitis virus genome RNA. Virology 1987, 161, 497–510. [Google Scholar] [CrossRef]
- Lin, C.W.; Wu, S.C. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 2003, 77, 2600–2606. [Google Scholar] [CrossRef][Green Version]
- Chen, H.L.; Chang, J.K.; Tang, R.B. Current recommendations for the Japanese encephalitis vaccine. J. Chin. Med. Assoc. 2015, 78, 271–275. [Google Scholar] [CrossRef]
- Monath, T.P.; Arroyo, J.; Levenbook, I.; Zhang, Z.X.; Catalan, J.; Draper, K.; Guirakhoo, F. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: Relevance to development and safety testing of live, attenuated vaccines. J. Virol. 2002, 76, 1932–1943. [Google Scholar] [CrossRef]
- Saedi, T.A.; Moeini, H.; Tan, W.S.; Yusoff, K.; Daud, H.M.; Chu, K.B.; Tan, S.G.; Bhassu, S. Detection and phylogenetic profiling of nodavirus associated with white tail disease in Malaysian Macrobrachium rosenbergii de Man. Mol. Biol. Rep. 2012, 39, 5785–5790. [Google Scholar] [CrossRef]
- Goh, Z.H.; Tan, S.G.; Bhassu, S.; Tan, W.S. Virus-like particles of Macrobrachium rosenbergii nodavirus produced in bacteria. J. Virol. Methods 2011, 175, 74–79. [Google Scholar] [CrossRef]
- Citarasu, T.; Lelin, C.; Thirumalaikumar, E.; Michael Babu, M.; Vakharia, V.N. Macrobrachium rosenbergii nodavirus (MrNV)-CP-RNA-2 DNA vaccine confers protective immunity in giant freshwater prawn Macrobrachium rosenbergii against MrNV infection. Fish Shellfish Immunol. 2019, 86, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Jariyapong, P.; Chotwiwatthanakun, C.; Somrit, M.; Jitrapakdee, S.; Xing, L.; Cheng, H.R.; Weerachatyanukul, W. Encapsulation and delivery of plasmid DNA by virus-like nanoparticles engineered from Macrobrachium rosenbergii nodavirus. Virus Res. 2014, 179, 140–146. [Google Scholar] [CrossRef]
- Thong, Q.X.; Biabanikhankahdani, R.; Ho, K.L.; Alitheen, N.B.; Tan, W.S. Thermally-responsive virus-like particle for targeted delivery of cancer drug. Sci. Rep. 2019, 9, 3945. [Google Scholar] [CrossRef]
- Ninyio, N.N.; Ho, K.L.; Ong, H.K.; Yong, C.Y.; Chee, H.Y.; Hamid, M.; Tan, W.S. Immunological analysis of the hepatitis B virus “a” determinant displayed on chimeric virus-like particles of Macrobrachium rosenbergii nodavirus capsid protein produced in Sf9 cells. Vaccines 2020, 8, 275. [Google Scholar] [CrossRef] [PubMed]
- Ong, H.K.; Yong, C.Y.; Tan, W.S.; Yeap, S.K.; Omar, A.R.; Razak, M.A.; Ho, K.L. An influenza A vaccine based on the extracellular domain of matrix 2 protein protects BALB/c mice against H1N1 and H3N2. Vaccines 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Y.; Yeap, S.K.; Goh, Z.H.; Ho, K.L.; Omar, A.R.; Tan, W.S. Induction of humoral and cell-mediated immune responses by hepatitis B virus epitope displayed on the virus-like particles of prawn nodavirus. Appl. Environ. Microbiol. 2015, 81, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Yong, C.Y.; Yeap, S.K.; Ho, K.L.; Omar, A.R.; Tan, W.S. Potential recombinant vaccine against influenza A virus based on M2e displayed on nodaviral capsid nanoparticles. Int. J. Nanomed. 2015, 10, 2751–2763. [Google Scholar] [CrossRef]
- Ura, T.; Okuda, K.; Shimada, M. Developments in viral vector-based vaccines. Vaccines 2014, 2, 624–641. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Arshad, S.S.; Selvarajah, G.T.; Abu, J.; Toung, O.P.; Abba, Y.; Bande, F.; Yasmin, A.R.; Sharma, R.; Ong, B.L.; et al. Prevalence and risk factors of Japanese encephalitis virus (JEV) in livestock and companion animal in high-risk areas in Malaysia. Trop. Anim. Health Prod. 2018, 50, 741–752. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Andersen, M.M.; Ronne, T. Side-effects with Japanese encephalitis vaccine. Lancet 1991, 337, 1044. [Google Scholar] [CrossRef]
- Plesner, A.M.; Arlien-Soborg, P.; Herning, M. Neurological complications and Japanese encephalitis vaccination. Lancet 1996, 348, 202–203. [Google Scholar] [CrossRef]
- Sakaguchi, M.; Nakashima, K.; Takahashi, H.; Nakayama, T.; Fujita, H.; Inouye, S. Anaphylaxis to Japanese encephalitis vaccine. Allergy 2001, 56, 804–805. [Google Scholar] [CrossRef]
- Global Advisory Committee on Vaccine Safety, 11–12 December 2013. Wkly. Epidemiol. Rec. 2014, 89, 53–60.
- Ho, K.L.; Gabrielsen, M.; Beh, P.L.; Kueh, C.L.; Thong, Q.X.; Streetley, J.; Tan, W.S.; Bhella, D. Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae? PLoS Biol. 2018, 16, e3000038. [Google Scholar] [CrossRef]
- Ho, K.L.; Kueh, C.L.; Beh, P.L.; Tan, W.S.; Bhella, D. Cryo-Electron microscopy structure of the Macrobrachium rosenbergii nodavirus capsid at 7 angstroms resolution. Sci. Rep. 2017, 7, 2083. [Google Scholar] [CrossRef] [PubMed]
- Pabisch, S.; Feichtenschlager, B.; Kickelbick, G.; Peterlik, H. Effect of interparticle interactions on size determination of zirconia and silica based systems- A comparison of SAXS, DLS, BET, XRD and TEM. Chem. Phys. Lett. 2012, 521, 91–97. [Google Scholar] [CrossRef]
- Eaton, P.; Quaresma, P.; Soares, C.; Neves, C.; de Almeida, M.P.; Pereira, E.; West, P. A direct comparison of experimental methods to measure dimensions of synthetic nanoparticles. Ultramicroscopy 2017, 182, 179–190. [Google Scholar] [CrossRef]
- Ishizu, K.I.; Watanabe, H.; Han, S.I.; Kanesashi, S.N.; Hoque, M.; Yajima, H.; Kataoka, K.; Handa, H. Roles of disulfide linkage and calcium ion-mediated interactions in assembly and disassembly of virus-like particles composed of simian virus 40 VP1 capsid protein. J. Virol. 2001, 75, 61–72. [Google Scholar] [CrossRef]
- Li, P.P.; Nakanishi, A.; Clark, S.W.; Kasamatsu, H. Formation of transitory intrachain and interchain disulfide bonds accompanies the folding and oligomerization of simian virus 40 Vp1 in the cytoplasm. Proc. Natl. Acad. Sci. USA 2002, 99, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Li, P.P.; Nakanishi, A.; Fontanes, V.; Kasamatsu, H. Pairs of Vp1 cysteine residues essential for simian virus 40 infection. J. Virol. 2005, 79, 3859–3864. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Q.; Jiang, L.; Li, M.; Li, P.; Zhang, Q.; Song, R.; Xu, Z. Morphology and stability changes of recombinant TMV particles caused by a cysteine residue in the foreign peptide fused to the coat protein. J. Virol. Methods 2007, 140, 212–217. [Google Scholar] [CrossRef]
- Guillén, G.; Aguilar, J.C.; Dueñas, S.; Hermida, L.; Guzmán, M.G.; Penton, E.; Iglesias, E.; Junco, J.; Torrens, I.; Lobaina, Y.; et al. Virus-Like Particles as vaccine antigens and adjuvants: Application to chronic disease, cancer immunotherapy and infectious disease preventive strategies. Procedia Vaccinol. 2010, 2, 128–133. [Google Scholar] [CrossRef]
- Gilbert, S.C. Virus-like particles as vaccine adjuvants. Mol. Biotechnol. 2001, 19, 169–177. [Google Scholar] [CrossRef]
- Lowenadler, B.; Lycke, N.; Svanholm, C.; Svennerholm, A.M.; Krook, K.; Gidlund, M. T and B cell responses to chimeric proteins containing heterologous T helper epitopes inserted at different positions. Mol. Immunol. 1992, 29, 1185–1190. [Google Scholar] [CrossRef]
- Phares, T.W.; Stohlman, S.A.; Hwang, M.; Min, B.; Hinton, D.R.; Bergmann, C.C. CD4 T cells promote CD8 T cell immunity at the priming and effector site during viral encephalitis. J. Virol. 2012, 86, 2416–2427. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Oswal, N.; Chawla, A.S.; Agrawal, T.; Biswas, M.; Vrati, S.; Rath, S.; George, A.; Bal, V.; Medigeshi, G.R. CD8 T cells protect adult naive mice from JEV-induced morbidity via lytic function. PLoS Negl. Trop. Dis. 2017, 11, e0005329. [Google Scholar] [CrossRef]
- Wei, J.C.; Huang, Y.Z.; Zhong, D.K.; Kang, L.; Ishag, H.; Mao, X.; Cao, R.B.; Zhou, B.; Chen, P.Y. Design and evaluation of a multi-epitope peptide against Japanese encephalitis virus infection in BALB/c mice. Biochem. Biophys. Res. Commun. 2010, 396, 787–792. [Google Scholar] [CrossRef]
- Biswas, S.M.; Ayachit, V.M.; Sapkal, G.N.; Mahamuni, S.A.; Gore, M.M. Japanese encephalitis virus produces a CD4+ Th2 response and associated immunoprotection in an adoptive-transfer murine model. J. Gen. Virol. 2009, 90, 818–826. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Zhen, Z.D.; Fan, D.Y.; Wang, P.G.; An, J. Transcriptomic analysis suggests the M1 polarization and launch of diverse programmed cell death pathways in Japanese encephalitis virus-infected macrophages. Viruses 2020, 12, 356. [Google Scholar] [CrossRef] [PubMed]
- Sailaja, G.; Skountzou, I.; Quan, F.S.; Compans, R.W.; Kang, S.M. Human immunodeficiency virus-like particles activate multiple types of immune cells. Virology 2007, 362, 331–341. [Google Scholar] [CrossRef] [PubMed]
- Wagstaffe, H.R.; Mooney, J.P.; Riley, E.M.; Goodier, M.R. Vaccinating for natural killer cell effector functions. Clin. Transl. Immunol. 2018, 7, e1010. [Google Scholar] [CrossRef] [PubMed]
- Warfield, K.L.; Perkins, J.G.; Swenson, D.L.; Deal, E.M.; Bosio, C.M.; Aman, M.J.; Yokoyama, W.M.; Young, H.A.; Bavari, S. Role of natural killer cells in innate protection against lethal ebola virus infection. J. Exp. Med. 2004, 200, 169–179. [Google Scholar] [CrossRef]
- Brandstadter, J.D.; Yang, Y. Natural killer cell responses to viral infection. J. Innate Immun. 2011, 3, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Cevik, H.; Feldman, H.A.; Canaday, L.M.; Lakes, N.; Waggoner, S.N. Targeting natural killer cells to enhance vaccine responses. Trends Pharmacol. Sci. 2021, 42, 789–801. [Google Scholar] [CrossRef]
- Burke, D.S.; Morill, J.C. Levels of interferon in the plasma and cerebrospinal fluid of patients with acute Japanese encephalitis. J. Infect. Dis. 1987, 155, 797–799. [Google Scholar] [CrossRef]
- Ravi, V.; Parida, S.; Desai, A.; Chandramuki, A.; Gourie-Devi, M.; Grau, G.E. Correlation of tumor necrosis factor levels in the serum and cerebrospinal fluid with clinical outcome in Japanese encephalitis patients. J. Med. Virol. 1997, 51, 132–136. [Google Scholar] [CrossRef]
- Winter, P.M.; Dung, N.M.; Loan, H.T.; Kneen, R.; Wills, B.; Thu le, T.; House, D.; White, N.J.; Farrar, J.J.; Hart, C.A.; et al. Proinflammatory cytokines and chemokines in humans with Japanese encephalitis. J. Infect. Dis. 2004, 190, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S.; Winthrop, K.; Calabrese, L. The role of IL-6 in host defence against infections: Immunobiology and clinical implications. Nat. Rev. Rheumatol. 2017, 13, 399–409. [Google Scholar] [CrossRef]
- Kalinski, P.; Hilkens, C.M.; Snijders, A.; Snijdewint, F.G.; Kapsenberg, M.L. IL-12-deficient dendritic cells, generated in the presence of prostaglandin E2, promote type 2 cytokine production in maturing human naive T helper cells. J. Immunol. 1997, 159, 28–35. [Google Scholar] [PubMed]
- Tau, G.; Rothman, P. Biologic functions of the IFN-gamma receptors. Allergy 1999, 54, 1233–1251. [Google Scholar] [CrossRef]
- Spellberg, B.; Edwards, J.E., Jr. Type 1/Type 2 immunity in infectious diseases. Clin. Infect. Dis. 2001, 32, 76–102. [Google Scholar] [CrossRef] [PubMed]
- Swarup, V.; Ghosh, J.; Duseja, R.; Ghosh, S.; Basu, A. Japanese encephalitis virus infection decrease endogenous IL-10 production: Correlation with microglial activation and neuronal death. Neurosci. Lett. 2007, 420, 144–149. [Google Scholar] [CrossRef] [PubMed]





| Groups | Percentage of Cell Gated (%) | |
|---|---|---|
| CD3+CD4+ | CD3+CD8+ | |
| HEPES | 14.01 ± 0.41 a | 8.56 ± 0.15 v |
| IMOJEV | 16.80 ± 0.55 b | 8.70 ± 0.49 w |
| MrNV-CP | 17.03 ± 0.44 c | 10.11 ± 0.58 x |
| MrNV-CP with alum | 14.41 ± 0.40 d | 7.67 ± 0.15 y |
| MrNV-CPJEV-DIII | 18.20 ± 0.1 e | 11.00 ± 0.36 z |
| MrNV-CPJEV-DIII with alum | 17.70 ± 0.56 f | 11.06 ± 0.58 z |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, K.; Ong, H.K.; Tan, W.S.; Arshad, S.S.; Ho, K.L. Immunological Analysis of Nodavirus Capsid Displaying the Domain III of Japanese Encephalitis Virus Envelope Protein. Pharmaceutics 2021, 13, 1826. https://doi.org/10.3390/pharmaceutics13111826
Kumar K, Ong HK, Tan WS, Arshad SS, Ho KL. Immunological Analysis of Nodavirus Capsid Displaying the Domain III of Japanese Encephalitis Virus Envelope Protein. Pharmaceutics. 2021; 13(11):1826. https://doi.org/10.3390/pharmaceutics13111826
Chicago/Turabian StyleKumar, Kiven, Hui Kian Ong, Wen Siang Tan, Siti Suri Arshad, and Kok Lian Ho. 2021. "Immunological Analysis of Nodavirus Capsid Displaying the Domain III of Japanese Encephalitis Virus Envelope Protein" Pharmaceutics 13, no. 11: 1826. https://doi.org/10.3390/pharmaceutics13111826
APA StyleKumar, K., Ong, H. K., Tan, W. S., Arshad, S. S., & Ho, K. L. (2021). Immunological Analysis of Nodavirus Capsid Displaying the Domain III of Japanese Encephalitis Virus Envelope Protein. Pharmaceutics, 13(11), 1826. https://doi.org/10.3390/pharmaceutics13111826







