Thermostable and Long-Circulating Albumin-Conjugated Arthrobacter globiformis Urate Oxidase
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
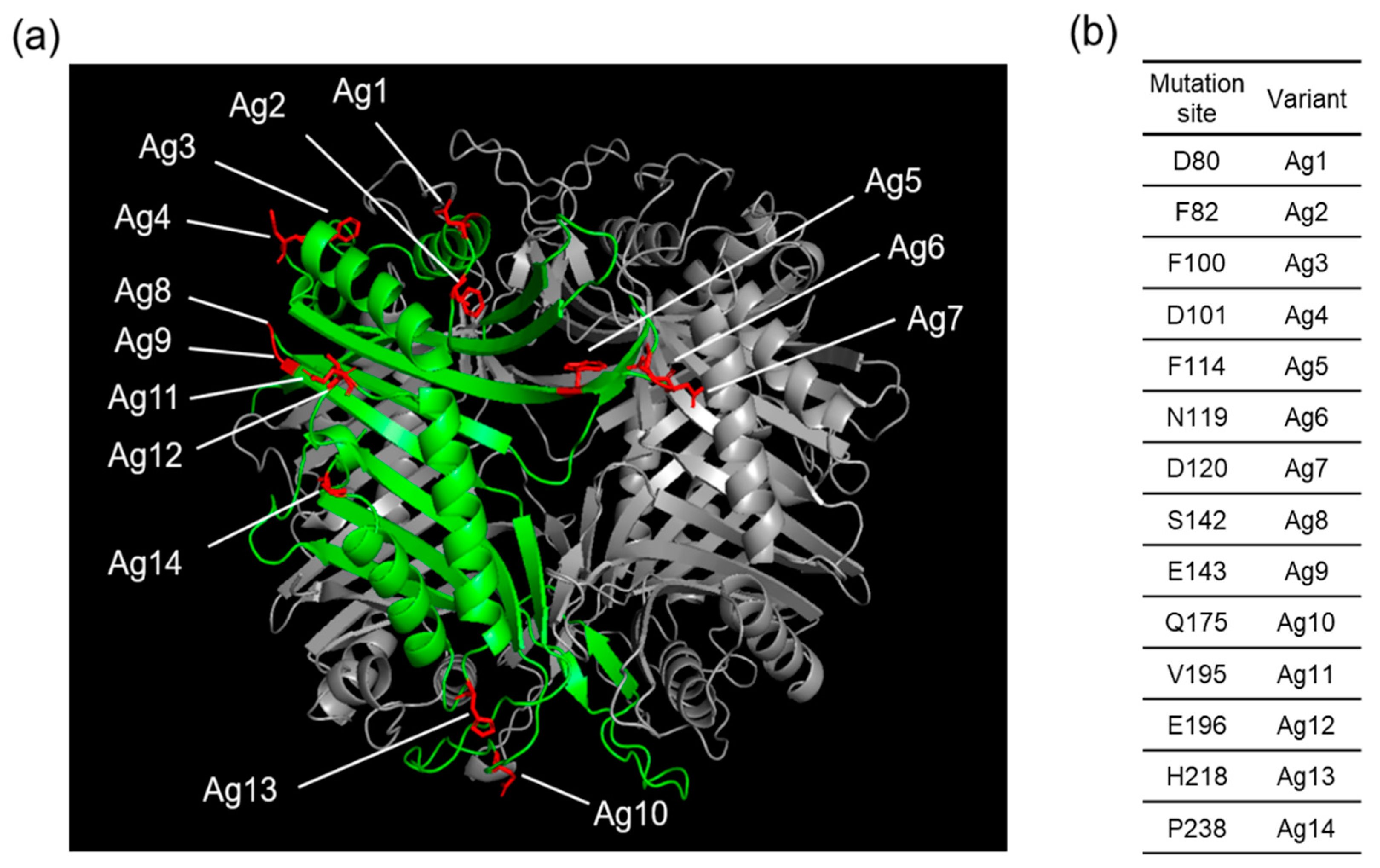
2.2. Computational Analysis of the frTet Incorporation Site in AgUox
2.3. Construction of Plasmids for Expression of AgUox-WT and AgUox-frTet Variants
2.4. Expression and Purification of AgUox-WT and AgUox-frTet Variants
2.5. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) and Dye Labeling Analysis of AgUox Variants
2.6. Enzymatic Activity Assay and Thermostability Assessment of AgUox-WT and AgUox-frTet Variants
2.7. Generation of HSA-Conjugated AgUox-frTet Variants
2.8. Pharmacokinetic Studies
3. Results and Discussion
3.1. Preparation of AgUox-WT and AgUox Containing frTet (AgUox-frTet) Variants
3.2. Enzymatic Activity and Thermostability Assays of AgUox Variants
3.3. Confirmation of the Site-Specific frTet Incorporation to AgUox
3.4. Site-Specific HSA-Conjugation to AgUox-frTet
3.5. Enzymatic Activity of the AgUox-HSA Conjugate
3.6. Pharmacokinetic Study of AgUox-WT and Ag12-HSA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mccarty, D.J.; Hollander, J.L. Identification of urate crystals in gouty synovial fluid. Ann. Intern. Med. 1961, 54, 452–460. [Google Scholar] [CrossRef]
- Harris, M.D.; Siegel, L.B.; Alloway, J.A. Gout and Hyperuricemia. Am. Fam. Physician 1999, 59, 925. [Google Scholar] [PubMed]
- Burns, C.M.; Wortmann, R.L. Gout therapeutics: New drugs for an old disease. Lancet 2011, 377, 165–177. [Google Scholar] [CrossRef]
- Nyborg, A.C.; Ward, C.; Zacco, A.; Chacko, B.; Grinberg, L.; Geoghegan, J.C.; Bean, R.; Wendeler, M.; Bartnik, F.; O’Connor, E.; et al. A therapeutic uricase with reduced immunogenicity risk and improved development properties. PLoS ONE 2016, 11, e0167935. [Google Scholar] [CrossRef] [PubMed]
- Ramazzina, I.; Folli, C.; Secchi, A.; Berni, R.; Percudani, R. Completing the uric acid degradation pathway through phylogenetic comparison of whole genomes. Nat. Chem. Biol. 2006, 2, 144–148. [Google Scholar] [CrossRef] [PubMed]
- Kahn, K.; Serfozo, P.; Tipton, P.A. Identification of the true product of the urate oxidase reaction. J. Am. Chem. Soc. 1997, 119, 5435–5442. [Google Scholar] [CrossRef]
- Iwata, H.; Nishio, S.; Yokoyama, M.; Matsumoto, A.; Takeuchi, M. Solubility of uric acid and supersaturation of monosodium urate: Why is uric acid so highly soluble in urine? J. Urol. 1989, 142, 1095–1098. [Google Scholar] [CrossRef]
- Tetko, I.V.; Tanchuk, V.Y.; Kasheva, T.N.; Villa, A.E. Estimation of aqueous solubility of chemical compounds using E-state indices. J. Chem. Inf. Comput. Sci. 2001, 41, 1488–1493. [Google Scholar] [CrossRef]
- Vogt, B. Urate oxidase (rasburicase) for treatment of severe tophaceous gout. Nephrol. Dial. Transplant. 2005, 20, 431–433. [Google Scholar] [CrossRef] [Green Version]
- Coiffier, B.; Mounier, N.; Bologna, S.; Fermé, C.; Tilly, H.; Sonet, A.; Christian, B.; Casasnovas, O.; Jourdan, E.; Belhadj, K.; et al. Efficacy and safety of rasburicase (recombinant urate oxidase) for the prevention and treatment of hyperuricemia during induction chemotherapy of aggressive non-Hodgkin’s lymphoma: Results of the GRAAL1 (Grouped’Etude des Lymphomes de l’Adulte Trial on Rasburicase Activity in Adult Lymphoma) study. J. Clin. Oncol. 2003, 21, 4402–4406. [Google Scholar] [CrossRef]
- Ryu, J.K.; Kim, H.S.; Nam, D.H. Current status and perspectives of biopharmaceutical drugs. Biotechnol. Bioprocess Eng. 2012, 17, 900–911. [Google Scholar] [CrossRef]
- Schlesinger, N.; Yasothan, U.; Kirkpatrick, P. Pegloticase. Nat. Rev. Drug Discov. 2011, 10, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Sundy, J.S.; Baraf, H.S.B.; Yood, R.A.; Edwards, N.L.; Gutierrez-Urena, S.R.; Treadwell, E.L.; Vázquez-Mellado, J.; White, W.B.; Lipsky, P.E.; Horowitz, Z.; et al. Efficacy and tolerability of pegloticase for the treatment of chronic gout in patients refractory to conventional treatment: Two randomized controlled trials. J. Am. Med. Assoc. 2011, 306, 711–720. [Google Scholar] [CrossRef] [Green Version]
- Sundy, J.S.; Becker, M.A.; Baraf, H.S.B.; Barkhuizen, A.; Moreland, L.W.; Huang, W.; Waltrip, R.W.; Maroli, A.N.; Horowitz, Z. Reduction of plasma urate levels following treatment with multiple doses of pegloticase (polyethylene glycol-conjugated uricase) in patients with treatment-failure gout: Results of a phase II randomized study. Arthritis Rheum. 2008, 58, 2882–2891. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Li, D.; Li, Y.; Shi, X.; Wang, J.; Rao, C.; Zhang, Y. Designing a mutant Candida uricase with improved polymerization state and enzymatic activity. Protein Eng. Des. Sel. 2017, 30, 753–759. [Google Scholar] [CrossRef]
- Kratz, F. Albumin as a drug carrier: Design of prodrugs, drug conjugates and nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef]
- Sleep, D. Albumin and its application in drug delivery. Expert Opin. Drug Deliv. 2015, 12, 793–812. [Google Scholar] [CrossRef]
- Bern, M.; Sand, K.M.K.; Nilsen, J.; Sandlie, I.; Andersen, J.T. The role of albumin receptors in regulation of albumin homeostasis: Implications for drug delivery. J. Control. Release 2015, 211, 144–162. [Google Scholar] [CrossRef]
- Curry, S.; Mandelkow, H.; Brick, P.; Franks, N. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat. Struct. Biol. 1998, 5, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Elsadek, B.; Kratz, F. Impact of albumin on drug delivery—New applications on the horizon. J. Control. Release 2012, 157, 4–28. [Google Scholar] [CrossRef]
- Lim, S.I.; Hahn, Y.S.; Kwon, I. Site-specific albumination of a therapeutic protein with multi-subunit to prolong activity in vivo. J. Control. Release 2015, 207, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Park, J.; Kim, S.; Kim, J.C.; Tae, G.; Jin, M.S.; Kwon, I. Intramolecular distance in the conjugate of urate oxidase and fatty acid governs FcRn binding and serum half-life in vivo. J. Control. Release 2020, 321, 49–58. [Google Scholar] [CrossRef]
- Deehan, M.; Garcês, S.; Kramer, D.; Baker, M.P.; Rat, D.; Roettger, Y.; Kromminga, A. Managing unwanted immunogenicity of biologicals. Autoimmun. Rev. 2015, 14, 569–574. [Google Scholar] [CrossRef]
- Suzuki, K.; Sakasegawa, S.I.; Misaki, H.; Sugiyama, M. Molecular cloning and expression of uricase gene from Arthrobacter globiformis in Escherichia coli and characterization of the gene product. J. Biosci. Bioeng. 2004, 98, 153–158. [Google Scholar] [CrossRef]
- Andersen, J.T.; Dalhus, B.; Viuff, D.; Ravn, B.T.; Gunnarsen, K.S.; Plumridge, A.; Bunting, K.; Antunes, F.; Williamson, R.; Athwal, S.; et al. Extending Serum Half-life of Albumin by Engineering Neonatal Fc Receptor (FcRn) Binding. J. Biol. Chem. 2014, 289, 13492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duttaroy, A.; Kanakaraj, P.; Osborn, B.L.; Schneider, H.; Pickeral, O.K.; Chen, C.; Zhang, G.; Kaithamana, S.; Singh, M.; Schulingkamp, R.; et al. Development of a Long-Acting Insulin Analog Using Albumin Fusion Technology. Diabetes 2005, 54, 251–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Wang, G.; Lang, L.; Jacobson, O.; Kiesewetter, D.O.; Liu, Y.; Ma, Y.; Zhang, X.; Wu, H.; Zhu, L.; et al. Chemical conjugation of evans blue derivative: A strategy to develop long-acting therapeutics through albumin binding. Theranostics 2016, 6, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, S.; Lyskov, S.; Gray, J.J. PyRosetta: A script-based interface for implementing molecular modeling algorithms using Rosetta. Bioinformatics 2010, 26, 689–691. [Google Scholar] [CrossRef]
- Alford, R.F.; Leaver-Fay, A.; Jeliazkov, J.R.; O’Meara, M.J.; DiMaio, F.P.; Park, H.; Shapovalov, M.V.; Renfrew, P.D.; Mulligan, V.K.; Kappel, K.; et al. The Rosetta All-Atom Energy Function for Macromolecular Modeling and Design. J. Chem. Theory Comput. 2017, 13, 3031–3048. [Google Scholar] [CrossRef]
- Yang, B.; Kwon, I. Multivalent albumin-FcRn interactions mediate a prominent extension of the serum half-life of a therapeutic protein. Mol. Pharm. 2021, 18, 2397–2405. [Google Scholar] [CrossRef]
- Yang, B.; Lim, S.I.; Kim, J.C.; Tae, G.; Kwon, I. Site-Specific Albumination as an Alternative to PEGylation for the Enhanced Serum Half-Life in Vivo. Biomacromolecules 2016, 17, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- O’Meara, M.J.; Leaver-Fay, A.; Tyka, M.D.; Stein, A.; Houlihan, K.; Dimaio, F.; Bradley, P.; Kortemme, T.; Baker, D.; Snoeyink, J.; et al. Combined covalent-electrostatic model of hydrogen bonding improves structure prediction with Rosetta. J. Chem. Theory Comput. 2015, 11, 609–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.; Bradley, P.; Greisen, P.; Liu, Y.; Mulligan, V.K.; Kim, D.E.; Baker, D.; Dimaio, F. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J. Chem. Theory Comput. 2016, 12, 6201–6212. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.-R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Kwon, K.; Jana, S.; Kim, S.; Avila-Crump, S.; Tae, G.; Mehl, R.A.; Kwon, I. Temporal control of efficient in vivo bioconjugation using a genetically encoded tetrazine-mediated inverse-electron-demand Diels-Alder reaction. Bioconjug. Chem. 2020, 31, 2456–2464. [Google Scholar] [CrossRef] [PubMed]
- Juan, E.C.M.; Hoque, M.M.; Shimizu, S.; Hossain, M.T.; Yamamoto, T.; Imamura, S.; Suzuki, K.; Tsunoda, M.; Amano, H.; Sekiguchi, T.; et al. Structures of Arthrobacterglobiformis urate oxidase-ligand complexes. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 815–822. [Google Scholar] [CrossRef]
- Schmidt, M.M.; Townson, S.A.; Andreucci, A.J.; King, B.M.; Schirmer, E.B.; Murillo, A.J.; Dombrowski, C.; Tisdale, A.W.; Lowden, P.A.; Masci, A.L.; et al. Crystal structure of an HSA/FcRn complex reveals recycling by competitive mimicry of HSA ligands at a pH-dependent hydrophobic interface. Structure 2013, 21, 1966–1978. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.J.; Holford, N.H.G. Mechanistic basis of using body size and maturation to predict clearance in humans. Drug Metab. Pharmacokinet. 2009, 24, 25–36. [Google Scholar] [CrossRef]
- Andersen, J.T.; Dalhus, B.; Cameron, J.; Daba, M.B.; Plumridge, A.; Evans, L.; Brennan, S.O.; Gunnarsen, K.S.; Bjørås, M.; Sleep, D.; et al. Structure-based mutagenesis reveals the albumin-binding site of the neonatal Fc receptor. Nat. Commun. 2012, 3, 610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahimizadeh, P.; Yang, S.; Lim, S.I. Albumin: An emerging opportunity in drug delivery. Biotechnol. Bioprocess Eng. 2020, 25, 985–995. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Kwon, I. Thermostable and Long-Circulating Albumin-Conjugated Arthrobacter globiformis Urate Oxidase. Pharmaceutics 2021, 13, 1298. https://doi.org/10.3390/pharmaceutics13081298
Yang B, Kwon I. Thermostable and Long-Circulating Albumin-Conjugated Arthrobacter globiformis Urate Oxidase. Pharmaceutics. 2021; 13(8):1298. https://doi.org/10.3390/pharmaceutics13081298
Chicago/Turabian StyleYang, Byungseop, and Inchan Kwon. 2021. "Thermostable and Long-Circulating Albumin-Conjugated Arthrobacter globiformis Urate Oxidase" Pharmaceutics 13, no. 8: 1298. https://doi.org/10.3390/pharmaceutics13081298
APA StyleYang, B., & Kwon, I. (2021). Thermostable and Long-Circulating Albumin-Conjugated Arthrobacter globiformis Urate Oxidase. Pharmaceutics, 13(8), 1298. https://doi.org/10.3390/pharmaceutics13081298





