Improving the Efficacy and Accessibility of Intracranial Viral Vector Delivery in Non-Human Primates
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Animal Procedures and MRI Analysis
2.2.1. Cortical Infusion
2.2.2. Thalamus Infusion
2.2.3. MTL Infusion
2.2.4. MRI Volume Extraction
2.3. Bench-Side Modeling
2.3.1. Cannula Production for Agar Infusions
2.3.2. Agar Phantom Infusion
2.3.3. Agar Phantom Image Processing
2.3.4. Color Component Selection
2.3.5. Threshold Value Selection
2.3.6. Statistical Methods
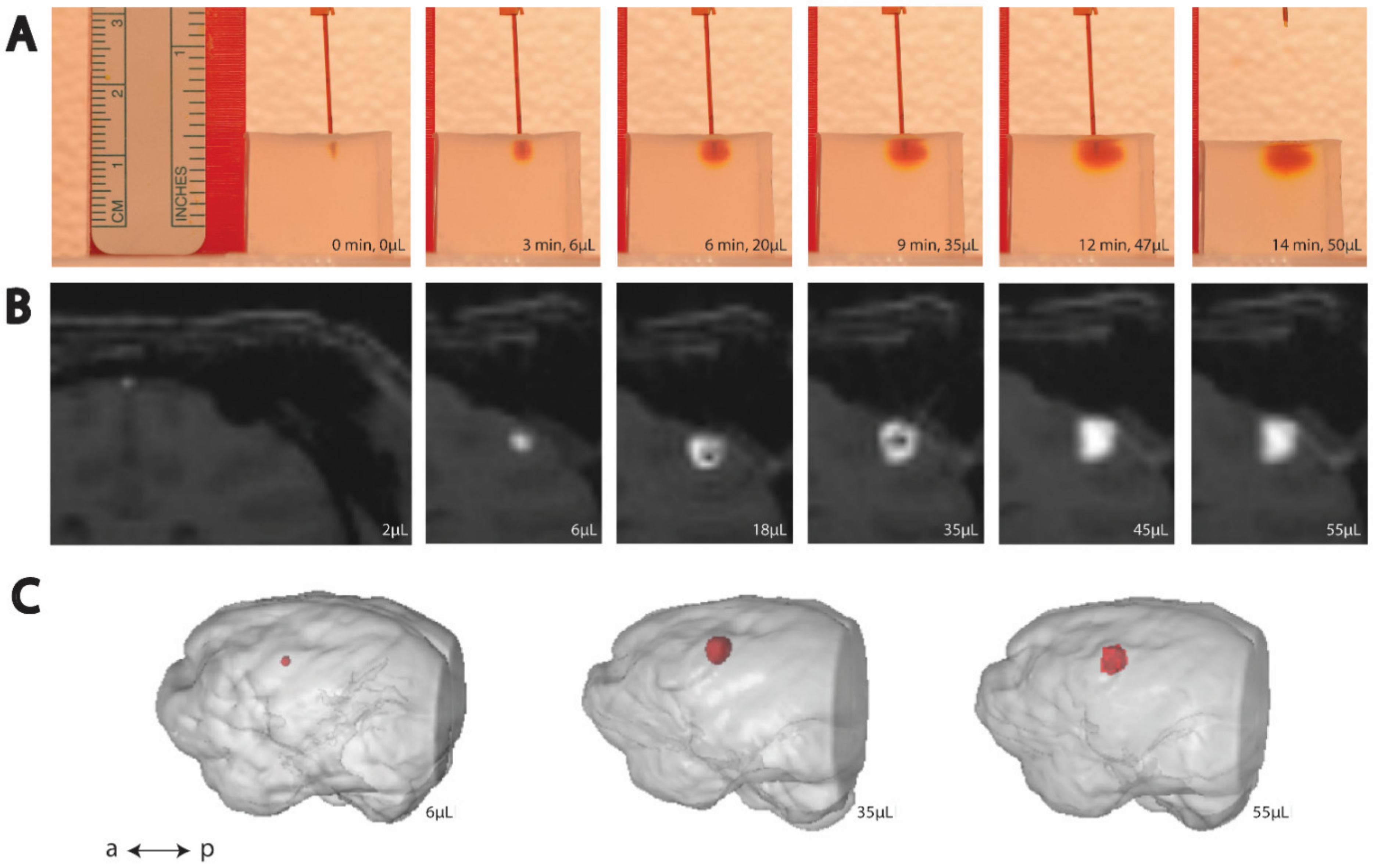
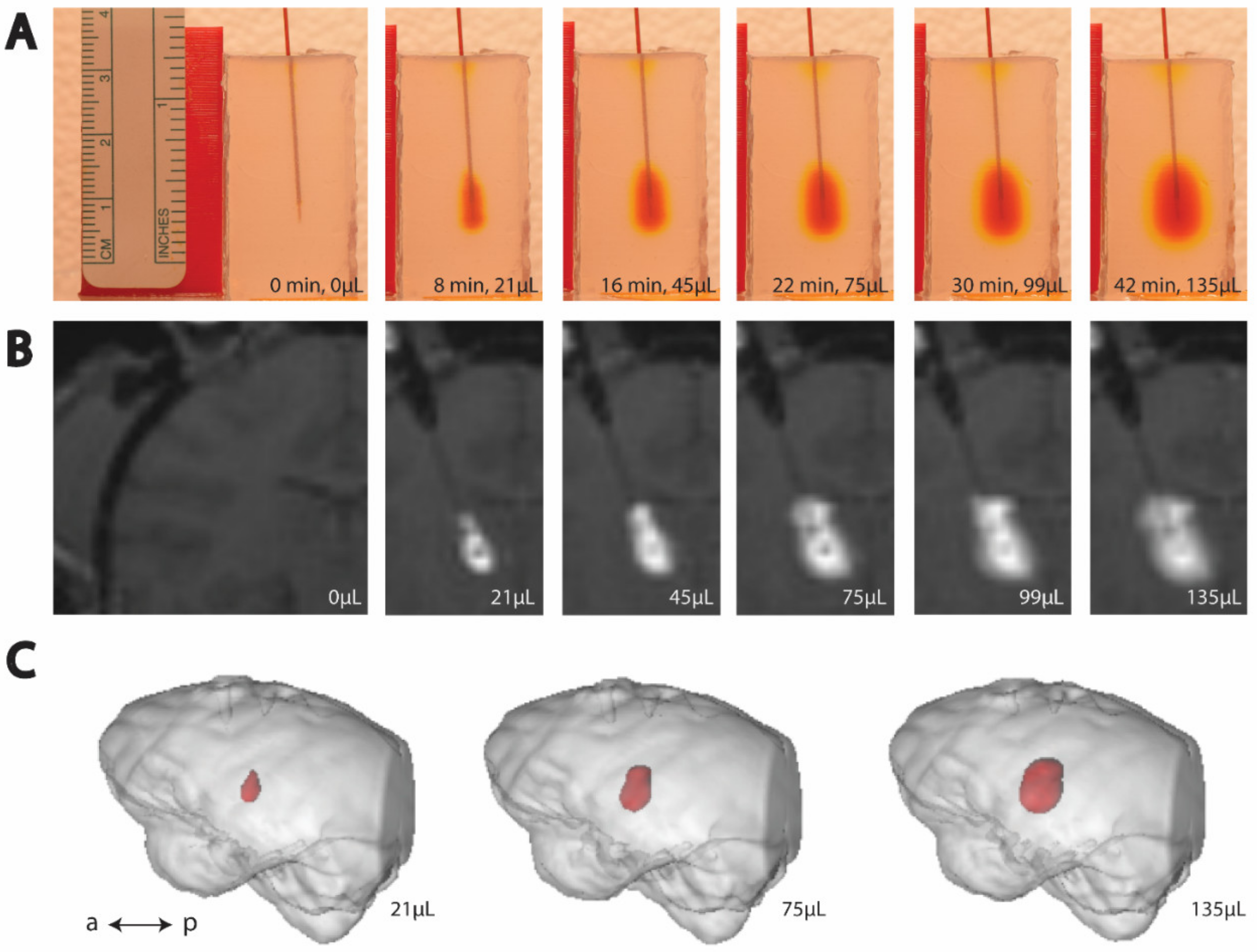
3. Results
3.1. Cortical and Thalamic Infusions
3.2. MTL Infusions
3.2.1. MTL Data Omitted from Agar Modeling
3.3. Histological Analysis
4. Discussion
4.1. Diversity of Modeled Structures
4.2. Insights from MTL Infusions
4.3. MRI Scan Parameters for Successful Contrast Label Visualization
4.4. Diffusion versus Convection Using Agar
4.5. Technical Considerations
4.6. Ethical Considerations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyden, E.S.; Zhang, F.; Bamberg, E.; Nagel, G.; Deisseroth, K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci. 2005, 8, 1263–1268. [Google Scholar] [CrossRef] [PubMed]
- Yizhar, O.; Fenno, L.E.; Davidson, T.J.; Mogri, M.; Deisseroth, K. Optogenetics in neural systems. Neuron 2011, 71, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Ting, J.T.; Feng, G. Recombineering strategies for developing next generation BAC transgenic tools for optogenetics and beyond. Front. Behav. Neurosci. 2014, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.M.; Sonabend, A.M.; Bruce, J.N. Convection-Enhanced Delivery. Neurotherapeutics 2017, 14, 358–371. [Google Scholar] [CrossRef]
- René, C.A.; Parks, R.J. Delivery of therapeutic agents to the central nervous system and the promise of extracellular vesicles. Pharmaceutics 2021, 13, 492. [Google Scholar] [CrossRef]
- Bobo, R.H.; Laske, D.W.; Akbasak, A.; Morrison, P.F.; Dedrick, R.L.; Oldfield, E.H. Convection-enhanced delivery of macromolecules in the brain. Proc. Natl. Acad. Sci. USA 1994, 91, 2076–2080. [Google Scholar] [CrossRef]
- Krauze, M.T.; Saito, R.; Noble, C.; Tamas, M.; Bringas, J.; Park, J.W.; Berger, M.S.; Bankiewicz, K. Reflux-free cannula for convection-enhanced high-speed delivery of therapeutic agents. J. Neurosurg. 2005, 103, 923–929. [Google Scholar] [CrossRef]
- Yazdan-Shahmorad, A.; Diaz-Botia, C.; Hanson, T.L.; Kharazia, V.; Ledochowitsch, P.; Maharbiz, M.M.; Sabes, P.N. “A Large-Scale Interface for Optogenetic Stimulation and Recording in Nonhuman Primates. Neuron 2016, 89, 927–939. [Google Scholar] [CrossRef]
- Yazdan-Shahmorad, A.; Tian, N.; Kharazia, V.; Samaranch, L.; Kells, A.; Bringas, J.; He, J.; Bankiewicz, K.; Sabes, P.N. Widespread optogenetic expression in macaque cortex obtained with MR-guided, convection enhanced delivery (CED) of AAV vector to the thalamus. J. Neurosci. Methods 2018, 293, 347–358. [Google Scholar] [CrossRef]
- Lieberman, D.M.; Laske, D.W.; Morrison, P.F.; Bankiewicz, K.S.; Oldfield, E.H. Convection-enhanced distribution of large molecules in gray matter during interstitial drug infusion. J. Neurosurg. 1995, 82, 1021–1029. [Google Scholar] [CrossRef]
- Lonser, R.R.; Gogate, N.; Morrison, P.F.; Wood, J.D.; Oldfield, E.H. Direct convective delivery of macromolecules to the spinal cord. J. Neurosurg. 1998, 89, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Lonser, R.R.; Walbridge, S.; Garmestani, K.; Butman, J.A.; Walters, H.A.; Vortmeyer, O.; Morrison, P.F.; Brechbiel, M.W.; Oldfield, E.H. Successful and safe perfusion of the primate brainstem: In vivo magnetic resonance imaging of macromolecular distribution during infusion. J. Neurosurg. 2002, 97, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Sanftner, L.M.; Sommer, J.M.; Suzuki, B.M.; Smith, P.H.; Vijay, S.; Vargas, J.A.; Forsayeth, J.R.; Cunningham, J.; Bankiewicz, K.S.; Kao, H.; et al. AAV2-mediated gene delivery to monkey putamen: Evaluation of an infusion device and delivery parameters. Exp. Neurol. 2005, 194, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Szerlip, N.J.; Walbridge, S.; Yang, L.; Morrison, P.F.; Degen, J.W.; Jarrell, S.T.; Kouri, J.; Kerr, P.B.; Kotin, R.; Oldfield, E.H.; et al. Real-time imaging of convection-enhanced delivery of viruses and virus-sized particles. J. Neurosurg. 2007, 107, 560–567. [Google Scholar] [CrossRef]
- Kells, A.P.; Hadaczek, P.; Yin, D.; Bringas, J.; Varenika, V.; Forsayeth, J.; Bankiewicz, K.S. Efficient gene therapy-based method for the delivery of therapeutics to primate cortex. Proc. Natl. Acad. Sci. USA 2009, 106, 2407–2411. [Google Scholar] [CrossRef]
- Yazdan-Shahmorad, A.; Diaz-Botia, C.; Hanson, T.; Ledochowitsch, P.; Maharabiz, M.M.; Sabes, P.N. Demonstration of a setup for chronic optogenetic stimulation and recording across cortical areas in non-human primates. SPIE BiOS 2015, 9305, 231–236. [Google Scholar] [CrossRef]
- Macknik, S.L.; Alexander, R.G.; Caballero, O.; Chanovas, J.; Nielsen, K.J.; Nishimura, N.; Schaffer, C.B.; Slovin, H.; Babayoff, A.; Barak, R.; et al. Advanced Circuit and Cellular Imaging Methods in Nonhuman Primates. J. Neurosci. 2019, 39, 8267–8274. [Google Scholar] [CrossRef]
- Khateeb, K.; Griggs, D.J.; Sabes, P.N.; Yazdan-Shahmorad, A. Convection Enhanced Delivery of Optogenetic Adeno-associated Viral Vector to the Cortex of Rhesus Macaque Under Guidance of Online MRI Images. J. Vis. Exp. 2019, 147, e59232. [Google Scholar] [CrossRef]
- Ojemann, W.K.; Griggs, D.J.; Ip, Z.; Caballero, O.; Jahanian, H.; Martinez-Conde, S.; Macknik, S.; Yazdan-Shahmorad, A. A mri-based toolbox for neurosurgical planning in nonhuman primates. J. Vis. Exp. 2020, 2020, e61098. [Google Scholar] [CrossRef]
- Chen, Z.J.; Gillies, G.T.; Broaddus, W.C.; Prabhu, S.S.; Fillmore, H.; Mitchell, R.M.; Corwin, F.D.; Fatouros, P.P. A realistic brain tissue phantom for intraparenchymal infusion studies. J. Neurosurg. 2004, 101, 314–322. [Google Scholar] [CrossRef]
- Pomfret, R.; Miranpuri, G.; Sillay, K. The substitute brain and the potential of the gel model. Ann. Neurosci. 2013, 20, 118–122. [Google Scholar] [CrossRef] [PubMed]
- Yazdan-Shahmorad, A.; Silversmith, D.B.; Kharazia, V.; Sabes, P.N. Targeted cortical reorganization using optogenetics in non-human primates. eLife 2018, 7, e31034. [Google Scholar] [CrossRef]
- Yazdan-Shahmorad, A.; Hanson, T.; Tian, N.; He, J.; Sabes, P. A novel technique for infusion of optogenetics viral vectors in nonhuman primates (NHPs) cortex using MR-guided convection enhanced delivery (CED). In Proceedings of the 6th International IEEE/EMBS Conference on Neural Engineering (NER), San Diego, CA, USA, 6–8 November 2013; pp. 5–8. [Google Scholar]
- Klapoetke, N.C.; Murata, Y.; Kim, S.S.; Pulver, S.R.; Birdsey-Benson, A.; Cho, Y.K.; Morimoto, T.K.; Chuong, A.S.; Carpenter, E.J.; Tian, Z.; et al. Independent optical excitation of distinct neural populations. Nat. Methods 2014, 11, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Mao, T.; Kusefoglu, D.; Hooks, B.M.; Huber, D.; Petreanu, L.; Svoboda, K. Long-Range Neuronal Circuits Underlying the Interaction between Sensory and Motor Cortex. Neuron 2011, 72, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Fredericks, J.M.; Dash, K.E.; Jaskot, E.M.; Bennett, T.W.; Lerchner, W.; Dold, G.; Ide, D.; Cummins, A.C.; Der Minassian, V.H.; Turchi, J.N.; et al. Methods for mechanical delivery of viral vectors into rhesus monkey brain. J. Neurosci. Methods 2020, 339, 108730. [Google Scholar] [CrossRef]
- Prezelski, K.; Keiser, M.; Stein, J.M.; Lucas, T.H.; Davidson, B.; Gonzalez-Alegre, P.; Vitale, F. Design and Validation of a Multi-Point Injection Technology for MR-Guided Convection Enhanced Delivery in the Brain. Front. Med. Technol. 2021, 3, 1–12. [Google Scholar] [CrossRef]
- Seunguk, O.H.; Odland, R.; Wilson, S.R.; Kroeger, K.M.; Liu, C.; Lowenstein, P.R.; Castro, M.G.; Hall, W.A.; Ohlfest, J.R. “Improved distribution of small molecules and viral vectors in the murine brain using a hollow fiber catheter. J. Neurosurg. 2007, 107, 568–577. [Google Scholar] [CrossRef]
- Tremblay, S.; Acker, L.; Afraz, A.; Albaugh, D.L.; Amita, H.; Andrei, A.R.; Angelucci, A.; Aschner, A.; Balan, P.F.; Basso, M.A.; et al. An Open Resource for Non-human Primate Optogenetics. Neuron 2020, 108, 1075–1090. [Google Scholar] [CrossRef]
- Belloir, T.; Montalgo Vargo, S.; Ahmed, Z.; Griggs, D.; Fisher, S.; Brown, T.; Chamanzar, M.; Yazdan-Shahmorad, A. Large-scale multimodal surface neural interfaces for non-human primates. iScience. under revision.
- Bloch, J.; Shea-brown, E.; Harchaoui, Z.; Shojai, A. E Network structure mediates functional reorganization induced by optogenetic stimulation of non-human primate sensorimotor cortex. iScience 2022, 25, 104285. [Google Scholar] [CrossRef]
- Griggs, D.J.; Belloir, T.; Yazdan-Shahmorad, A. Large-scale neural interfaces for optogenetic actuators and sensors in non-human primates. SPIE BiOS 2021, 1166305, 17. [Google Scholar] [CrossRef]
- Griggs, D.J.; Bloch, J.; Fisher, S.; Ojemann, W.K.S.; Coubrough, K.M.; Khateeb, K.; Chu, M.; Yazdan-Shahmorad, A. Demonstration of an Optimized Large-scale Optogenetic Cortical Interface for Non-human Primates. IEEE EMBC 2022, 119395, 7–15. [Google Scholar]
- Griggs, D.J.; Khateeb, K.; Philips, S.; Chan, J.W.; Ojemann, W.K.S.; Yazdan-Shahmorad, A. Optimized large-scale optogenetic interface for non-human primates. SPIE BiOS 2019, 1086605, 3. [Google Scholar] [CrossRef]
- Griggs, D.J.; Khateeb, K.; Zhou, J.; Liu, T.; Wang, R.; Yazdan-Shahmorad, A. Multi-modal artificial dura for simultaneous large-scale optical access and large-scale electrophysiology in non-human primate cortex. J. Neural Eng. 2021, 18, 055006. [Google Scholar] [CrossRef]
- Komatsu, M.; Sugano, E.; Tomita, H.; Fujii, N. A chronically implantable bidirectional neural interface for non-human primates. Front. Neurosci. 2017, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- Ledochowitsch, P.; Yazdan-Shahmorad, A.; Bouchard, K.E.; Diaz-Botia, C.; Hanson, T.L.; He, J.W.; Seybold, B.A.; Olivero, E.; Phillips, E.A.; Blanche, T.J.; et al. Strategies for optical control and simultaneous electrical readout of extended cortical circuits. J. Neurosci. Methods 2015, 256, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Rajalingham, R.; Sorenson, M.; Azadi, R.; Bohn, S.; DiCarlo, J.J.; Afraz, A. Chronically implantable LED arrays for behavioral optogenetics in primates. Nat. Methods 2021, 18, 1112–1116. [Google Scholar] [CrossRef]
- Yazdan-Shahmorad, A.; Silversmith, D.B.; Sabes, P.N. Novel techniques for large-scale manipulations of cortical networks in non-human primates. In Proceedings of the 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 17–21 July 2018; pp. 5479–5482. [Google Scholar] [CrossRef]
- Strange, B.A.; Witter, M.P.; Lein, E.S.; Moser, E.I. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 2014, 15, 655–669. [Google Scholar] [CrossRef]
- Zola, S.M.; Squire, L.R.; Teng, E.; Stefanacci, L.; Buffalo, E.A.; Clark, R.E. Impaired recognition memory in monkeys after damage limited to the hippocampal region. J. Neurosci. 2000, 20, 451–463. [Google Scholar] [CrossRef]
- Hampton, R.R.; Buckmaster, C.A.; Anuszkiewicz-Lundgren, D.; Murray, E.A. Method for making selective lesions of the hippocampus in Macaque monkeys using NMDA and a longitudinal surgical approach. Hippocampus 2004, 14, 9–18. [Google Scholar] [CrossRef]
- Simmons, J.M.; Saad, Z.S.; Lizak, M.J.; Ortiz, M.; Koretsky, A.P.; Richmond, B.J. Mapping prefrontal circuits in vivo with manganese-enhanced magnetic resonance imaging in monkeys. J. Neurosci. 2008, 28, 7637–7647. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Russell, W.; Burch, R. The Principles of Humane Experimental Technique; Methuen: London, UK, 1959. [Google Scholar]
- Prescott, M.J.; Poirier, C. The role of MRI in applying the 3Rs to non-human primate neuroscience. Neuroimage 2021, 225, 117521. [Google Scholar] [CrossRef] [PubMed]






| NHP Name | C1 | C2 | CT | T1 | T2 | MTL1 | MTL2 | MTL3 | MTL4 | MTL5 |
|---|---|---|---|---|---|---|---|---|---|---|
| Infusion Target | Cortex | Cortex, Thalamus | Thalamus | MTL (HPC+C) | MTL (EC) | MTL (HPC+C) | MTL (HPC) | |||
| NHP Variety | Rhesus Macaque | Pigtail Macaque | ||||||||
| Sex | M | M | M | F | F | F | F | F | M | M |
| Age (y) | 7 | 8 | 8 | 9 | 11 | 8 | 9 | 8 | 8 | 5 |
| Weight (kg) | 16.5 | 17.5 | 8.7 | 7.5 | 6.5 | 8.2 | 6.4 | 5.7 | 8.88 | 7.2 |
| Cannula Step Tip Length (mm) | 1 | 1 (Cortex) and 3 (Thalamus) | 3 | |||||||
| Left Hemisphere Infusions (µL) | 50, 50, 50, 50 | 50, 50, 50, 50 | 50 (Cortex) | 140, 120 | 115, 85 | 15 | 20 | 10, 10 | 15 | N/A |
| Right Hemisphere Infusions (µL) | - | - | 246 (Thalamus) | 152, 111 | - | 15 | 20 | 10, 10 | 15 | 20 |
| Contrast Agent | Gadoteridol | Manganese | ||||||||
| MRI Timing | Live | Live (2 infusions) | Live | Next-day | ||||||
| Institution | University of California, San Francisco | University of Washington, Seattle | ||||||||
| NHP Alias | Monkey J [8] | Monkey G [8] | NHP-H [9,23] | NHP-A [9] | NHP-B [9] | N/A | ||||
| Animal Name | C1 | C2 | CT | T1 | T2 | MTL1 | MTL2 | MTL3 | MTL4 | MTL5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hemisphere | Left | Right | Left | Right | Left | Left | Right | Left | Right | Left | Right | Left | Right | Left | Right | Right |
| Target | S1+M1 | S1 | MT | AT+PT | HPC | C | HPC | EC | HPC | C | HPC | |||||
| Vector Serotype | AAV5 | AAV2.9 | AAV2.2 | AAV2.9 | AAVRetrograde | PHP.eB | AAV2 | PHP.eB | ||||||||
| Vector | AAV-CamKIIa-C1V1-EYFP | AAV-CaMKII-ChR2(H134R)-YFP | AAV-CAG-hChR2-H134R-tdTomato | AAV-Syn-Chronos-GFP | AAV-CAG-hChR2-H134R-tdTomato | AAV-CAG-hChR2-H134R-tdTomato | AAV-3xhI56i(core)-minBG-ChR2(CRC)-EYFP-WPRE3-BGHpA | AAV-mDLX5/6-ChrimsonR-tdTomato | ||||||||
| Titer (vm/mL) | 2.5 × 1012 | 5.26 × 1012 | 1.02 × 1013 | 5.26 × 1012 | 7.6 × 1012 | 9 × 1012 | 7.6 × 1012 | 9 × 1012 | 3.25 × 1013 | 8.7 × 1012 | 3.55 × 1012 | |||||
| Cortical Infusion Rates (µL/min) | Duration (min) | Thalamic Infusion Rates (µL/min) | Duration (min) | MTL Infusion Rates (µL/min) | Duration (min) |
|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 |
| 2 | 1 | 2 | 1 | 2 | 1 |
| 3 | 1 | 3 | 80 | 3 | 3 |
| 4 | 1 | 2 | 1 | 2 | 1 |
| 5 | 6 | 1 | 1 | 1 | 1 |
| 4 | 1 | ||||
| 3 | 1 | ||||
| 2 | 1 | ||||
| 1 | 1 | ||||
| Total infused: 50 µL | Total time: 14 min | Total infused: 246 µL | Total time: 84 min | Total infused: 15 µL | Total time: 7 min |
| Cortical Infusion Slopes | |||
|---|---|---|---|
| NHP | MRI Slopes (µL/µL) | Trial | Gel Slopes (µL/µL) |
| C1 | 2.86 | 1 | 3.03 |
| C1 | 3.90 | 2 | 2.69 |
| C1 | 3.07 | 3 | 2.72 |
| C1 | 4.61 | 4 | 4.74 |
| C2 | 2.67 | 5 | 4.11 |
| C2 | 3.05 | 6 | 4.67 |
| CT | 3.07 | 7 | 4.23 |
| 8 | 3.10 | ||
| 9 | 4.81 | ||
| 10 | 3.82 | ||
| Thalamic Infusion Slopes | |||
| NHP | MRI Slopes (µL/µL) | Trial | Gel Slopes (µL/µL) |
| T1 | 3.48 | 1 | 2.63 |
| T2 | 3.59 | 2 | 3.71 |
| T2 | 3.30 | 3 | 4.02 |
| CT | 6.38 | 4 | 2.66 |
| 5 | 3.17 | ||
| NHP, Hemisphere | Infusion Volume (µL) | Measured Volume (µL) |
|---|---|---|
| MTL1, left | 15 | 221.1 |
| MTL1, right | 15 | 212.2 |
| MTL2, left | 20 | 285.3 |
| MTL3, right | 20 | 204.7 |
| Average: | 230.8 | |
| Gel Trial | Infusion Volume (µL) | Measured Volume (µL) |
| 1 | 15 | 196.7 |
| 2 | 15 | 229.2 |
| 3 | 15 | 260.2 |
| 4 | 15 | 220.7 |
| 5 | 15 | 207.0 |
| Average: | 222.8 | |
| % Error: | 3.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Griggs, D.J.; Garcia, A.D.; Au, W.Y.; Ojemann, W.K.S.; Johnson, A.G.; Ting, J.T.; Buffalo, E.A.; Yazdan-Shahmorad, A. Improving the Efficacy and Accessibility of Intracranial Viral Vector Delivery in Non-Human Primates. Pharmaceutics 2022, 14, 1435. https://doi.org/10.3390/pharmaceutics14071435
Griggs DJ, Garcia AD, Au WY, Ojemann WKS, Johnson AG, Ting JT, Buffalo EA, Yazdan-Shahmorad A. Improving the Efficacy and Accessibility of Intracranial Viral Vector Delivery in Non-Human Primates. Pharmaceutics. 2022; 14(7):1435. https://doi.org/10.3390/pharmaceutics14071435
Chicago/Turabian StyleGriggs, Devon J., Aaron D. Garcia, Wing Yun Au, William K. S. Ojemann, Andrew Graham Johnson, Jonathan T. Ting, Elizabeth A. Buffalo, and Azadeh Yazdan-Shahmorad. 2022. "Improving the Efficacy and Accessibility of Intracranial Viral Vector Delivery in Non-Human Primates" Pharmaceutics 14, no. 7: 1435. https://doi.org/10.3390/pharmaceutics14071435
APA StyleGriggs, D. J., Garcia, A. D., Au, W. Y., Ojemann, W. K. S., Johnson, A. G., Ting, J. T., Buffalo, E. A., & Yazdan-Shahmorad, A. (2022). Improving the Efficacy and Accessibility of Intracranial Viral Vector Delivery in Non-Human Primates. Pharmaceutics, 14(7), 1435. https://doi.org/10.3390/pharmaceutics14071435






