Synthesis, Molecular Docking, and Bioactivity Study of Novel Hybrid Benzimidazole Urea Derivatives: A Promising α-Amylase and α-Glucosidase Inhibitor Candidate with Antioxidant Activity
Abstract
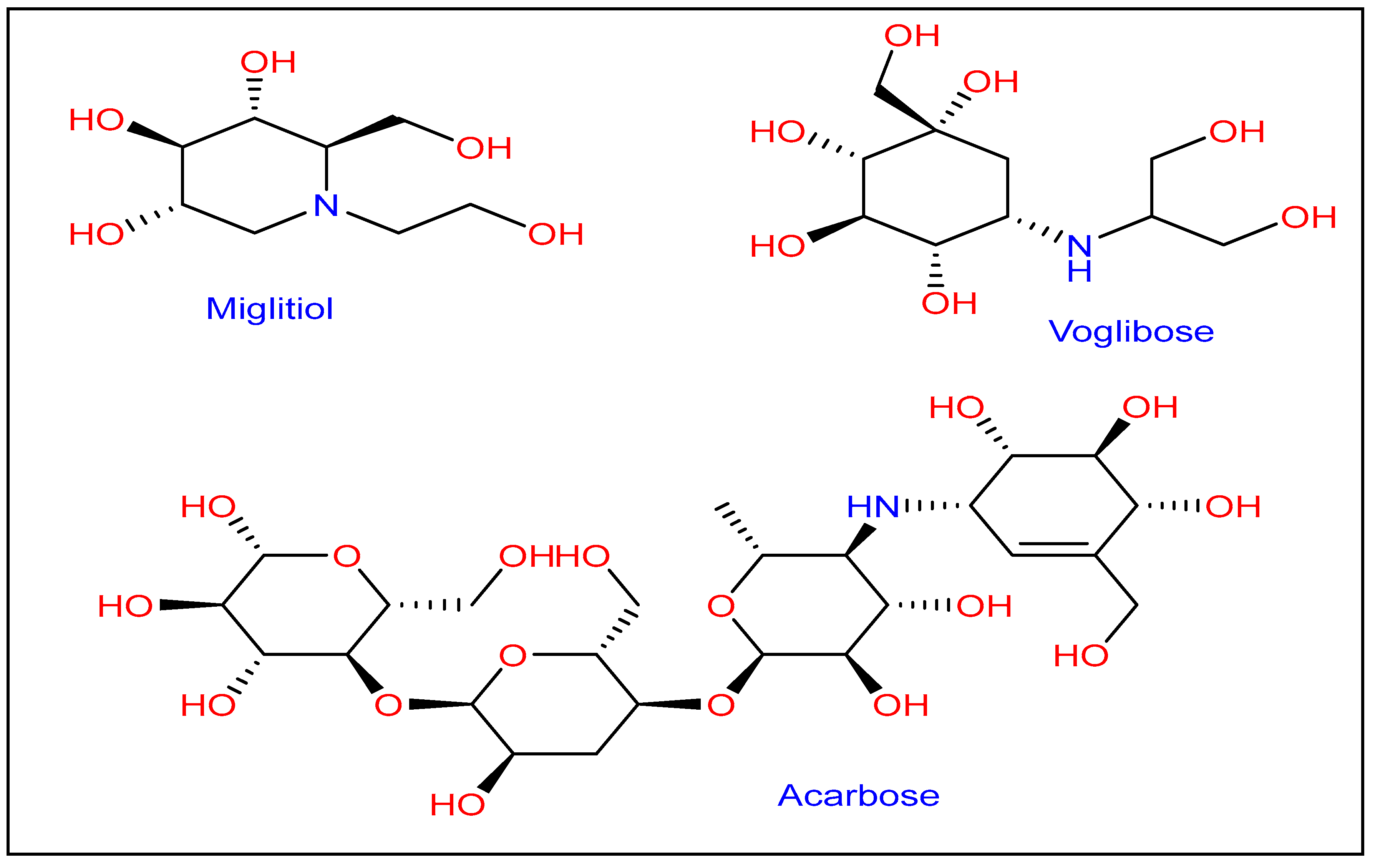
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. Biology
2.2.1. Antioxidant Activity
2.2.2. α-Amylase and α-Glucosidase Inhibition Activity
2.3. In Silico Docking Study
3. Conclusions
4. Experimental
4.1. General Details
4.2. Chemistry: Benzimidazole-Ureas 3a–h Synthesis: General Procedure
4.3. Biology
4.3.1. Antioxidant Study
Total Antioxidant Capacity (TAC)
DPPH Scavenging Activity (DPPH-SA)
Ferric Reducing Antioxidant Power (FRAP) Assay
Iron Chelating Activity Assay (MCA)
4.3.2. α-Amylase and α-Glucosidase Inhibition Assay
α-Amylase Inhibition Assay
α-Glucosidase Inhibition Assay
4.4. Statistical Analysis
4.5. Molecular Docking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shahidpour, S.; Panahi, F.; Yousefi, R.; Nourisefat, M.; Nabipoor, M.; Khalafi-Nezhad, A. Design and synthesis of new antidiabetic α-glucosidase and α-amylase inhibitors based on pyrimidine-fused heterocycles. Med. Chem. Res. 2015, 24, 3086–3096. [Google Scholar] [CrossRef]
- Cheng, H.C.; Chang, T.K.; Su, W.C.; Tsai, H.L.; Wang, J.Y. Narrative review of the influence of diabetes mellitus and hyperglycemia on colorectal cancer risk and oncological outcomes. Transl. Oncol. 2021, 14, 101089. [Google Scholar] [CrossRef]
- Ozougwu, J.; Obimba, K.; Belonwu, C.; Unakalamba, K. The pathogenesis and pathophysiology of type 1 and type 2 diabetes mellitus. J. Physiol. Pathophysiol. 2013, 4, 46–57. [Google Scholar] [CrossRef] [Green Version]
- Saeedi, P.; Petersohn, I.; Salpea, P.; Malanda, B.; Karuranga, S.; Unwin, N.; Colagiuri, S.; Guariguata, L.; Motala, A.A.; Ogurtsova, K.; et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2019, 157, 107843. [Google Scholar] [CrossRef] [Green Version]
- Al-Omar, M.S.; Mohammed, H.A.; Mohammed, S.A.A.; Abd-Elmoniem, E.; Kandil, Y.I.; Eldeeb, H.M.; Chigurupati, S.; Sulaiman, G.M.; Al-Khurayyif, H.K.; Almansour, B.S. Anti-Microbial, Anti-Oxidant, and α-Amylase Inhibitory Activity of Traditionally-Used Medicinal Herbs: A Comparative Analyses of Pharmacology, and Phytoconstituents of Regional Halophytic Plants’ Diaspora. Molecules 2020, 25, 5457. [Google Scholar] [CrossRef] [PubMed]
- Mani, V.; Arfeen, M.; Mohammed, H.A.; Elsisi, H.A.; Sajid, S.; Almogbel, Y.; Aldubayan, M.; Dhanasekaran, M.; Alhowail, A. Sukkari Dates Seed Improves Type-2 Diabetes Mellitus-Induced Memory Impairment by Reducing Blood Glucose Levels and Enhancing Brain Cholinergic Transmission: In Vivo and Molecular Modeling Studies. Saudi Pharm. J. 2022, 30, 750–763. [Google Scholar] [CrossRef]
- Asgari, M.S.; Mohammadi-Khanaposhtani, M.; Kiani, M.; Ranjbar, P.R.; Zabihi, E.; Pourbagher, R.; Larijani, B. Biscoumarin-1,2,3-triazole hybrids as novel anti-diabetic agents: Design, synthesis, in vitro α-glucosidase inhibition, kinetic, and docking studies. Bioorg. Chem. 2019, 92, 103206. [Google Scholar] [CrossRef]
- Piconi, L.; Quagliaro, L.; Ceriello, A. Oxidative stress in diabetes. Clin. Chem. Lab. Med. 2003, 41, 1144–1149. [Google Scholar] [CrossRef] [PubMed]
- Oberley, L.W. Free radicals and diabetes. Free Radic. Biol. Med. 1988, 5, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, Y.; Zhan, L.; Rayat, G.R.; Xiao, J.; Jiang, H.; Li, X.; Chen, K. Anti-diabetic effects of natural antioxidants from fruits. Trends Food Sci. Technol. 2021, 117, 3–14. [Google Scholar] [CrossRef]
- Amin, E.; Abdel-Bakky, M.S.; Darwish, M.A.; Mohammed, H.A.; Chigurupati, S.; Qureshi, K.A.; Hassan, M.H.A. The Glycemic Control Potential of Some Amaranthaceae Plants, with Particular Reference to In Vivo Antidiabetic Potential of Agathophora alopecuroides. Molecules 2022, 27, 973. [Google Scholar] [CrossRef] [PubMed]
- Yorek, M.A. The role of oxidative stress in diabetic vascular and neural disease. Free Radic. Res. 2003, 37, 471–480. [Google Scholar] [CrossRef]
- Morcoss, M.M.; El Shimaa, M.N.; Ibrahem, R.A.; Abdel-Rahman, H.M.; Abdel-Aziz, M.; Abou El-Ella, D.A. Design, Synthesis, Mechanistic Studies and In Silico ADME predictions of Benzimidazole derivatives as Novel Antifungal Agents. Bioorg. Chem. 2020, 22, 103956. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.M.; Patel, N.B.; Chan-Bacab, M.J.; Rivera, G. N-Mannich bases of benzimidazole as a potent antitubercular and antiprotozoal agents: Their synthesis and computational studies. Synth. Commun. 2020, 50, 858–878. [Google Scholar] [CrossRef]
- Onajole, O.K.; Lun, S.; Yun, Y.J.; Langue, D.Y.; Jaskula-Dybka, M.; Flores, A.; Frazier, E.; Scurry, A.C.; Zavala, A.; Arreola, K.R.; et al. Design, Synthesis and biological evaluation of novel imidazo [1,2-a] pyridinecarboxamides as potent anti-tuberculosis agents. Chem. Biol. Drug Des. 2020, 96, 1362–1371. [Google Scholar] [CrossRef]
- Ganie, A.M.; Dar, A.M.; Khan, F.A.; Dar, B.A. Benzimidazole derivatives as potential antimicrobial and antiulcer agents: A mini review. Mini Rev. Med. Chem. 2019, 19, 1292–1297. [Google Scholar] [CrossRef]
- Tonelli, M.; Gabriele, E.; Piazza, F.; Basilico, N.; Parapini, S.; Tasso, B.; Loddo, R.; Sparatore, F.; Sparatore, A. Benzimidazole derivatives endowed with potent antileishmanial activity. J. Enzym. Inhib. Med. Chem. 2018, 33, 210–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taha, M.; Mosaddik, A.; Rahim, F.; Ali, S.; Ibrahim, M.; Almandil, N.B. Synthesis, antiglycation and antioxidant potentials of benzimidazole derivatives. J. King Saud Univ. Sci. 2020, 32, 191–194. [Google Scholar] [CrossRef]
- Sirim, M.M.; Krishna, V.S.; Sriram, D.; Tan, O.U. Novel benzimidazole-acrylonitrile hybrids and their derivatives: Design, synthesis and antimycobacterial activity. Eur. J. Med. Chem. 2020, 188, 112010. [Google Scholar] [CrossRef]
- Pan, T.; He, X.; Chen, B.; Chen, H.; Geng, G.; Luo, H.; Zhang, H.; Bai, C. Development of benzimidazole derivatives to inhibit HIV-1 replication through protecting APOBEC3G protein. Eur. J. Med. Chem. 2015, 95, 500–513. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, C.; Tang, L.; Lu, K.; Zhao, H.; Wu, W.; Jiang, Y. Synthesis and biological evaluation of piperidyl benzimidazole carboxamide derivatives as potent PARP-1 inhibitors and antitumor agents. Chin. Chem. Lett. 2020, 31, 136–140. [Google Scholar] [CrossRef]
- Ashok, D.; Reddy, M.R.; Nagaraju, N.; Dharavath, R.; Ramakrishna, K.; Gundu, S.; Shravani, P.; Sarasija, M. Microwave-assisted synthesis and in vitro antiproliferative activity of some novel 1,2,3-triazole-based pyrazole aldehydes and their benzimidazole derivatives. Med. Chem. Res. 2020, 29, 699–706. [Google Scholar] [CrossRef]
- Sharma, S.; Kumar, D.; Singh, G.; Monga, V.; Kumar, B. Recent advancements in the development of heterocyclic anti-inflammatory agents. Eur. J. Med. Chem. 2020, 16, 112438. [Google Scholar] [CrossRef] [PubMed]
- Achar, K.C.; Hosamani, K.M.; Seetharamareddy, H.R. In-vivo analgesic and anti-inflammatory activities of newly synthesized benzimidazole derivatives. Eur. J. Med. Chem. 2010, 45, 2048–2054. [Google Scholar] [CrossRef] [PubMed]
- Francesconi, V.; Cichero, E.; Schenone, S.; Naesens, L.; Tonelli, M. Synthesis and Biological Evaluation of Novel (thio) semicarbazone-Based Benzimidazoles as Antiviral Agents against Human Respiratory Viruses. Molecules 2020, 25, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mavrova, A.T.; Anichina, K.K.; Vuchev, D.I.; Tsenov, J.A.; Denkova, P.S.; Kondeva, M.S.; Micheva, M.K. Antihelminthic activity of some newly synthesized 5 (6)-(un)substituted-1H-benzimidazol-2-ylthioacetylpiperazine derivatives. Eur. J. Med. Chem. 2006, 41, 1412–1420. [Google Scholar] [CrossRef] [PubMed]
- Hori, A.; Imaeda, Y.; Kubo, K.; Kusaka, M. Novel benzimidazole derivatives selectively inhibit endothelial cell growth and suppress angiogenesis in vitro and in vivo. Cancer Lett. 2002, 183, 53. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Mohsen, H.T.; Ragab, F.A.F.; Ramla, M.M.; El Diwani, H.I. Novel benzimidazole-pyrimidine conjugates as potent antitumor agents. Eur. J. Med. Chem. 2010, 45, 2336. [Google Scholar] [CrossRef]
- Velaparthi, U.; Liu, P.; Balasubramanian, B.; Carboni, J.; Attar, R.; Gottardis, M.; Li, A.; Greer, A.; Zoeckler, M.; Wittman, M.D.; et al. Imidazole moiety replacements in the 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-one inhibitors of insulin-like growth factor receptor-1 (IGF-1R) to improve cytochrome P450 profile. Med. Chem. Lett. 2007, 17, 3072. [Google Scholar] [CrossRef] [PubMed]
- Pagano, M.A.; Andrzejewska, M.; Ruzzene, M.; Sarno, S.; Cesaro, L.; Bain, J.; Elliott, M.; Meggio, F.; Kazimierczuk, Z.; Pinna, L.A. Optimization of protein kinase CK2 inhibitors derived from 4,5,6,7-tetrabromobenzimidazole. J. Med. Chem. 2004, 47, 6239. [Google Scholar] [CrossRef]
- Neff, D.K.; LeeDutra, A.; Blevitt, J.M.; Axe, F.U.; Hack, M.D.; Buma, J.C.; Rynberg, R.; Brunmark, A.; Karlsson, L.; Breitenbucher, G. 2-Aryl benzimidazoles featuring alkyl-linked pendant alcohols and amines as inhibitors of checkpoint kinase Chk2. Bioorg. Med. Chem. Lett. 2007, 17, 6467. [Google Scholar] [CrossRef]
- Arienti, K.L.; Brunmark, A.; Axe, F.U.; McClure, K.; Lee, A.; Blevitt, J.; Neff, D.K.; Huang, L.; Crawford, S.; Pandit, C.R.; et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J. Med. Chem. 2005, 48, 1873. [Google Scholar] [CrossRef] [PubMed]
- Hajduk, J.P.; Boyd, S.; Nettesheim, D.; Nienaber, V.; Severin, J.; Smith, R.; Davidson, D.; Rockway, T.; Fesik, S.W. Identification of novel inhibitors of urokinase via NMR-based screening. J. Med. Chem. 2000, 43, 3862–3866. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Taha, M.; Rahim, F.; Hayat, S.; Zaman, K.; Iqbal, N.; Selvaraj, M.; Sajid, M.; Bangesh, M.A.; Khan, F.; et al. Synthesis of benzimidazole derivatives as potent inhibitors for α-amylase and their molecular docking study in management of type-II diabetes. J. Mol. Struct. 2021, 1232, 130029. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, S. Recent Advances in Synthetic α-Glucosidase Inhibitors. ChemMedChem 2017, 12, 819–829. [Google Scholar] [CrossRef]
- Ullah, H.; Ullah, H.; Taha, M.; Khan, F.; Rahim, F.; Uddin, I.; Sarfraz, M.; Ali Shah, S.A.; Aziz, A.; Mubeen, S. Synthesis, In Vitro α-Amylase Activity, and Molecular Docking Study of New Benzimidazole Derivatives. Russ. J. Org. Chem. 2021, 57, 968–975. [Google Scholar] [CrossRef]
- Cakmak, U.; Oz-Tuncay, F.; Basoglu-Ozdemir, S.; Ayazoglu-Demir, E.; Demir, I.; Colak, A.; Celik-Uzuner, S.; Erdem, S.S.; Yildirim, N. Synthesis of hydrazine containing piperazine or benzimidazole derivatives and their potential as α-amylase inhibitors by molecular docking, inhibition kinetics and in vitro cytotoxicity activity studies. Med. Chem. Res. 2021, 30, 1886–1904. [Google Scholar] [CrossRef]
- Khan, S.; Ullah, H.; Rahim, F.; Nawaz, M.; Hussain, R.; Rasheed, L. Synthesis, in vitro α-Amylase, α-Glucosidase Activities and Molecular Docking Study of New Benzimidazole Bearing Thiazolidinone Derivatives. J. Mol. Struct. 2022, 1269, 133812. [Google Scholar] [CrossRef]
- Taha, M.; Imran, S.; Ismail, N.H.; Selvaraj, M.; Rahim, F.; Chigurupati, S.; Ullah, H.; Khan, F.; Salar, U.; Javid, M.T.; et al. Biology-oriented drug synthesis (BIODS) of 2-(2-methyl-5-nitro-1H-imidazol-1-yl) ethyl aryl ether derivatives, in vitro α-amylase inhibitory activity and in silico studies. Bioorg. Chem. 2017, 74, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Ling, N.; Che, Q.T.; Zhang, Y.W.; Yang, H.X.; Ruan, Y.; Zhao, T.T. Synthesis, structure and biological properties of benzimidazole-based Cu (II)/Zn (II) complexes. Inorg. Chem. Commun. 2019, 105, 97–101. [Google Scholar] [CrossRef]
- Sivaramakarthikeyan, R.; Iniyaval, S.; Saravanan, V.; Lim, W.M.; Mai, C.W.; Ramalingan, C. Molecular Hybrids Integrated with Benzimidazole and Pyrazole Structural Motifs: Design, Synthesis, Biological Evaluation, and Molecular Docking Studies. ACS Omega 2020, 5, 10089–10098. [Google Scholar] [CrossRef]
- Arshad, T.; Khan, K.M.; Rasool, N.; Salar, U.; Hussain, S.; Asghar, H.; Ashraf, M.; Wadood, A.; Riaz, M.; Perveen, S.; et al. 5-Bromo-2-aryl benzimidazole derivatives as non-cytotoxic potential dual inhibitors of α-glucosidase and urease enzymes. Bioorg. Chem. 2017, 72, 21–31. [Google Scholar] [CrossRef]
- Ozil, M.; Parlak, C.; Baltas, N.A. Simple and efficient synthesis of benzimidazoles containing piperazine or morpholine skeleton at C-6 position as glucosidase inhibitors with antioxidant activity. Bioorg. Chem. 2018, 6, 468–477. [Google Scholar] [CrossRef] [PubMed]
- Taha, M.; Rahim, F.; Zaman, K.; Selvaraj, M.; Uddin, N.; Farooq, R.K.; Nawaz, M.; Sajid, M.; Nawaz, F.; Ibrahim, M.; et al. Synthesis, α-glycosidase inhibitory potential and molecular docking study of benzimidazole derivatives. Bioorg. Chem. 2020, 95, 103555. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Singh, A.; Singh, V.; Verma, R.K.; Tomar, J.; Mall, R. Synthesis, molecular docking, α-glucosidase inhibition, and antioxidant activity studies of novel benzimidazole derivatives. Med. Chem. Res. 2020, 29, 1846–1866. [Google Scholar] [CrossRef]
- Rahim, F.; Zaman, K.; Taha, M.; Ullah, H.; Ghufran, M.; Wadood, A.; Rehman, W.; Uddin, N.; Shah, S.A.A.; Sajid, M.; et al. Synthesis, in vitro alphaglucosidase inhibitory potential of benzimidazole bearing bis-Schiff bases and their molecular docking study. Bioorg. Chem. 2020, 94, 103394. [Google Scholar] [CrossRef]
- Asemanipoor, N.; Mohammadi-Khanaposhtani, M.; Moradi, S.; Vahidi, M.; Asadi, M.; Faramarzi, M.A.; Mahdavi, M.; Biglar, M.; Larijani, B.; Hamedifar, H.; et al. Synthesis and biological evaluation of new benzimidazole-1,2,3-triazole hybrids as potential α-glucosidase inhibitors. Bioorg. Chem. 2020, 95, 103482. [Google Scholar] [CrossRef]
- Bharadwaj, S.S.; Poojary, B.; Nandish, S.K.M.; Kengaiah, J.; Kirana, M.P.; Shankar, M.K.; Das, A.J.; Kulal, A.; Sannaningaiah, D. Efficient synthesis and in silico studies of the benzimidazole hybrid scaffold with the quinolinyloxadiazole skeleton with potential α-glucosidase inhibitory, anticoagulant, and antiplatelet activities for type-II diabetes mellitus management and treating thrombotic disorders. ACS Omega 2018, 3, 12562–12574. [Google Scholar] [CrossRef]
- Zawawi, N.K.N.A.; Taha, M.; Ahmat, N.; Ismail, N.H.; Wadood, A.; Rahim, F. Synthesis, molecular docking studies of hybrid benzimidazole as α-glucosidase inhibitor. Bioorg. Chem. 2017, 70, 184–191. [Google Scholar] [CrossRef]
- Menteşe, E.; Baltas, N.; Emirik, M. Synthesis, α-Glucosidase Inhibition and in Silico Studies of Some 4-(5-Fluoro-2-substituted-1H-benzimidazol-6-yl) morpholine Derivatives. Bioorg. Chem. 2020, 101, 104002. [Google Scholar] [CrossRef]
- El Bakri, Y.; Anouar, E.H.; Marmouzi, I.; Sayah, K.; Ramli, Y.; El Abbes Faouzi, M.; Essassi, E.M.; Mague, J.T. Potential antidiabetic activity and molecular docking studies of novel synthesized benzimidazole and 10-amino-2-methyl-4-oxo pyrimido [1,2-a] benzimidazole derivatives. J. Mol. Model. 2018, 24, 1–10. [Google Scholar] [CrossRef]
- Aroua, L.M.; Almuhaylan, H.R.; Alminderej, F.M.; Messaoudi, S.; Chigurupati, S.; Al-Mahmoud, S.; Mohammed, H.A. A facile approach synthesis of benzoylaryl benzimidazole as potential α-amylase and α-glucosidase inhibitor with antioxidant activity. Bioorg. Chem. 2021, 114, 105073. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Singh, A.; Verma, R.K.; Mall, R.; Azeem, U. Synthesis, biological evaluation and molecular docking studies of novel benzimidazole derivatives. Comput. Biol. Chem. 2018, 72, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Deswal, L.; Verma, V.; Kumar, D.; Kaushik, C.P.; Kumar, A.; Deswal, Y.; Punia, S. Synthesis and antidiabetic evaluation of benzimidazole-tethered 1,2,3-triazoles. Arch. Pharm. 2020, 353, 2000090. [Google Scholar] [CrossRef] [PubMed]
- Gündüz, M.G.; Ugur, S.B.; Güney, F.; Özkul, C.; Krishna, V.S.; Kaya, S.; Sriramd, D.; Dogan, S.D. 1,3-Disubstituted urea derivatives: Synthesis, antimicrobial activity evaluation and in silico studies. Bioorg. Chem. 2020, 102, 104104. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Brindisi, M. Urea derivatives in modern drug discovery and medicinal chemistry. J. Med. Chem. 2019, 63, 2751–2788. [Google Scholar] [CrossRef]
- Mishra, C.B.; Mongre, R.K.; Kumari, S.; Jeong, D.K.; Tiwari, M. Synthesis, in vitro and in vivo anticancer activity of novel 1-(4-imino-1-substituted-1 H-pyrazolo [3,4-d] pyrimidin-5-(4 H)-yl) urea derivatives. RSC Adv. 2016, 6, 24491–24500. [Google Scholar] [CrossRef]
- Kilic-Kurt, Z.; Ozmen, N.; Bakar-Ates, F. Synthesis and anticancer activity of some pyrimidine derivatives with aryl urea moieties as apoptosis-inducing agents. Bioorg. Chem. 2020, 101, 104028. [Google Scholar] [CrossRef]
- Ma, L.Y.; Wang, B.; Pang, L.P.; Zhang, M.; Wang, S.Q.; Zheng, Y.C.; Shao, K.P.; Xue, D.Q.; Liu, H.M. Design and synthesis of novel 1,2,3-triazole-pyrimidine-urea hybrids as potential anticancer agents. Bioorg. Med. Chem. Lett. 2015, 25, 1124–1128. [Google Scholar] [CrossRef]
- Faraji, A.; Motahari, R.; Hasanvand, Z.; Bakhshaiesh, T.O.; Toolabi, M.; Moghimi, S.; Firoozpour, L.; Boshagh, M.A.; Rahmani, R.; Ketabforoosh, S.H.M.E.; et al. Quinazolin-4(3H)-one based agents bearing thiadiazole-urea: Synthesis and evaluation of anti-proliferative and antiangiogenic activity. Bioorg. Chem. 2021, 108, 104553. [Google Scholar] [CrossRef]
- Xie, X.X.; Li, H.; Wang, J.; Mao, S.; Xin, M.H.; Lu, S.M.; Mei, Q.B.; Zhang, S.Q. Synthesis and anticancer effects evaluation of 1-alkyl-3-(6-(2-methoxy-3-sulfonylaminopyridin-5-yl) benzo[d]thiazol-2-yl) urea as anticancer agents with low toxicity. Bioorg. Med. Chem. 2015, 23, 6477–6485. [Google Scholar] [CrossRef]
- Ture, A.; Kahraman, D.C.; Cetin-Atalay, R.; Helvacioglu, S.; Charehsaz, M.; Küçükgüzel, I. Synthesis, anticancer activity, toxicity evaluation and molecular docking studies of novel phenylaminopyrimidine-(thio) urea hybrids as potential kinase inhibitors. Comput. Biol. Chem. 2019, 78, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.N.; Wang, X.F.; Li, T.; Wu, D.W.; Fu, X.B.; Zhang, G.J.; Shen, X.C.; Wang, H.S. Design, synthesis, and biological evaluation of novel quinazolinyl-diaryl urea derivatives as potential anticancer agents. Eur. J. Med. Chem. 2016, 107, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Kurt, B.Z.; Kandas, N.O.; Dag, A.; Sonmez, F.; Kucukislamoglu, M. Synthesis and biological evaluation of novel coumarin-chalcone derivatives containing urea moiety as potential anticancer agents. Arab. J. Chem. 2020, 13, 1120–1129. [Google Scholar] [CrossRef]
- Zarei, O.; Azimian, F.; Hamzeh-Mivehroud, M.; Mojarrad, J.S.; Hemmati, S.; Dastmalchi, S. Design, synthesis, and biological evaluation of novel benzo[b]thiophene-diaryl urea derivatives as potential anticancer agents. Med. Chem. Res. 2020, 29, 1438–1448. [Google Scholar] [CrossRef]
- Aroua, L.M.; Al-Hakimi, A.N.; Abdulghani, M.A.M.; Alhag, S.K. Cytotoxic urea Schiff base complexes for multidrug discovery as anticancer activity and low in vivo oral assessing toxicity. Arab. J. Chem. 2022, 15, 103986. [Google Scholar] [CrossRef]
- Al-Hakimi, A.N.; Alminderej, F.; Aroua, L.; Alhag, S.K.; Alfaifi, M.Y.; Mahyoub, J.A.; Elbehairi, S.I.; Alnafisah, A.S. Design, synthesis, characterization of zirconium (IV), cadmium (II) and iron (III) complexes derived from Schiff base 2-aminomethylbenzimidazole, 2-hydroxynaphtadehyde and evaluation of their biological activity. Arab. J. Chem. 2020, 13, 7378–7389. [Google Scholar] [CrossRef]
- Alminderej, F.M.; Aroua, L. Design, Synthesis, Characterization and Anticancer Evaluation of Novel Mixed Complexes Derived from 2-(1H-Benzimizadol-2-yl) aniline Schiff base and 2-Mercaptobenzimidazole or 2-Aminobenzothiazole. Egypt. J. Chem. 2021, 64, 3351–3364. [Google Scholar] [CrossRef]
- Aroua, L.M.; Al-Hakimi, A.N.; Abdulghani, M.A.M.; Alhag, S.K. Elaboration of novel urea bearing schiff bases as potent in vitro anticancer candidates with low in vivo acute oral toxicity. Main Group Met. Chem. 2022, 21, 953–973. [Google Scholar]
- Ghrab, S.; Aroua, L.; Mekni, N.; Beji, M. Simple approach for the regioselective synthesis of a bis (β-aminoalcohol) derived from polyoxyethylene: First report of fast ring-opening of polyoxyethylene diglycidyl ethers with sodium amide. Res. Chem. Intermed. 2018, 44, 3537–3548. [Google Scholar] [CrossRef]
- Ghrab, S.; Aroua, L.; Beji, M. One-pot Three Component Synthesis of ω-(oxathiolan-2-thion-5-yl)-α-oxazolidin-2-ones. J. Heterocycl. Chem. 2017, 54, 2397–2404. [Google Scholar] [CrossRef]
- Khan, I.A.; Saddique, F.A.; Aslam, S.; Ashfaq, U.A.; Ahmad, M.; Al-Hussain, S.A.; Zaki, M.E.A. Synthesis of Novel N-Methylmorpholine-Substituted Benzimidazolium Salts as Potential α-Glucosidase Inhibitors. Molecules 2022, 27, 6012. [Google Scholar] [CrossRef] [PubMed]
- Ramasubbu, N.; Paloth, V.; Luo, Y.; Brayer, G.D.; Levine, M.J. Structure of human salivary α-amylase at 1.6 Å resolution: Implications for its role in the oral cavity. Acta Crystallogr. Sect. 1996, D52, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.K.; Li, C.; Withers, S.G.; Brayer, G.D. Order and Disorder: Differential structural impacts of myricetin and ethyl caffeate on human amylase, an antidiabetic target. J. Med. Chem. 2012, 55, 10177–10186. [Google Scholar] [PubMed]
- Roig-Zamboni, V.; Cobucci-Ponzano, B.; Iacono, R.; Ferrara, M.C.; Germany, S.; Bourne, Y.; Parenti, G.; Moracci, M.; Sulzenbacher, G. Structure of human lysosomal acid α-glucosidase–a guide for the treatment of Pompe disease. Nat. Commun. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, H.A.; Al-Omar, M.S.; Khan, R.A.; Mohammed, S.A.A.; Qureshi, K.A.; Abbas, M.M.; Al Rugaie, O.; Abd-Elmoniem, E.; Ahmad, A.M.; Kandil, Y.I. Chemical Profile, Antioxidant, Antimicrobial, and Anticancer Activities of the Water-Ethanol Extract of Pulicaria Undulata Growing in the Oasis of Central Saudi Arabian Desert. Plants 2021, 10, 1811. [Google Scholar] [CrossRef]
- Benzie, I.F.F.; Strain, J.J. The ferric reducing ability of plasma (FRAP) as a measure of “antioxidant power”: The FRAP assay. Anal. Biochem. 1996, 239, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, H.A. Phytochemical Analysis, Antioxidant Potential, and Cytotoxicity Evaluation of Traditionally Used Artemisia Absinthium L. (Wormwood) Growing in the Central Region of Saudi Arabia. Plants 2022, 11, 1028. [Google Scholar] [CrossRef]
- Martin, A.E.; Montgomery, P.A. Acarbose: An Alpha-Glucosidase Inhibitor. Am. J. Health-Syst. Pharm. 1996, 53, 2277–2290. [Google Scholar] [CrossRef]
- Yousuf, S.; Khan, K.M.; Salar, U.; Chigurupati, S.; Muhammad, M.T.; Wadood, A.; Aldubayan, M.; Vijayan, V.; Riaz, M.; Perveen, S. 2′-Aryl and 4′-arylidene substituted pyrazolones: As potential α-amylase inhibitors. Eur. J. Med. Chem. 2018, 159, 47–58. [Google Scholar] [CrossRef]
- Saleem, F.; Khan, K.M.; Chigurupati, S.; Solangi, M.; Nemala, A.R.; Mushtaq, M.; Ul-Haq, Z.; Taha, M.; Perveen, S. Synthesis of azachalcones, their α-amylase, α-glucosidase inhibitory activities, kinetics, and molecular docking studies. Bioorg. Chem. 2021, 106, 104489. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinform. 2008, 24, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Mseddi, K.; Alimi, F.; Noumi, E.; Veettil, V.N.; Deshpande, S.; Adnan, M.; Hamdi, A.; Elkahoui, S.; Ahmed, A.; Kadri, A.; et al. Thymus musilii Velen. as a promising source of potent bioactive compounds with its pharmacological properties: In vitro and in silico analysis. Arab. J. Chem. 2020, 13, 6782–6801. [Google Scholar]
- Pedretti, A.; Villa, L.; Vistoli, G. VEGA-An open platform to develop chemo-bioinformatics applications, using plug-in architecture and script programming. J. Comput. Aided Mol. Des. 2004, 18, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Dassault Systems BIOVIA. BIOVIA Discovery Studio Visualizer; v16.1.0.15350; Dassault Systems: San Diego, CA, USA, 2015. [Google Scholar]








| Entry | Isocyanate 2 | Benzimidazole Urea 3 | Time b (h) | Yield c (%) |
|---|---|---|---|---|
| 1 |  |  | 1 | 85 |
| 2 |  |  | 1 | 90 |
| 3 |  |  | 2 | 80 |
| 4 |  |  | 8 | 75 |
| 5 |  |  | 4 | 65 |
| 6 |  |  | 10 | 60 |
| 7 |  |  | 2 | 93 |
| 8 |  |  | 10 | 85 |
| Compounds 3 | TAC | DPPH-SA | FRAP | MCA |
|---|---|---|---|---|
| 3a | 1.33 ± 0.46 | 1.17 ± 0.07 | 6.53 ± 0.03 | 1.62 ± 0.10 |
| 3b | 5.16 ± 0.69 | 0.96 ± 0.06 | 9.25 ± 0.20 | 1.55 ± 0.07 |
| 3c | 4.43 ± 0.53 | 1.61 ± 0.02 | 8.65 ± 0.29 | 1.55 ± 0.22 |
| 3d | 2.47 ± 0.42 | 2.89 ± 1.07 | 5.67 ± 0.69 | 1.46 ± 0.10 |
| 3e | 2.52 ± 0.80 | 1.70 ± 0.01 | 2.75 ± 0.15 | 1.47 ± 0.09 |
| 3f | 8.50 ± 0.87 | 1.55 ± 0.03 | 5.99 ± 0.02 | 1.40 ± 0.07 |
| 3g | 10.06 ± 1.51 | 3.86 ± 0.04 | 16.12 ± 0.29 | 1.36 ± 0.09 |
| 3h | 2.25 ± 0.65 | 1.09 ± 0.03 | 6.79 ± 0.50 | 2.95 ± 0.21 |
| Compounds 3 | α-Amylase Inhibition IC50 ± SEM (µM) | α-Glucosidase Inhibition IC50 ± SEM (µM) |
|---|---|---|
| 3a | 40.7 ± 0.06 | 42.97 ± 0.19 |
| 3b | 41.7 ± 0.06 | 45.97 ± 0.19 |
| 3c | 18.65 ± 0.23 | 17.47 ± 0.03 |
| 3d | 25.65 ± 0.03 | 27.47 ± 0.13 |
| 3e | 20.7 ± 0.06 | 21.97 ± 0.19 |
| 3f | 28.33 ± 0.02 | 29.01 ± 0.12 |
| 3g | 22.33 ± 0.12 | 23.01 ± 0.12 |
| 3h | 43.04 ± 0.02 | 44.99 ± 0.09 |
| Acarbose | 14.21 ± 0.02 | 15.41 ± 0.32 |
| Compounds 3 | α-Amylase (E kcal/mol) | α-Glucosidase (E kcal/mol) |
|---|---|---|
| 3a | −8.8 | −8.7 |
| 3b | −10.3 | −9.9 |
| 3c | −8.8 | −8.2 |
| 3d | −8.6 | −8.6 |
| 3e | −8.0 | −7.6 |
| 3f | −7.8 | −7.6 |
| 3g | −11.2 | −10.0 |
| 3h | −10.4 | −10.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aroua, L.M.; Alosaimi, A.H.; Alminderej, F.M.; Messaoudi, S.; Mohammed, H.A.; Almahmoud, S.A.; Chigurupati, S.; Albadri, A.E.A.E.; Mekni, N.H. Synthesis, Molecular Docking, and Bioactivity Study of Novel Hybrid Benzimidazole Urea Derivatives: A Promising α-Amylase and α-Glucosidase Inhibitor Candidate with Antioxidant Activity. Pharmaceutics 2023, 15, 457. https://doi.org/10.3390/pharmaceutics15020457
Aroua LM, Alosaimi AH, Alminderej FM, Messaoudi S, Mohammed HA, Almahmoud SA, Chigurupati S, Albadri AEAE, Mekni NH. Synthesis, Molecular Docking, and Bioactivity Study of Novel Hybrid Benzimidazole Urea Derivatives: A Promising α-Amylase and α-Glucosidase Inhibitor Candidate with Antioxidant Activity. Pharmaceutics. 2023; 15(2):457. https://doi.org/10.3390/pharmaceutics15020457
Chicago/Turabian StyleAroua, Lotfi M., Abdulelah H. Alosaimi, Fahad M. Alminderej, Sabri Messaoudi, Hamdoon A. Mohammed, Suliman A. Almahmoud, Sridevi Chigurupati, Abuzar E. A. E. Albadri, and Nejib H. Mekni. 2023. "Synthesis, Molecular Docking, and Bioactivity Study of Novel Hybrid Benzimidazole Urea Derivatives: A Promising α-Amylase and α-Glucosidase Inhibitor Candidate with Antioxidant Activity" Pharmaceutics 15, no. 2: 457. https://doi.org/10.3390/pharmaceutics15020457
APA StyleAroua, L. M., Alosaimi, A. H., Alminderej, F. M., Messaoudi, S., Mohammed, H. A., Almahmoud, S. A., Chigurupati, S., Albadri, A. E. A. E., & Mekni, N. H. (2023). Synthesis, Molecular Docking, and Bioactivity Study of Novel Hybrid Benzimidazole Urea Derivatives: A Promising α-Amylase and α-Glucosidase Inhibitor Candidate with Antioxidant Activity. Pharmaceutics, 15(2), 457. https://doi.org/10.3390/pharmaceutics15020457








