Targeted Two-Step Delivery of Oncotheranostic Nano-PLGA for HER2-Positive Tumor Imaging and Therapy In Vivo: Improved Effectiveness Compared to One-Step Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. PLGA Nanoparticle Synthesis and Characterization
2.2. Cell Culture
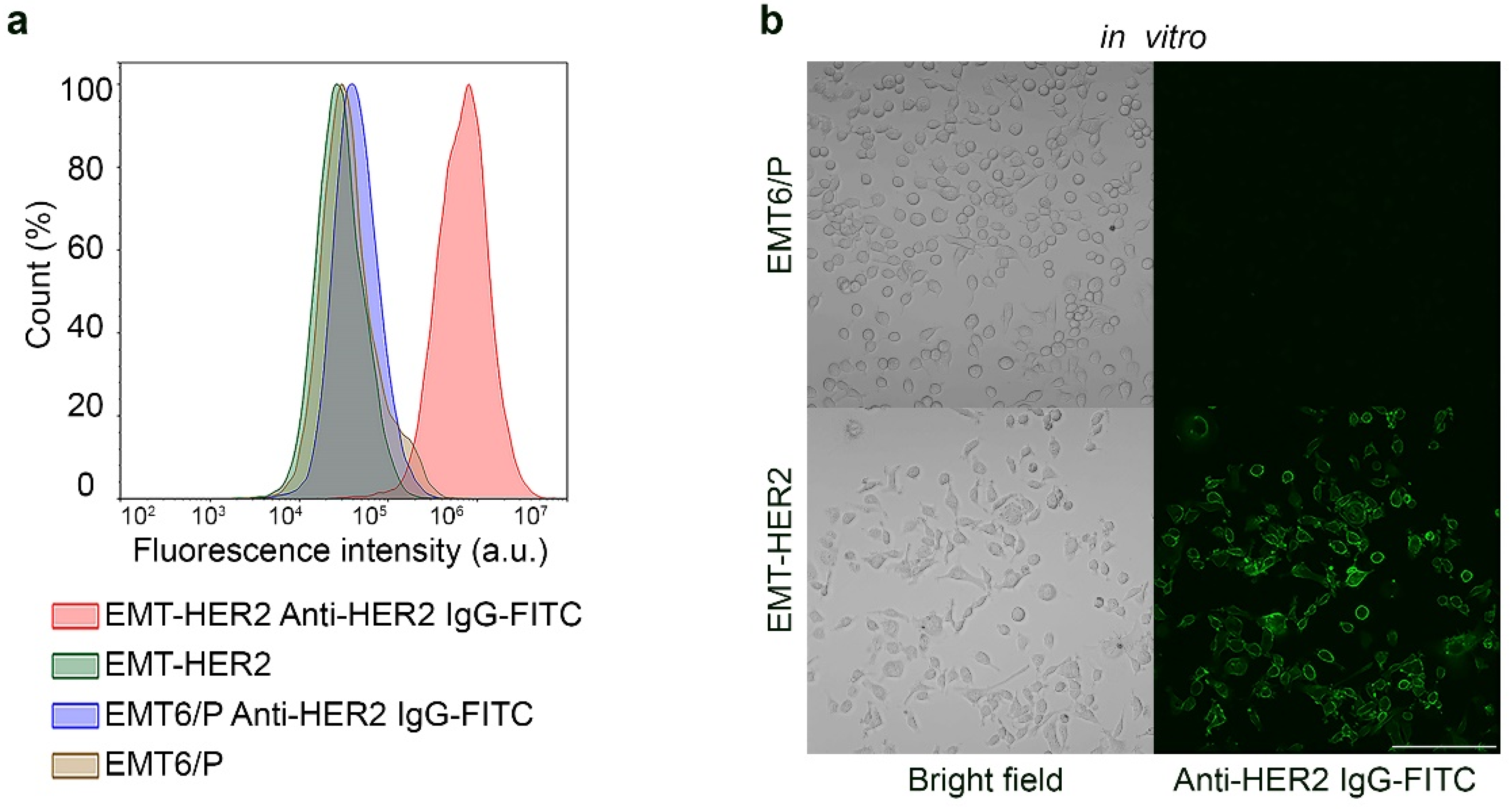
2.3. EMT-HER2 Cells
2.4. Flow Cytometry
2.5. Tumor-Bearing Mice
2.6. Cryosections
2.7. Confocal Laser Scanning Microscopy
2.8. In Vivo Imaging
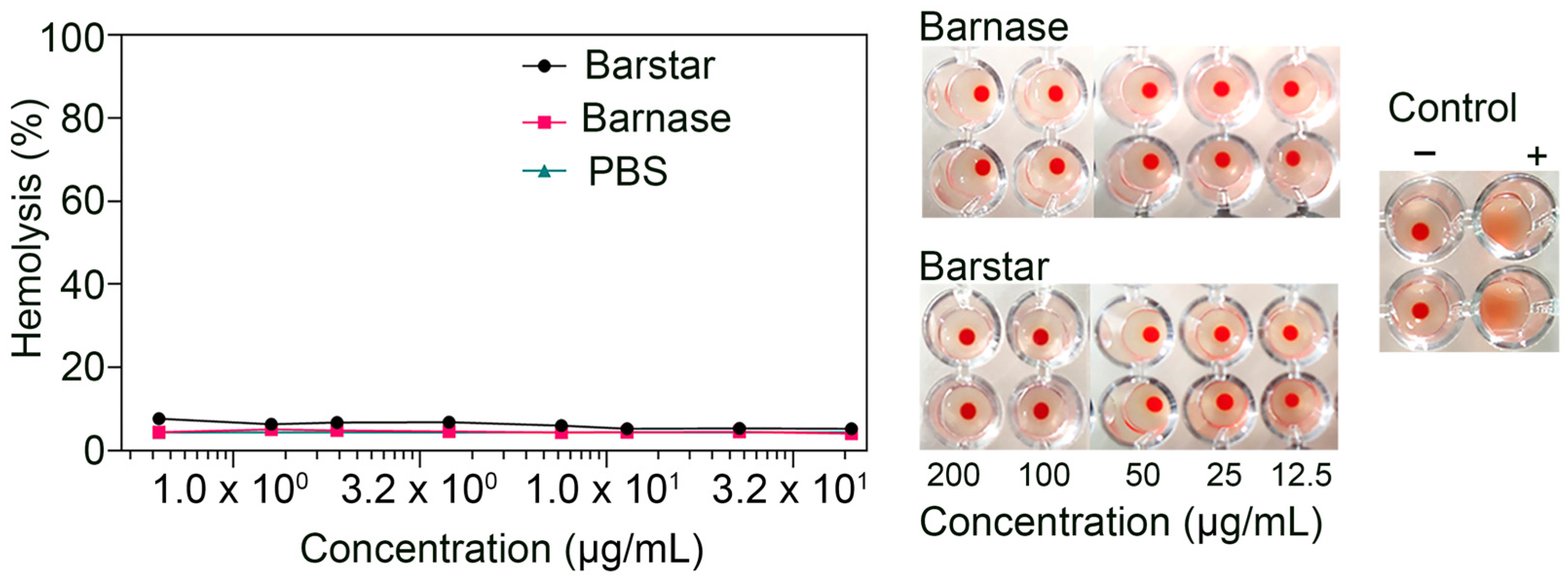
2.9. Red Blood Cell Hemolysis
2.10. Agglutination Study
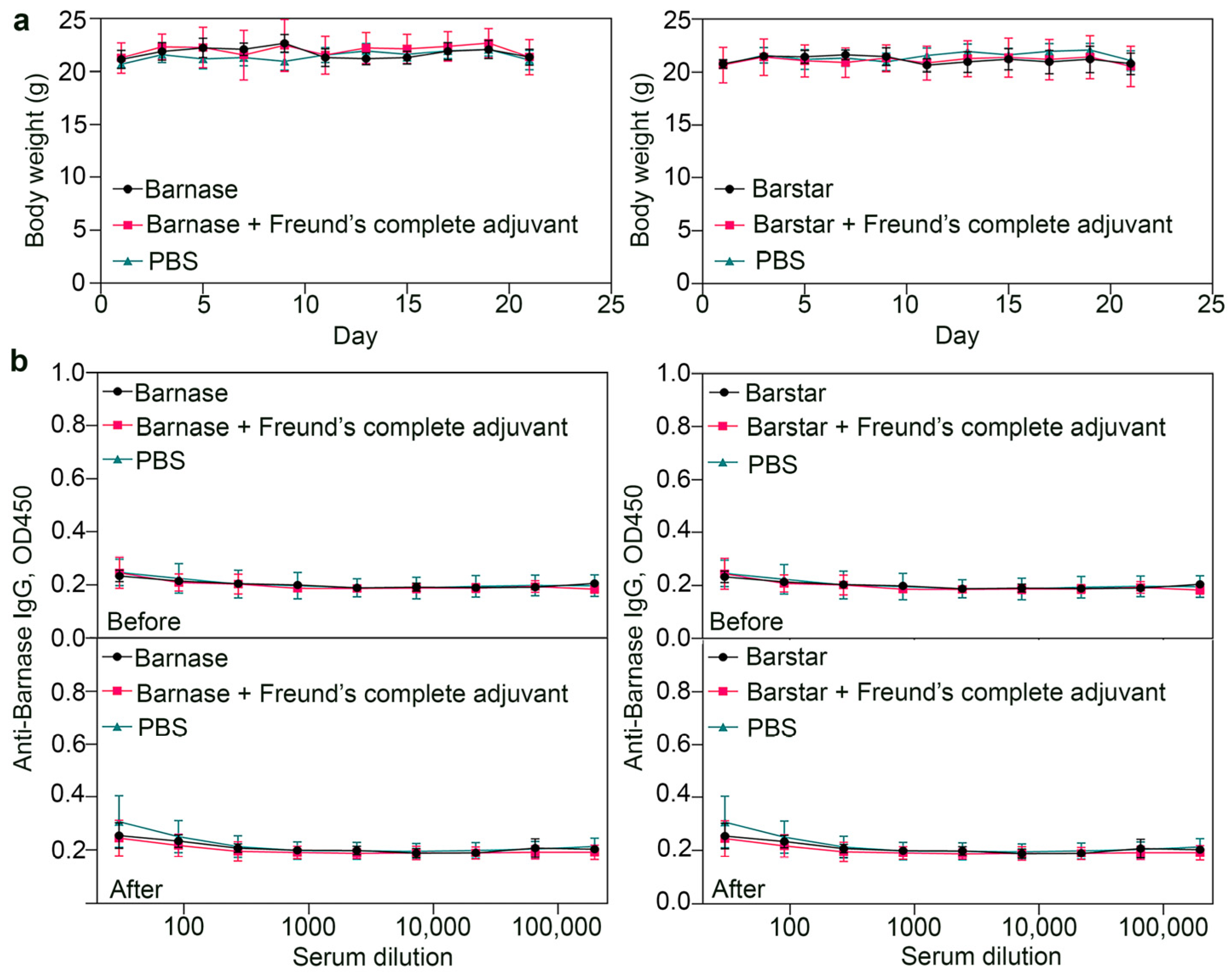
2.11. Immunogenicity Study
2.12. ELISA
3. Results
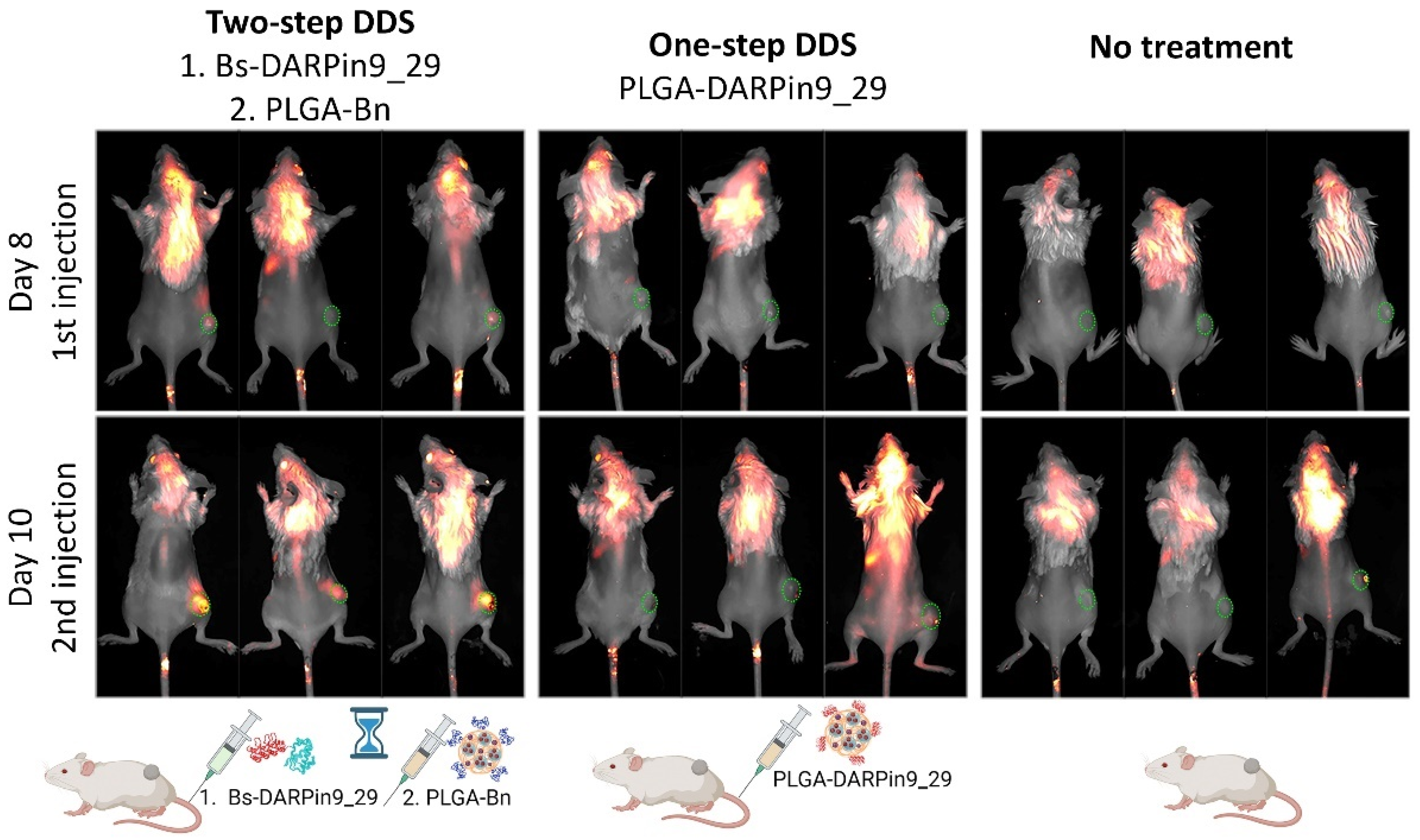
3.1. Design of the Experiment: One-Step DDS vs. Two-Step DDS In Vivo
3.2. Synthesis and Characterization of Polymer Nanocarriers for Targeted Delivery to HER2-Overexpressing Cancer Cells
3.3. Development of HER2-Overexpressing Allografts in Immunocompetent BALB/c Mice
3.4. In Vivo Bioimaging: HER2-Positive Allograft Visualization with One-Step and Two-Step DDS
3.5. In Vivo Therapy: One-Step DDS vs. Two-Step DDS for HER-Positive Tumor Growth Inhibition
3.6. Biosafety Aspects: Barnase and Barstar Hemotoxicity Study
3.7. Biosafety Aspects: Immunogenicity Study of Barnase and Barstar
4. Discussion
| Delivery System | Immunogenicity | Steric Hindrance | Ka | Representation in Mammals |
|---|---|---|---|---|
| Barnase*barstar | ✓ Both proteins are not immunogenic (this study). | ✓ Proteins are comparable in size (12 and 10 kDa), and therefore steric hindrances should not arise [28]. | 1014 M−1 [30] | ✓ Both proteins were isolated from bacteria and are not represented in mammals [30]. |
| Streptavidin*biotin | ✗ Streptavidin is highly immunogenic [59]. | ✗ The significant difference in the size (56 kDa and 244 Da) of the molecules can cause steric hindrance: if biotin is bound to a non-smooth surface, then streptavidin will not be able to recognize it. This imposes restrictions on the use of this system in nanomedicine [60,61,62]. | 1015 M−1 [63] | ✗ Biotin, or vitamin H, is presented in the blood of mammals, which may cause obstacles to the appropriate interaction of streptavidin*biotin [64,65,66]. |
| Hapten* antibody | ✗ Antibodies are immunogenic and may have effector functions, which make them not the best candidate for long-term treatment [67]. | ✗ IgG is 150 kDa protein, while hapten usually is a low-molecular compound, so steric difficulties may arise when the components interact [67,68]. | 105–1010 M−1 [69] | ✗ Antibodies are presented in blood and have effector functions as critical participants in the immune defense [41]. |
| Nucleic acids: (i) DNA*DNA (ii) RNA*RNA (iii) mirror-imaged oligonucleotides (iv) phosphorodiamidate morpholino oligomers (v) peptide nucleic acids (vi) locked nucleic acid | ✓ Nucleotides as natural molecules are not immunogenic [70]. ✓ Some mirror-imaged oligonucleotides, phosphorodiamidate morpholino oligomers, and peptide nucleic acids are not immunogenic [71,72,73,74]. | ✗ Nucleotides are small molecules (less than 500 Da). Since the entire sequence of nucleotides is essential for recognition purposes, steric hindrance is a common problem in recognition processes. | Depending on the base pair number | ✗ The presence of nucleases in the serum is an obstacle in the development of the system based on oligonucleotides due to the fast degradation [70]. ✓ Due to their artificial origin, mirror-imaged oligonucleotides, phosphorodiamidate morpholino oligomers and peptide nucleic acids are resistant to degradation by nucleases, and peptide and locked nucleic acids are also resistant to protease digestion [72,74,75,76,77]. |
| Click chemistry | ✓ Molecules used in bio-orthogonal chemistry are supposed to be not highly immunogenic [78]. | ✗ Molecules used in bio-orthogonal chemistry are small, which can cause steric hindrance. | ✓ ✗ Covalent bonding is stronger than affinity interaction but is not reversible. | ✓ Molecules used in bio-orthogonal chemistry are not represented in mammals [75]. |
| SpyTag/SpyCatcher | N/A | ✓ Steric hindrances should not arise [79]. | ✓ ✗ Covalent bonding is stronger than affinity interaction but is not reversible. | ✓ Both proteins were isolated from bacteria and modified by bioengineering and are not represented in mammals [80]. |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 2019, 4, e10143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Cheng, K.; Chen, K.; Xu, C.; Ma, P.; Dang, G.; Yang, Y.; Lei, Q.; Huang, H.; Yu, Y.; et al. Nanoparticle-based medicines in clinical cancer therapy. Nano Today 2022, 45, 101512. [Google Scholar] [CrossRef]
- Masood, F. Polymeric nanoparticles for targeted drug delivery system for cancer therapy. Mater. Sci. Eng. C Mater. Biol. Appl. 2016, 60, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.Z.; Langer, R.; Farokhzad, O.C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 2012, 63, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; Tan, W.S.; Ho, K.L.; Mariatulqabtiah, A.R.; Abu Kasim, N.H.; Abd Rahman, N.; Wong, T.W.; Chee, C.F. Challenges and Complications of Poly(lactic-co-glycolic acid)-Based Long-Acting Drug Product Development. Pharmaceutics 2022, 14, 614. [Google Scholar] [CrossRef] [PubMed]
- Krauss, A.C.; Gao, X.; Li, L.; Manning, M.L.; Patel, P.; Fu, W.; Janoria, K.G.; Gieser, G.; Bateman, D.A.; Przepiorka, D.; et al. FDA Approval Summary: (Daunorubicin and Cytarabine) Liposome for Injection for the Treatment of Adults with High-Risk Acute Myeloid Leukemia. Clin. Cancer Res. 2019, 25, 2685–2690. [Google Scholar] [CrossRef] [PubMed]
- Danhier, F. To exploit the tumor microenvironment: Since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 2016, 244, 108–121. [Google Scholar] [CrossRef]
- Ejigah, V.; Owoseni, O.; Bataille-Backer, P.; Ogundipe, O.D.; Fisusi, F.A.; Adesina, S.K. Approaches to Improve Macromolecule and Nanoparticle Accumulation in the Tumor Microenvironment by the Enhanced Permeability and Retention Effect. Polymers 2022, 14, 2601. [Google Scholar] [CrossRef]
- Iyer, A.K.; Khaled, G.; Fang, J.; Maeda, H. Exploiting the enhanced permeability and retention effect for tumor targeting. Drug Discov. Today 2006, 11, 812–818. [Google Scholar] [CrossRef]
- Nichols, J.W.; Bae, Y.H. EPR: Evidence and fallacy. J. Control. Release 2014, 190, 451–464. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, Q. Development of liposomal formulations: From concept to clinical investigations. Asian J. Pharm. Sci. 2013, 8, 81–87. [Google Scholar] [CrossRef] [Green Version]
- Swenson, C.E.; Perkins, W.R.; Roberts, P.; Janoff, A.S. Liposome technology and the development of Myocet™ (liposomal doxorubicin citrate). Breast 2001, 10, 1–7. [Google Scholar] [CrossRef]
- Drozdov, A.S.; Nikitin, P.I.; Rozenberg, J.M. Systematic Review of Cancer Targeting by Nanoparticles Revealed a Global Association between Accumulation in Tumors and Spleen. IJMS 2021, 22, 13011. [Google Scholar] [CrossRef]
- Rubin, I.; Yarden, Y. The basic biology of HER2. Ann. Oncol. 2001, 12, S3–S8. [Google Scholar] [CrossRef] [PubMed]
- Shipunova, V.O.; Komedchikova, E.N.; Kotelnikova, P.A.; Zelepukin, I.V.; Schulga, A.A.; Proshkina, G.M.; Shramova, E.I.; Kutscher, H.L.; Telegin, G.B.; Kabashin, A.V.; et al. Dual Regioselective Targeting the Same Receptor in Nanoparticle-Mediated Combination Immuno/Chemotherapy for Enhanced Image-Guided Cancer Treatment. ACS Nano 2020, 14, 12781–12795. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Sizikov, A.A.; Cherkasov, V.R. Light-activated polymer nanoparticles for image-guided breast cancer treatment. In 2022 International Conference Laser Optics (ICLO); Saint Petersburg, Russian, 20 June–24 June 2022, IEEE: New York, NY, USA, 2022; p. 1. ISBN 978-1-6654-6663-9. [Google Scholar]
- von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Nikitin, M.P.; Zdobnova, T.A.; Lukash, S.V.; Stremovskiy, O.A.; Deyev, S.M. Protein-assisted self-assembly of multifunctional nanoparticles. Proc. Natl. Acad. Sci. USA 2010, 107, 5827–5832. [Google Scholar] [CrossRef] [Green Version]
- Nikitin, M.P.; Shipunova, V.O.; Deyev, S.M.; Nikitin, P.I. Biocomputing based on particle disassembly. Nat. Nanotechnol. 2014, 9, 716–722. [Google Scholar] [CrossRef]
- Nikitin, M.P.; Zelepukin, I.V.; Shipunova, V.O.; Sokolov, I.L.; Deyev, S.M.; Nikitin, P.I. Enhancement of the blood-circulation time and performance of nanomedicines via the forced clearance of erythrocytes. Nat. Biomed. Eng. 2020, 4, 717–731. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Kovalenko, V.L.; Kotelnikova, P.A.; Sogomonyan, A.S.; Shilova, O.N.; Komedchikova, E.N.; Zvyagin, A.V.; Nikitin, M.P.; Deyev, S.M. Targeting Cancer Cell Tight Junctions Enhances PLGA-Based Photothermal Sensitizers’ Performance In Vitro and In Vivo. Pharmaceutics 2022, 14, 43. [Google Scholar] [CrossRef]
- Zelepukin, I.V.; Yaremenko, A.V.; Shipunova, V.O.; Babenyshev, A.V.; Balalaeva, I.V.; Nikitin, P.I.; Deyev, S.M.; Nikitin, M.P. Nanoparticle-based drug delivery via RBC-hitchhiking for the inhibition of lung metastases growth. Nanoscale 2019, 11, 1636–1646. [Google Scholar] [CrossRef]
- Kalinin, R.S.; Shipunova, V.O.; Rubtsov, Y.P.; Ukrainskay, V.M.; Schulga, A.; Konovalova, E.V.; Volkov, D.V.; Yaroshevich, I.A.; Moysenovich, A.M.; Belogurov, A.A.; et al. Barnase-barstar Specific Interaction Regulates Car-T Cells Cytotoxic Activity toward Malignancy. Dokl. Biochem. Biophys. 2023, 508, 1–4. [Google Scholar] [CrossRef]
- Obozina, A.S.; Komedchikova, E.N.; Kolesnikova, O.A.; Iureva, A.M.; Kovalenko, V.L.; Zavalko, F.A.; Rozhnikova, T.V.; Tereshina, E.D.; Mochalova, E.N.; Shipunova, V.O. Genetically Encoded Self-Assembling Protein Nanoparticles for the Targeted Delivery In Vitro and In Vivo. Pharmaceutics 2023, 15, 231. [Google Scholar] [CrossRef]
- Stepanov, A.V.; Kalinin, R.S.; Shipunova, V.O.; Zhang, D.; Xie, J.; Rubtsov, Y.P.; Ukrainskaya, V.M.; Schulga, A.; Konovalova, E.V.; Volkov, D.V.; et al. Switchable targeting of solid tumors by BsCAR T cells. Proc. Natl. Acad. Sci. USA 2022, 119, e2210562119. [Google Scholar] [CrossRef]
- Aghayeva, U.F.; Nikitin, M.P.; Lukash, S.V.; Deyev, S.M. Denaturation-resistant bifunctional colloidal superstructures assembled via the proteinaceous barnase-barstar interface. ACS Nano 2013, 7, 950–961. [Google Scholar] [CrossRef]
- Komedchikova, E.N.; Kolesnikova, O.A.; Tereshina, E.D.; Kotelnikova, P.A.; Sogomonyan, A.S.; Stepanov, A.V.; Deyev, S.M.; Nikitin, M.P.; Shipunova, V.O. Two-Step Targeted Drug Delivery via Proteinaceous Barnase-Barstar Interface and Doxorubicin-Loaded Nano-PLGA Outperforms One-Step Strategy for Targeted Delivery to HER2-Overexpressing Cells. Pharmaceutics 2022, 15, 52. [Google Scholar] [CrossRef]
- Deyev, S.M.; Waibel, R.; Lebedenko, E.N.; Schubiger, A.P.; Plückthun, A. Design of multivalent complexes using the barnase*barstar module. Nat. Biotechnol. 2003, 21, 1486–1492. [Google Scholar] [CrossRef]
- Schreiber, G.; Fersht, A.R. Energetics of protein-protein interactions: Analysis ofthe Barnase-Barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 1995, 248, 478–486. [Google Scholar] [CrossRef]
- Schreiber, G. Methods for studying the interaction of barnase with its inhibitor barstar. Methods Mol. Biol. 2001, 160, 213–226. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Sogomonyan, A.S.; Zelepukin, I.V.; Nikitin, M.P.; Deyev, S.M. PLGA Nanoparticles Decorated with Anti-HER2 Affibody for Targeted Delivery and Photoinduced Cell Death. Molecules 2021, 26, 3955. [Google Scholar] [CrossRef]
- Pillai, O.; Panchagnula, R. Polymers in drug delivery. Curr. Opin. Chem. Biol. 2001, 5, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-based nanoparticles: An overview of biomedical applications. J. Control. Release 2012, 161, 505–522. [Google Scholar] [CrossRef] [PubMed]
- El-Maghawry, E.; Tadros, M.I.; Elkheshen, S.A.; Abd-Elbary, A. Eudragit®-S100 Coated PLGA Nanoparticles for Colon Targeting of Etoricoxib: Optimization and Pharmacokinetic Assessments in Healthy Human Volunteers. Int. J. Nanomed. 2020, 15, 3965–3980. [Google Scholar] [CrossRef]
- Lecio, G.; Ribeiro, F.V.; Pimentel, S.P.; Reis, A.A.; Da Silva, R.V.C.; Nociti-Jr, F.; Moura, L.; Duek, E.; Casati, M.; Casarin, R.C.V. Novel 20% doxycycline-loaded PLGA nanospheres as adjunctive therapy in chronic periodontitis in type-2 diabetics: Randomized clinical, immune and microbiological trial. Clin. Oral Investig. 2020, 24, 1269–1279. [Google Scholar] [CrossRef]
- Martinez, V.; Henary, M. Nile Red and Nile Blue: Applications and Syntheses of Structural Analogues. Chemistry 2016, 22, 13764–13782. [Google Scholar] [CrossRef]
- Lin, C.W.; Shulok, J.R.; Kirley, S.D.; Cincotta, L.; Foley, J.W. Lysosomal localization and mechanism of uptake of Nile blue photosensitizers in tumor cells. Cancer Res 1991, 51, 2710–2719. [Google Scholar]
- LEWIS, M.R.; GOLAND, P.P.; SLOVITER, H.A. The action of oxazine dyes on tumors in mice. Cancer Res. 1949, 9, 736–740. [Google Scholar]
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic. Bioeng. Transl. Med. 2016, 1, 10–29. [Google Scholar] [CrossRef]
- Gebauer, M.; Skerra, A. Engineered protein scaffolds as next-generation antibody therapeutics. Curr. Opin. Chem. Biol. 2009, 13, 245–255. [Google Scholar] [CrossRef]
- Rosenblum, D.; Joshi, N.; Tao, W.; Karp, J.M.; Peer, D. Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 2018, 9, 1410. [Google Scholar] [CrossRef] [Green Version]
- Silverman, J.; Liu, Q.; Lu, Q.; Bakker, A.; To, W.; Duguay, A.; Alba, B.M.; Smith, R.; Rivas, A.; Li, P.; et al. Multivalent avimer proteins evolved by exon shuffling of a family of human receptor domains. Nat. Biotechnol. 2005, 23, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Parizek, P.; Kummer, L.; Rube, P.; Prinz, A.; Herberg, F.W.; Plückthun, A. Designed ankyrin repeat proteins (DARPins) as novel isoform-specific intracellular inhibitors of c-Jun N-terminal kinases. ACS Chem. Biol. 2012, 7, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Shipunova, V.O.; Deyev, S.M. Artificial scaffold polypeptides as an efficient tool for the targeted delivery of nanostructures in vitro and in vivo. Acta Nat. 2022, 14, 54–72. [Google Scholar] [CrossRef] [PubMed]
- Binz, H.K.; Amstutz, P.; Plückthun, A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat. Biotechnol. 2005, 23, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Shipunova, V.O.; Shramova, E.I.; Schulga, A.A.; Shilova, M.V.; Deyev, S.M.; Proshkina, G.M. Delivery of Barnase to Cells in Liposomes Functionalized by Her2-Specific DARPin Module. Russ. J. Bioorg. Chem. 2020, 46, 1156–1161. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Kolesnikova, O.A.; Kotelnikova, P.A.; Soloviev, V.D.; Popov, A.A.; Proshkina, G.M.; Nikitin, M.P.; Deyev, S.M. Comparative Evaluation of Engineered Polypeptide Scaffolds in HER2-Targeting Magnetic Nanocarrier Delivery. ACS Omega 2021, 6, 16000–16008. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Kotelnikova, P.A.; Aghayeva, U.F.; Stremovskiy, O.A.; Novikov, I.A.; Schulga, A.A.; Nikitin, M.P.; Deyev, S.M. Self-assembling nanoparticles biofunctionalized with magnetite-binding protein for the targeted delivery to HER2/neu overexpressing cancer cells. J. Magn. Magn. Mater. 2019, 469, 450–455. [Google Scholar] [CrossRef]
- Shramova, E.; Proshkina, G.; Shipunova, V.; Ryabova, A.; Kamyshinsky, R.; Konevega, A.; Schulga, A.; Konovalova, E.; Telegin, G.; Deyev, S. Dual Targeting of Cancer Cells with DARPin-Based Toxins for Overcoming Tumor Escape. Cancers 2020, 12, 3014. [Google Scholar] [CrossRef]
- Koide, A.; Wojcik, J.; Gilbreth, R.N.; Hoey, R.J.; Koide, S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J. Mol. Biol. 2012, 415, 393–405. [Google Scholar] [CrossRef] [Green Version]
- Beste, G.; Schmidt, F.S.; Stibora, T.; Skerra, A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc. Natl. Acad. Sci. USA 1999, 96, 1898–1903. [Google Scholar] [CrossRef] [Green Version]
- Steiner, D.; Forrer, P.; Plückthun, A. Efficient selection of DARPins with sub-nanomolar affinities using SRP phage display. J. Mol. Biol. 2008, 382, 1211–1227. [Google Scholar] [CrossRef] [Green Version]
- Deyev, S.; Vorobyeva, A.; Schulga, A.; Proshkina, G.; Güler, R.; Löfblom, J.; Mitran, B.; Garousi, J.; Altai, M.; Buijs, J.; et al. Comparative Evaluation of Two DARPin Variants: Effect of Affinity, Size, and Label on Tumor Targeting Properties. Mol. Pharm. 2019, 16, 995–1008. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Belova, M.M.; Kotelnikova, P.A.; Shilova, O.N.; Mirkasymov, A.B.; Danilova, N.V.; Komedchikova, E.N.; Popovtzer, R.; Deyev, S.M.; Nikitin, M.P. Photothermal Therapy with HER2-Targeted Silver Nanoparticles Leading to Cancer Remission. Pharmaceutics 2022, 14, 1013. [Google Scholar] [CrossRef]
- Novoselova, M.; Chernyshev, V.S.; Schulga, A.; Konovalova, E.V.; Chuprov-Netochin, R.N.; Abakumova, T.O.; German, S.; Shipunova, V.O.; Mokrousov, M.D.; Prikhozhdenko, E.; et al. Effect of Surface Modification of Multifunctional Nanocomposite Drug Delivery Carriers with DARPin on Their Biodistribution In Vitro and In Vivo. ACS Appl. Bio Mater. 2022, 5, 2976–2989. [Google Scholar] [CrossRef]
- Frejd, F.Y.; Kim, K.-T. Affibody molecules as engineered protein drugs. Exp. Mol. Med. 2017, 49, e306. [Google Scholar] [CrossRef] [Green Version]
- Kotelnikova, P.A.; Shipunova, V.O.; Aghayeva, U.F.; Stremovskiy, O.A.; Nikitin, M.P.; Novikov, I.A.; Schulga, A.A.; Deyev, S.M.; Petrov, R.V. Synthesis of Magnetic Nanoparticles Stabilized by Magnetite-Binding Protein for Targeted Delivery to Cancer Cells. Dokl. Biochem. Biophys. 2018, 481, 198–200. [Google Scholar] [CrossRef]
- Shipunova, V.O.; Zelepukin, I.V.; Stremovskiy, O.A.; Nikitin, M.P.; Care, A.; Sunna, A.; Zvyagin, A.V.; Deyev, S.M. Versatile Platform for Nanoparticle Surface Bioengineering Based on SiO2-Binding Peptide and Proteinaceous Barnase*Barstar Interface. ACS Appl. Mater. Interfaces 2018, 10, 17437–17447. [Google Scholar] [CrossRef]
- Förster, G.J.; Santos, E.B.; Smith-Jones, P.M.; Zanzonico, P.; Larson, S.M. Pretargeted radioimmunotherapy with a single-chain antibody/streptavidin construct and radiolabeled DOTA-biotin: Strategies for reduction of the renal dose. J. Nucl. Med. 2006, 47, 140–149. [Google Scholar]
- Dundas, C.M.; Demonte, D.; Park, S. Streptavidin-biotin technology: Improvements and innovations in chemical and biological applications. Appl. Microbiol. Biotechnol. 2013, 97, 9343–9353. [Google Scholar] [CrossRef]
- Huang, S.C.; Stump, M.D.; Weiss, R.; Caldwell, K.D. Binding of biotinylated DNA to streptavidin-coated polystyrene latex: Effects of chain length and particle size. Anal. Biochem. 1996, 237, 115–122. [Google Scholar] [CrossRef]
- Neish, C.S.; Martin, I.L.; Henderson, R.M.; Edwardson, J.M. Direct visualization of ligand-protein interactions using atomic force microscopy. Br. J. Pharmacol. 2002, 135, 1943–1950. [Google Scholar] [CrossRef] [Green Version]
- Wilchek, M.; Bayer, E.A. [2] Introduction to avidin-biotin technology. In Avidin-Biotin Technology; Elsevier: Amsterdam, The Netherlands, 1990; pp. 5–13. ISBN 9780121820855. [Google Scholar]
- Rusckowski, M.; Fogarasi, M.; Fritz, B.; Hnatowich, D.J. Effect of endogenous biotin on the applications of streptavidin and biotin in mice. Nucl. Med. Biol. 1997, 24, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Luong, J.H.T.; Male, K.B.; Glennon, J.D. Biotin interference in immunoassays based on biotin-strept(avidin) chemistry: An emerging threat. Biotechnol. Adv. 2019, 37, 634–641. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.-H.; Soda, Y.; Izawa, K.; Kobayashi, S.; Tani, K.; Maruyama, K.; Tojo, A.; Asano, S. A versatile drug delivery system using streptavidin-tagged pegylated liposomes and biotinylated biomaterials. Int. J. Pharm. 2013, 454, 478–485. [Google Scholar] [CrossRef]
- Goldenberg, D.M. Targeted therapy of cancer with radiolabeled antibodies. J. Nucl. Med. 2002, 43, 693–713. [Google Scholar] [PubMed]
- Stickney, D.R.; Anderson, L.D.; Slater, J.B.; Ahlem, C.N.; Kirk, G.A.; Schweighardt, S.A.; Frincke, J.M. Bifunctional antibody: A binary radiopharmaceutical delivery system for imaging colorectal carcinoma. Cancer Res. 1991, 51, 6650–6655. [Google Scholar]
- Stéen, E.J.L.; Edem, P.E.; Nørregaard, K.; Jørgensen, J.T.; Shalgunov, V.; Kjaer, A.; Herth, M.M. Pretargeting in nuclear imaging and radionuclide therapy: Improving efficacy of theranostics and nanomedicines. Biomaterials 2018, 179, 209–245. [Google Scholar] [CrossRef]
- Kuijpers, W.H.; Bos, E.S.; Kaspersen, F.M.; Veeneman, G.H.; van Boeckel, C.A. Specific recognition of antibody-oligonucleotide conjugates by radiolabeled antisense nucleotides: A novel approach for two-step radioimmunotherapy of cancer. Bioconjug. Chem. 1993, 4, 94–102. [Google Scholar] [CrossRef]
- Wlotzka, B.; Leva, S.; Eschgfäller, B.; Burmeister, J.; Kleinjung, F.; Kaduk, C.; Muhn, P.; Hess-Stumpp, H.; Klussmann, S. In vivo properties of an anti-GnRH Spiegelmer: An example of an oligonucleotide-based therapeutic substance class. Proc. Natl. Acad. Sci. USA 2002, 99, 8898–8902. [Google Scholar] [CrossRef] [Green Version]
- Young, B.E.; Kundu, N.; Sczepanski, J.T. Mirror-Image Oligonucleotides: History and Emerging Applications. Chemistry 2019, 25, 7981–7990. [Google Scholar] [CrossRef]
- Liu, G. A Revisit to the Pretargeting Concept-A Target Conversion. Front. Pharmacol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- Altai, M.; Membreno, R.; Cook, B.; Tolmachev, V.; Zeglis, B.M. Pretargeted Imaging and Therapy. J. Nucl. Med. 2017, 58, 1553–1559. [Google Scholar] [CrossRef]
- Patra, M.; Zarschler, K.; Pietzsch, H.-J.; Stephan, H.; Gasser, G. New insights into the pretargeting approach to image and treat tumours. Chem. Soc. Rev. 2016, 45, 6415–6431. [Google Scholar] [CrossRef] [Green Version]
- Karkare, S.; Bhatnagar, D. Promising nucleic acid analogs and mimics: Characteristic features and applications of PNA, LNA, and morpholino. Appl. Microbiol. Biotechnol. 2006, 71, 575–586. [Google Scholar] [CrossRef]
- Mallikaratchy, P.; Gardner, J.; Nordstrøm, L.U.R.; Veomett, N.J.; McDevitt, M.R.; Heaney, M.L.; Scheinberg, D.A. A self-assembling short oligonucleotide duplex suitable for pretargeting. Nucleic Acid Ther. 2013, 23, 289–299. [Google Scholar] [CrossRef] [Green Version]
- van de Watering, F.C.J.; Rijpkema, M.; Robillard, M.; Oyen, W.J.G.; Boerman, O.C. Pretargeted imaging and radioimmunotherapy of cancer using antibodies and bioorthogonal chemistry. Front. Med. 2014, 1, 44. [Google Scholar] [CrossRef] [Green Version]
- Bae, Y.; Kim, G.J.; Kim, H.; Park, S.G.; Jung, H.S.; Kang, S. Engineering Tunable Dual Functional Protein Cage Nanoparticles Using Bacterial Superglue. Biomacromolecules 2018, 19, 2896–2904. [Google Scholar] [CrossRef]
- Zakeri, B.; Fierer, J.O.; Celik, E.; Chittock, E.C.; Schwarz-Linek, U.; Moy, V.T.; Howarth, M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. USA 2012, 109, E690–E697. [Google Scholar] [CrossRef] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shipunova, V.O.; Komedchikova, E.N.; Kotelnikova, P.A.; Nikitin, M.P.; Deyev, S.M. Targeted Two-Step Delivery of Oncotheranostic Nano-PLGA for HER2-Positive Tumor Imaging and Therapy In Vivo: Improved Effectiveness Compared to One-Step Strategy. Pharmaceutics 2023, 15, 833. https://doi.org/10.3390/pharmaceutics15030833
Shipunova VO, Komedchikova EN, Kotelnikova PA, Nikitin MP, Deyev SM. Targeted Two-Step Delivery of Oncotheranostic Nano-PLGA for HER2-Positive Tumor Imaging and Therapy In Vivo: Improved Effectiveness Compared to One-Step Strategy. Pharmaceutics. 2023; 15(3):833. https://doi.org/10.3390/pharmaceutics15030833
Chicago/Turabian StyleShipunova, Victoria O., Elena N. Komedchikova, Polina A. Kotelnikova, Maxim P. Nikitin, and Sergey M. Deyev. 2023. "Targeted Two-Step Delivery of Oncotheranostic Nano-PLGA for HER2-Positive Tumor Imaging and Therapy In Vivo: Improved Effectiveness Compared to One-Step Strategy" Pharmaceutics 15, no. 3: 833. https://doi.org/10.3390/pharmaceutics15030833
APA StyleShipunova, V. O., Komedchikova, E. N., Kotelnikova, P. A., Nikitin, M. P., & Deyev, S. M. (2023). Targeted Two-Step Delivery of Oncotheranostic Nano-PLGA for HER2-Positive Tumor Imaging and Therapy In Vivo: Improved Effectiveness Compared to One-Step Strategy. Pharmaceutics, 15(3), 833. https://doi.org/10.3390/pharmaceutics15030833






