Analytical Insights into Protein–Alum Interactions and Their Impact on Conformational Epitope
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials Used in the Capillary Electrophoresys (CE)
- ▪
- To prepare the Tris Acetate buffers, a proper weight of Tris(hydroxymethyl)aminomethane was dissolved in ultra-purified water to reach the final desiderated molarity. A proper volume of glacial acetic acid was added to obtain the desired pH.
- ▪
- To prepare the Tris Acetate SDS buffers, proper weights of Tris(hydroxymethyl)aminomethane and SDS were dissolved in ultra-purified water to reach the final desiderated molarity. A proper volume of glacial acetic acid was added to obtain the desired pH.
2.2. Antigens and Adjuvants
2.2.1. Preparation of Antigen Formulation without Alum
2.2.2. Preparation of Alum Samples
2.3. Instrument and Separation Methods
- ▪
- Uncoated fused silica column (AB Sciex, Framingham, MA, USA); inner diameter (ID): 50 μm; total capillary length: 70.2 cm (60 cm to detector).
- ▪
- Neutral capillary linear polyacrylamide (LPA) coated capillary (AB Sciex, Framingham, MA, USA) with an inner diameter (ID): 50 µm; total capillary length: 50.2 cm (40 cm to detector).
2.4. Materials for In Vitro Relative Potency Assay (IVRP)
2.4.1. Monovalent Formulations
2.4.2. Multivalent Formulations
2.4.3. Antibodies
2.5. Luminex Assay Procedure
2.6. Hydroxyl Radical Footprinting (HRF) Assay
2.6.1. Materials
2.6.2. Mass Spectra Acquisition
2.6.3. UPLC (Ultra-Performance Liquid Chromatography) Chromatographic Method
2.6.4. HRF Sample Preparation Protocols
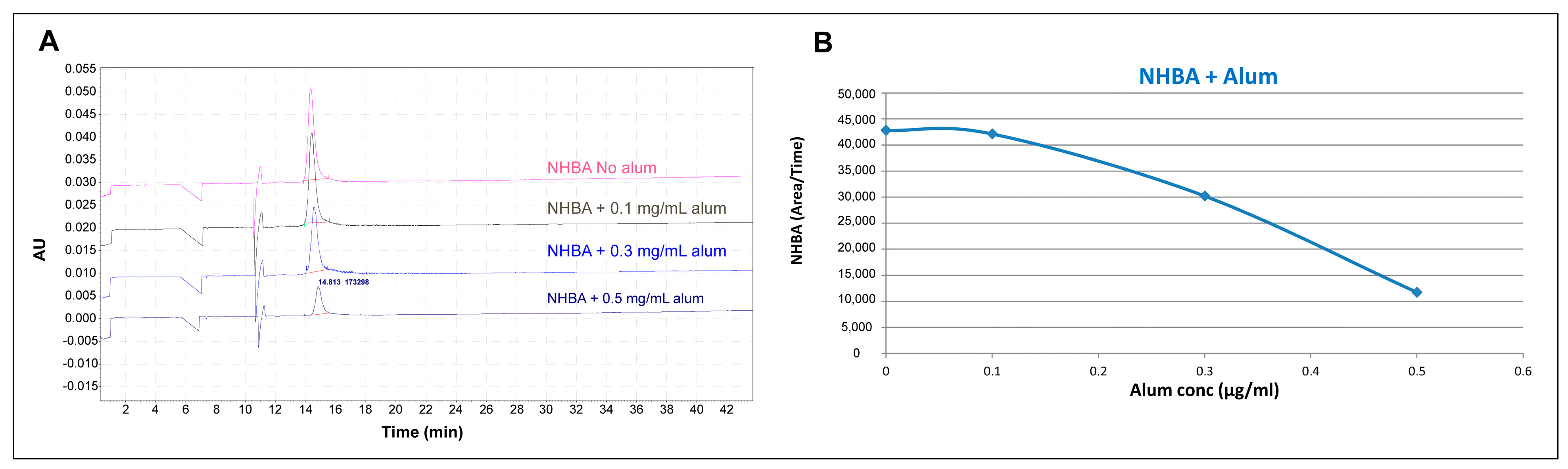
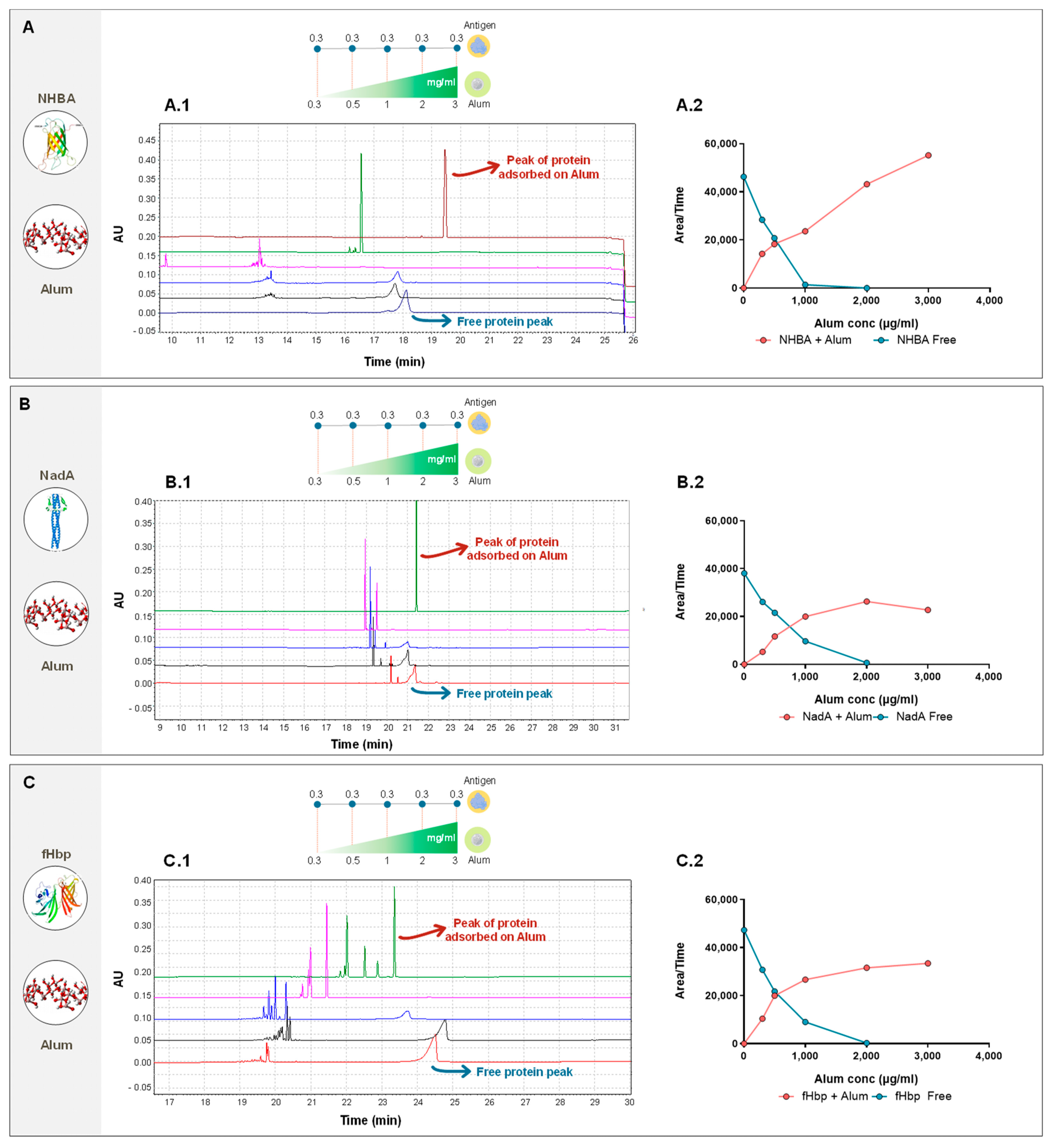
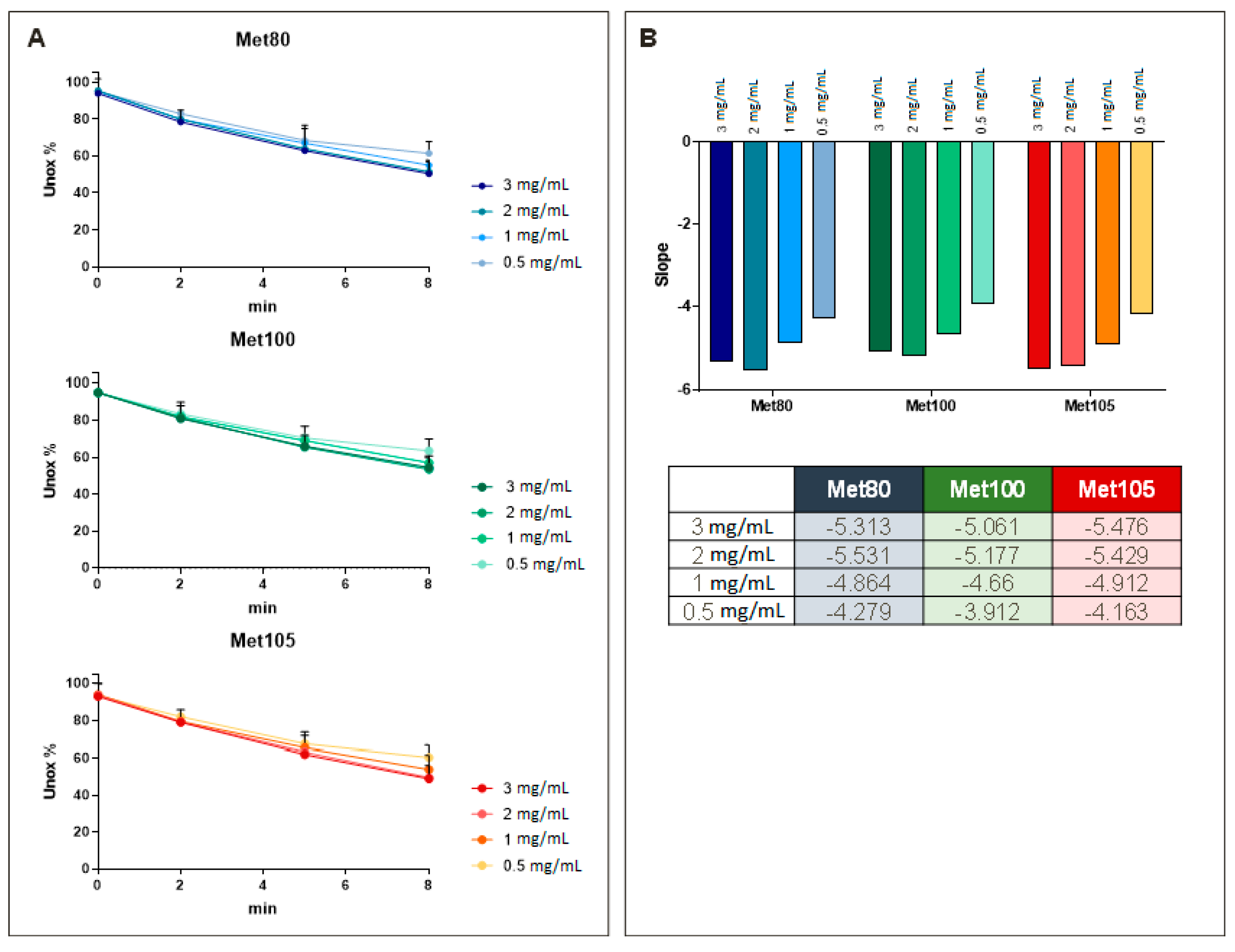
3. Results
- ▪
- TRIS Acetate pH 8.0
- ▪
- Molarity of TRIS Acetate buffer (50 mM)
- ▪
- SDS concentration (15 mM)
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Delany, I.; Rappuoli, R.; De Gregorio, E. Vaccines for the 21st century. EMBO Mol. Med. 2014, 6, 708–720. [Google Scholar] [CrossRef]
- Guimarães, L.E.; Baker, B.; Perricone, C.; Shoenfeld, Y. Vaccines, adjuvants and autoimmunity. Pharmacol. Res. 2015, 100, 190–209. [Google Scholar] [CrossRef] [PubMed]
- HogenEsch, H.; O’Hagan, D.T.; Fox, C.B. Optimizing the utilization of aluminum adjuvants in vaccines: You might just get what you want. NPJ Vaccines 2018, 3, 51. [Google Scholar] [CrossRef] [PubMed]
- Glenny, A.T.; Pope, C.G.; Waddington, H.; Wallace, U. Immunological notes. XVII–XXIV. J. Pathol. Bacteriol. 1926, 29, 31–40. [Google Scholar] [CrossRef]
- Facciolà, A.; Visalli, G.; Laganà, A.; Di Pietro, A. An Overview of Vaccine Adjuvants: Current Evidence and Future Perspectives. Vaccines 2022, 10, 819. [Google Scholar] [CrossRef] [PubMed]
- Levesque, P.M.; de Alwis, U. Mechanism of adsorption of three recombinant Streptococcus pneumoniae (Sp) vaccine antigens by an aluminum adjuvant. Hum. Vaccines 2005, 1, 70–73. [Google Scholar] [CrossRef] [PubMed]
- Hem, S.L.; Hogenesch, H. Relationship between physical and chemical properties of aluminum-containing adjuvants and immunopotentiation. Expert. Rev. Vaccines 2007, 6, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Clapp, T.; Siebert, P.; Chen, D.; Jones Braun, L. Vaccines with aluminum-containing adjuvants: Optimizing vaccine efficacy and thermal stability. J. Pharm. Sci. 2011, 100, 388–401. [Google Scholar] [CrossRef]
- Rappuoli, R.a.L.V. I Vaccini Dell’era Globale; Zanichelli: Bologna, Italy, 2021. [Google Scholar]
- Skibinski, D.A.; Baudner, B.C.; Singh, M.; O’Hagan, D.T. Combination vaccines. J. Glob. Infect. Dis. 2011, 3, 63–72. [Google Scholar] [CrossRef]
- Smith, J.; Lipsitch, M.; Almond, J.W. Vaccine production, distribution, access, and uptake. Lancet 2011, 378, 428–438. [Google Scholar] [CrossRef]
- Hansen, B.; Sokolovska, A.; HogenEsch, H.; Hem, S.L. Relationship between the strength of antigen adsorption to an aluminum-containing adjuvant and the immune response. Vaccine 2007, 25, 6618–6624. [Google Scholar] [CrossRef] [PubMed]
- Laera, D.; HogenEsch, H.; O’Hagan, D.T. Aluminum Adjuvants-‘Back to the Future’. Pharmaceutics 2023, 15, 1884. [Google Scholar] [CrossRef] [PubMed]
- Laera, D.; Scarpellini, C.; Tavarini, S.; Baudner, B.; Marcelli, A.; Pergola, C.; Meppen, M.; O’Hagan, D.T. Maturation of Aluminium Adsorbed Antigens Contributes to the Creation of Homogeneous Vaccine Formulations. Vaccines 2023, 11, 155. [Google Scholar] [CrossRef]
- Kumru, O.S.; Joshi, S.B.; Smith, D.E.; Middaugh, C.R.; Prusik, T.; Volkin, D.B. Vaccine instability in the cold chain: Mechanisms, analysis and formulation strategies. Biologicals 2014, 42, 237–259. [Google Scholar] [CrossRef] [PubMed]
- Hamborg, M.; Foged, C. Characterizing the Association Between Antigens and Adjuvants. In Subunit Vaccine Delivery; Foged, C., Rades, T., Perrie, Y., Hook, S., Eds.; Springer: New York, NY, USA, 2015; pp. 413–426. [Google Scholar] [CrossRef]
- Serruto, D.; Bottomley, M.J.; Ram, S.; Giuliani, M.M.; Rappuoli, R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine 2012, 30 (Suppl. S2), B87–B97. [Google Scholar] [CrossRef] [PubMed]
- Malito, E.; Biancucci, M.; Faleri, A.; Ferlenghi, I.; Scarselli, M.; Maruggi, G.; Lo Surdo, P.; Veggi, D.; Liguori, A.; Santini, L.; et al. Structure of the meningococcal vaccine antigen NadA and epitope mapping of a bactericidal antibody. Proc. Natl. Acad. Sci. USA 2014, 111, 17128–17133. [Google Scholar] [CrossRef] [PubMed]
- Pizza, M.; Rappuoli, R. Neisseria meningitidis: Pathogenesis and immunity. Curr. Opin. Microbiol. 2015, 23, 68–72. [Google Scholar] [CrossRef]
- Tettelin, H.; Saunders, N.J.; Heidelberg, J.; Jeffries, A.C.; Nelson, K.E.; Eisen, J.A.; Ketchum, K.A.; Hood, D.W.; Peden, J.F.; Dodson, R.J.; et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 2000, 287, 1809–1815. [Google Scholar] [CrossRef]
- Oszwałdowski, S.; Zawistowska-Gibuła, K.; Roberts, K.P. Capillary electrophoretic separation of nanoparticles. Anal. Bioanal. Chem. 2011, 399, 2831–2842. [Google Scholar] [CrossRef]
- Vessely, C.; Estey, T.; Randolph, T.W.; Henderson, I.; Cooper, J.; Nayar, R.; Braun, L.J.; Carpenter, J.F. Stability of a trivalent recombinant protein vaccine formulation against botulinum neurotoxin during storage in aqueous solution. J. Pharm. Sci. 2009, 98, 2970–2993. [Google Scholar] [CrossRef]
- Kim, H.R.; Andrieux, K.; Delomenie, C.; Chacun, H.; Appel, M.; Desmaële, D.; Taran, F.; Georgin, D.; Couvreur, P.; Taverna, M. Analysis of plasma protein adsorption onto PEGylated nanoparticles by complementary methods: 2-DE, CE and Protein Lab-on-chip system. Electrophoresis 2007, 28, 2252–2261. [Google Scholar] [CrossRef]
- Schmitt-Kopplin, P. Capillary Electrophoresis; Methods and Protocols. Preface; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2008; Volume 384. [Google Scholar] [CrossRef]
- Hoiczyk, E.; Roggenkamp, A.; Reichenbecher, M.; Lupas, A.; Heesemann, J. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. Embo J. 2000, 19, 5989–5999. [Google Scholar] [CrossRef]
- Shank-Retzlaff, M.; Wang, F.; Morley, T.; Anderson, C.; Hamm, M.; Brown, M.; Rowland, K.; Pancari, G.; Zorman, J.; Lowe, R.; et al. Correlation between mouse potency and in vitro relative potency for human papillomavirus Type 16 virus-like particles and Gardasil vaccine samples. Hum. Vaccines 2005, 1, 191–197. [Google Scholar] [CrossRef]
- Hem, S.L.; HogenEsch, H.; Middaugh, C.R.; Volkin, D.B. Preformulation studies—The next advance in aluminum adjuvant-containing vaccines. Vaccine 2010, 28, 4868–4870. [Google Scholar] [CrossRef]
- Morefield, G.L.; Sokolovska, A.; Jiang, D.; HogenEsch, H.; Robinson, J.P.; Hem, S.L. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine 2005, 23, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Domina, M.; Lanza Cariccio, V.; Benfatto, S.; Venza, M.; Venza, I.; Donnarumma, D.; Bartolini, E.; Borgogni, E.; Bruttini, M.; Santini, L.; et al. Epitope Mapping of a Monoclonal Antibody Directed against Neisserial Heparin Binding Antigen Using Next Generation Sequencing of Antigen-Specific Libraries. PLoS ONE 2016, 11, e0160702. [Google Scholar] [CrossRef] [PubMed]
- Vacca, I.; Del Tordello, E.; Gasperini, G.; Pezzicoli, A.; Di Fede, M.; Rossi Paccani, S.; Marchi, S.; Mubaiwa, T.D.; Hartley-Tassell, L.E.; Jennings, M.P.; et al. Neisserial Heparin Binding Antigen (NHBA) Contributes to the Adhesion of Neisseria meningitidis to Human Epithelial Cells. PLoS ONE 2016, 11, e0162878. [Google Scholar] [CrossRef]
- Tullius, T.D.; Dombroski, B.A. Hydroxyl radical “footprinting”: High-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. USA 1986, 83, 5469–5473. [Google Scholar] [CrossRef] [PubMed]
- Chance, M.R.; Sclavi, B.; Woodson, S.A.; Brenowitz, M. Examining the conformational dynamics of macromolecules with time-resolved synchrotron X-ray ‘footprinting’. Structure 1997, 5, 865–869. [Google Scholar] [CrossRef] [PubMed]
- Hanai, R.; Wang, J.C. Protein footprinting by the combined use of reversible and irreversible lysine modifications. Proc. Natl. Acad. Sci. USA 1994, 91, 11904–11908. [Google Scholar] [CrossRef]
- Horinishi, H.; Hachimori, Y.; Kurihara, K.; Shibata, K. States of Amino Acid Residues in Proteins. 3. Histidine Residues in Insulin, Lysozyme, Albumin and Proteinases as Determined with a New Reagent of Diazo-i-h-Tetrazole. Biochim. Biophys. Acta 1964, 86, 477–489. [Google Scholar] [PubMed]
- Wang, L.; Chance, M.R. Protein Footprinting Comes of Age: Mass Spectrometry for Biophysical Structure Assessment. Mol. Cell. Proteom. 2017, 16, 706–716. [Google Scholar] [CrossRef] [PubMed]
- Fenton, H.J.H. LXXIII.—Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 1894, 65, 899–910. [Google Scholar] [CrossRef]
- Jones, L.S.; Peek, L.J.; Power, J.; Markham, A.; Yazzie, B.; Middaugh, C.R. Effects of Adsorption to Aluminum Salt Adjuvants on the Structure and Stability of Model Protein Antigens. J. Biol. Chem. 2005, 280, 13406–13414. [Google Scholar] [CrossRef]






| Antigen | mAb Clone | Isotype | Source Purity | Epitope Recognized |
|---|---|---|---|---|
| fHbp | 12C1/D7 | IgG2b | Hybridoma purified | Conformational |
| NadA | 6E3/29 | IgG1 | Hybridoma purified | Conformational |
| NHBA | 10E8/A5 | IgG2b | Hybridoma purified | Conformational |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Corrado, A.; Toppazzini, M.; Vadi, A.; Malzone, C.; Galasso, R.; Donati, A.; De Ricco, R.; Berti, F. Analytical Insights into Protein–Alum Interactions and Their Impact on Conformational Epitope. Pharmaceutics 2024, 16, 420. https://doi.org/10.3390/pharmaceutics16030420
Corrado A, Toppazzini M, Vadi A, Malzone C, Galasso R, Donati A, De Ricco R, Berti F. Analytical Insights into Protein–Alum Interactions and Their Impact on Conformational Epitope. Pharmaceutics. 2024; 16(3):420. https://doi.org/10.3390/pharmaceutics16030420
Chicago/Turabian StyleCorrado, Alessio, Mila Toppazzini, Alessandro Vadi, Carmine Malzone, Rosy Galasso, Alessandro Donati, Riccardo De Ricco, and Francesco Berti. 2024. "Analytical Insights into Protein–Alum Interactions and Their Impact on Conformational Epitope" Pharmaceutics 16, no. 3: 420. https://doi.org/10.3390/pharmaceutics16030420






