L-Citrullinato-Bipyridine and L-Citrullinato-Phenanthroline Mixed Copper Complexes: Synthesis, Characterization and Potential Anticancer Activity
Abstract
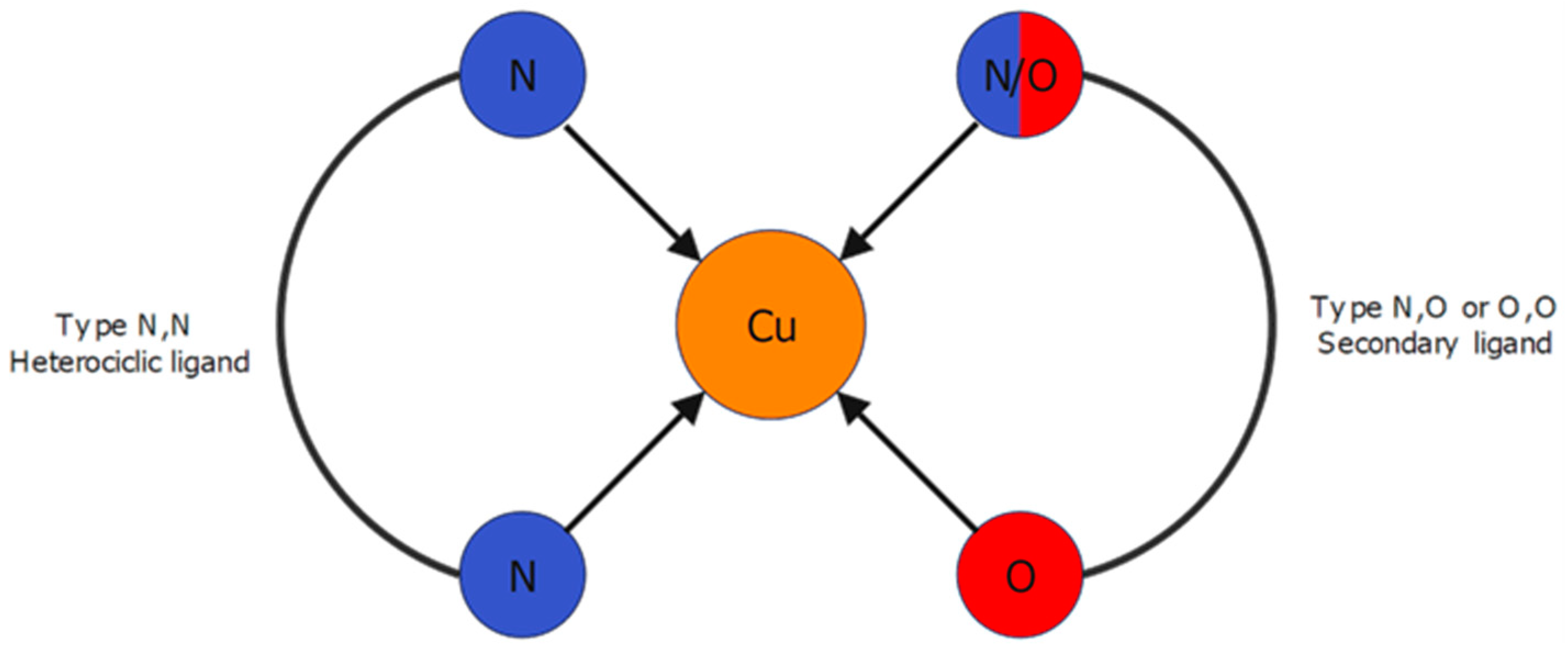
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Equipment
2.3. Software Crystallography
2.4. Synthesis of Complexes
2.4.1. Synthesis of [Cu(L-Citr)(bipy)(NO3)]n (Complex 1)
2.4.2. Synthesis of [Cu(L-Citr)(phen)(NO3)]n (Complex 2)
2.5. Quantum Mechanical Calculations
2.6. Docking Studies with AutoDock 4.2
2.7. In-Vitro Assays
2.7.1. Affinity for CT-DNA
2.7.2. Fluorometric Assays
2.7.3. Interaction with pUC19 Plasmid
2.7.4. Cytotoxic Assays
3. Results
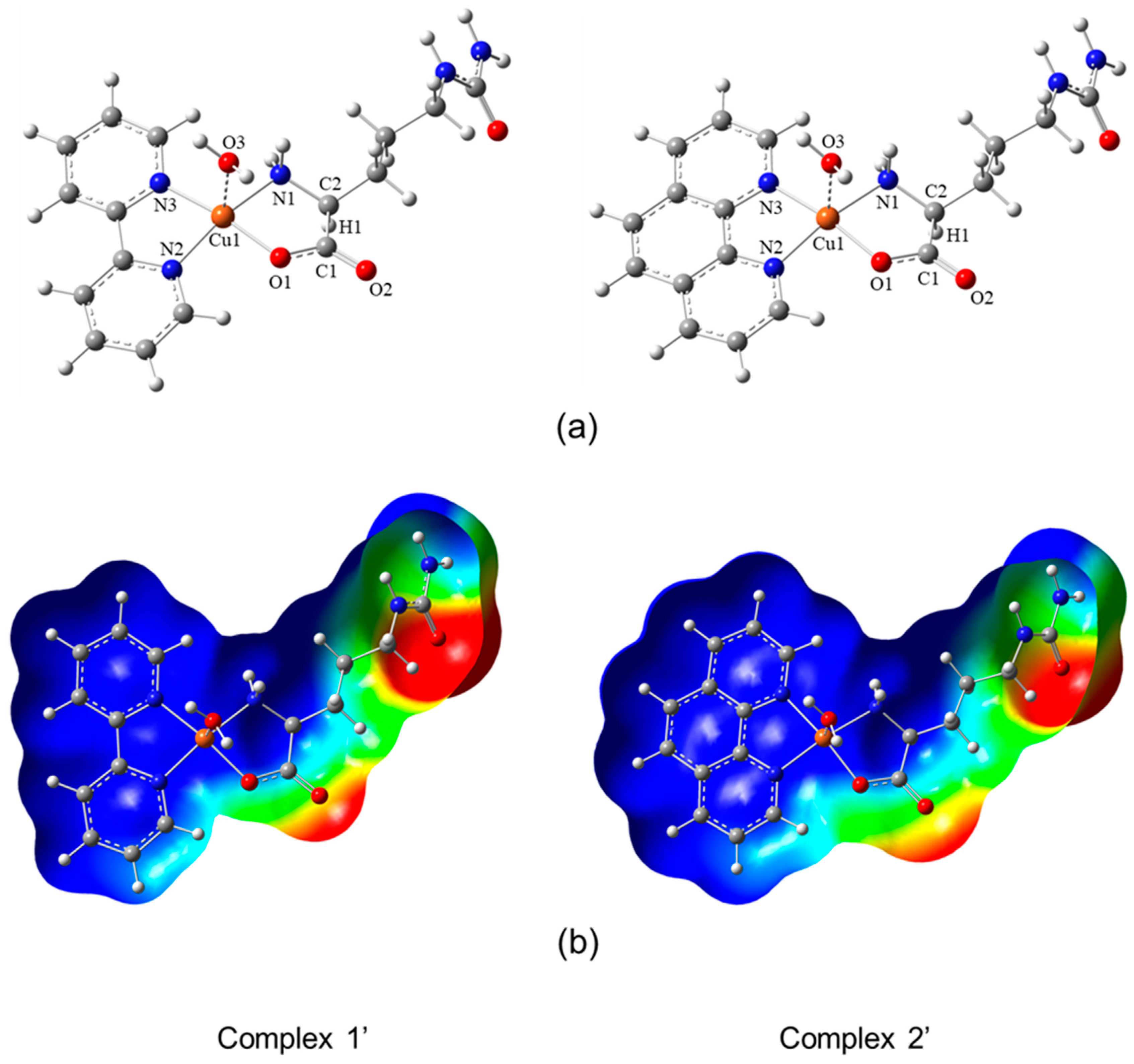
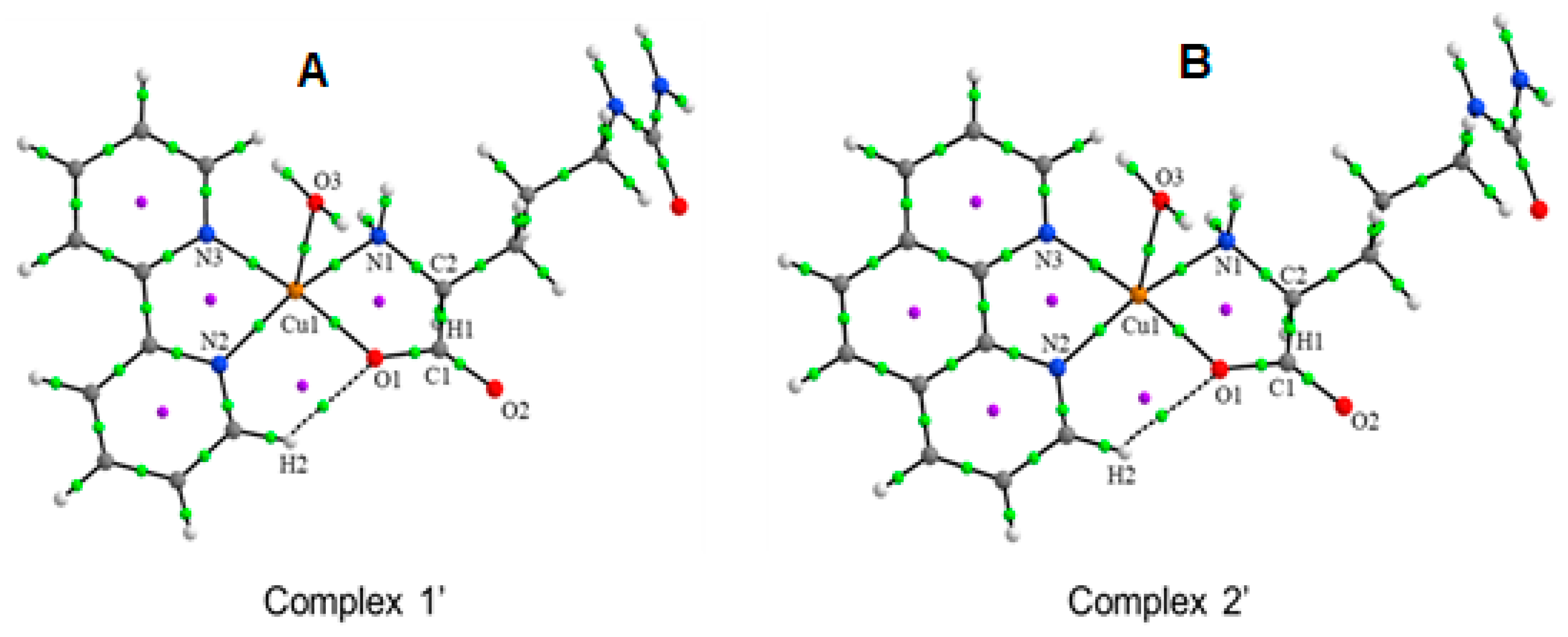
3.1. Molecular Structures
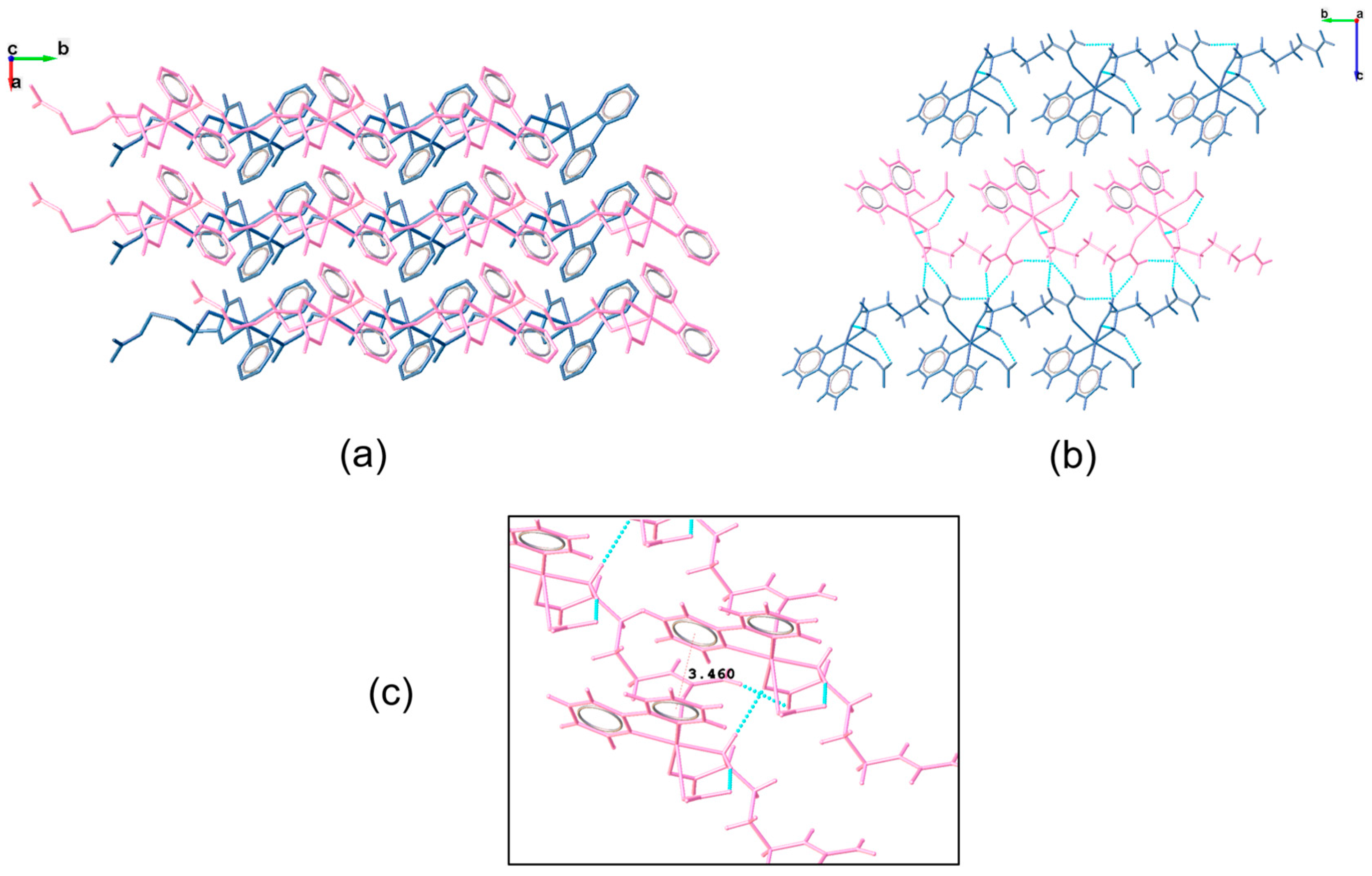
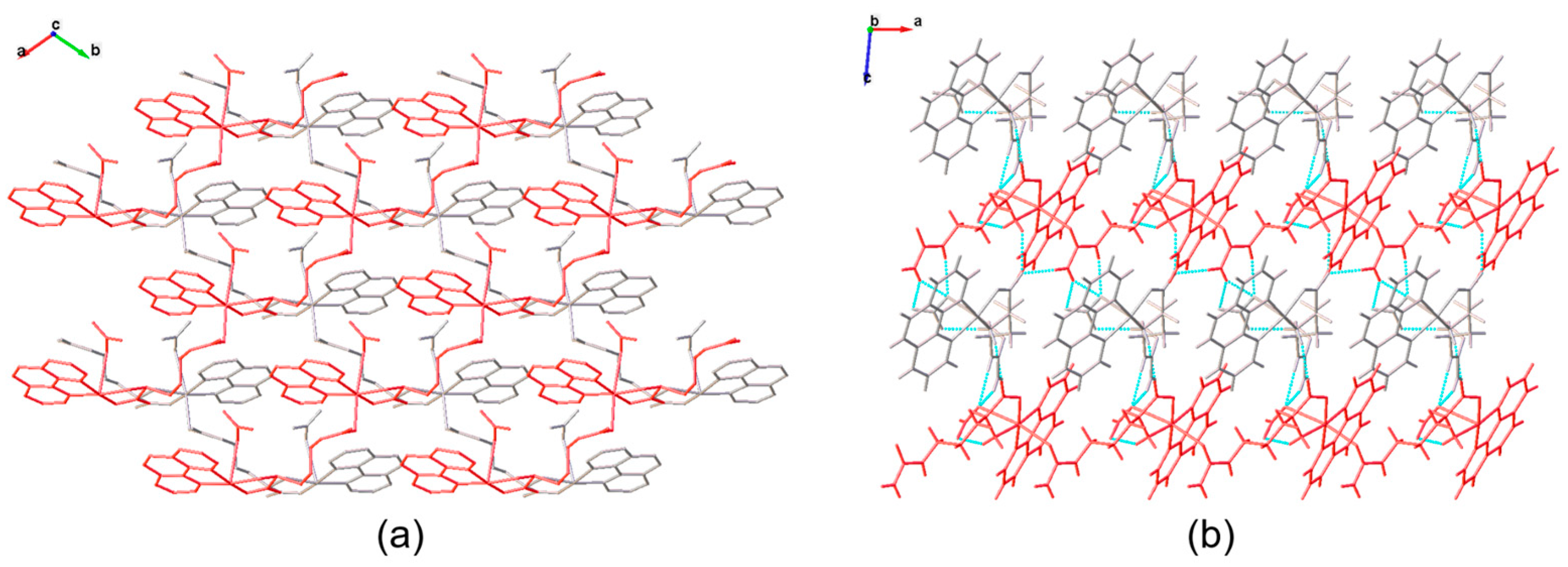
3.2. Molecular Structure and Non-Covalent Interactions
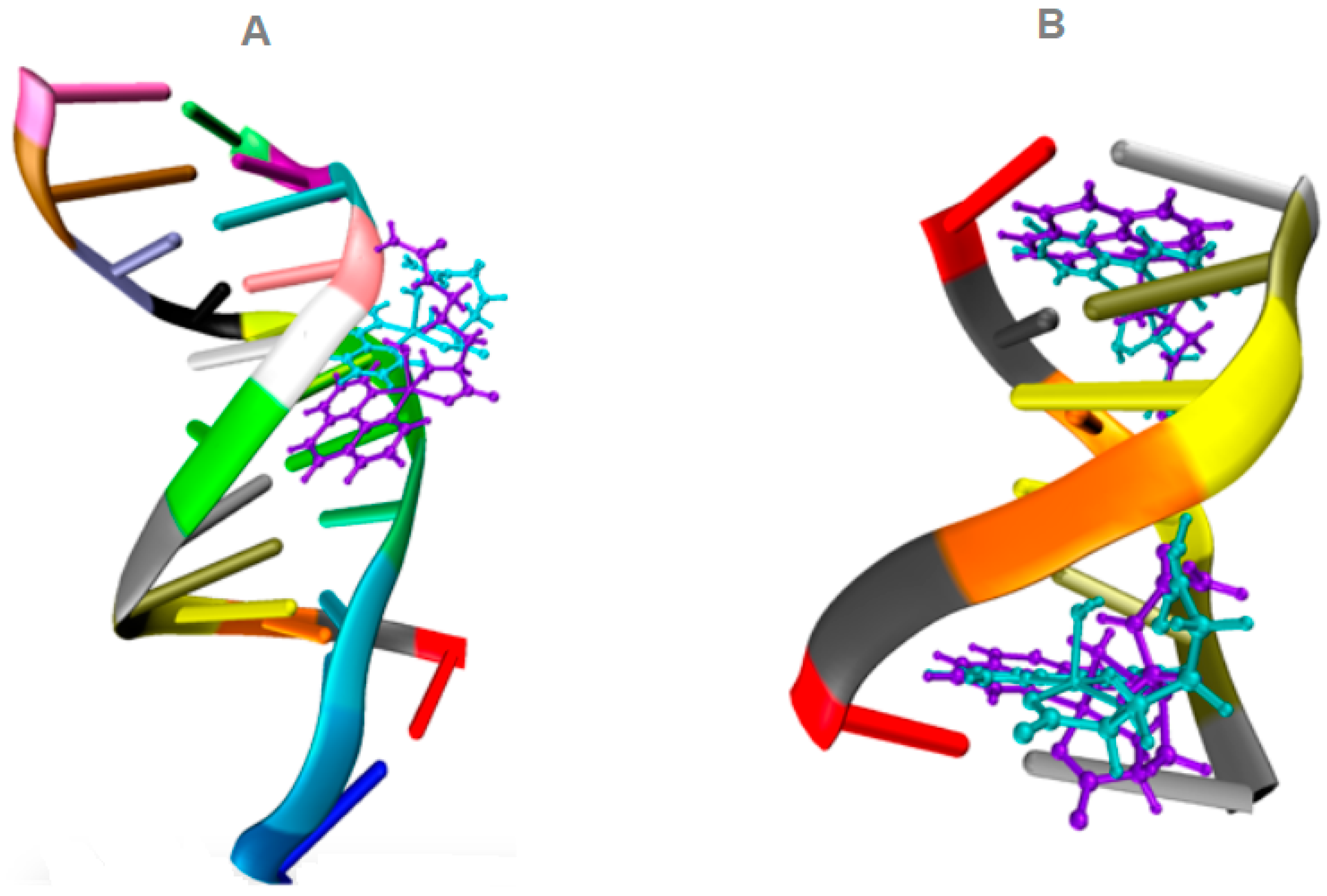
3.3. Molecular Docking
3.4. In-Vitro Experiments
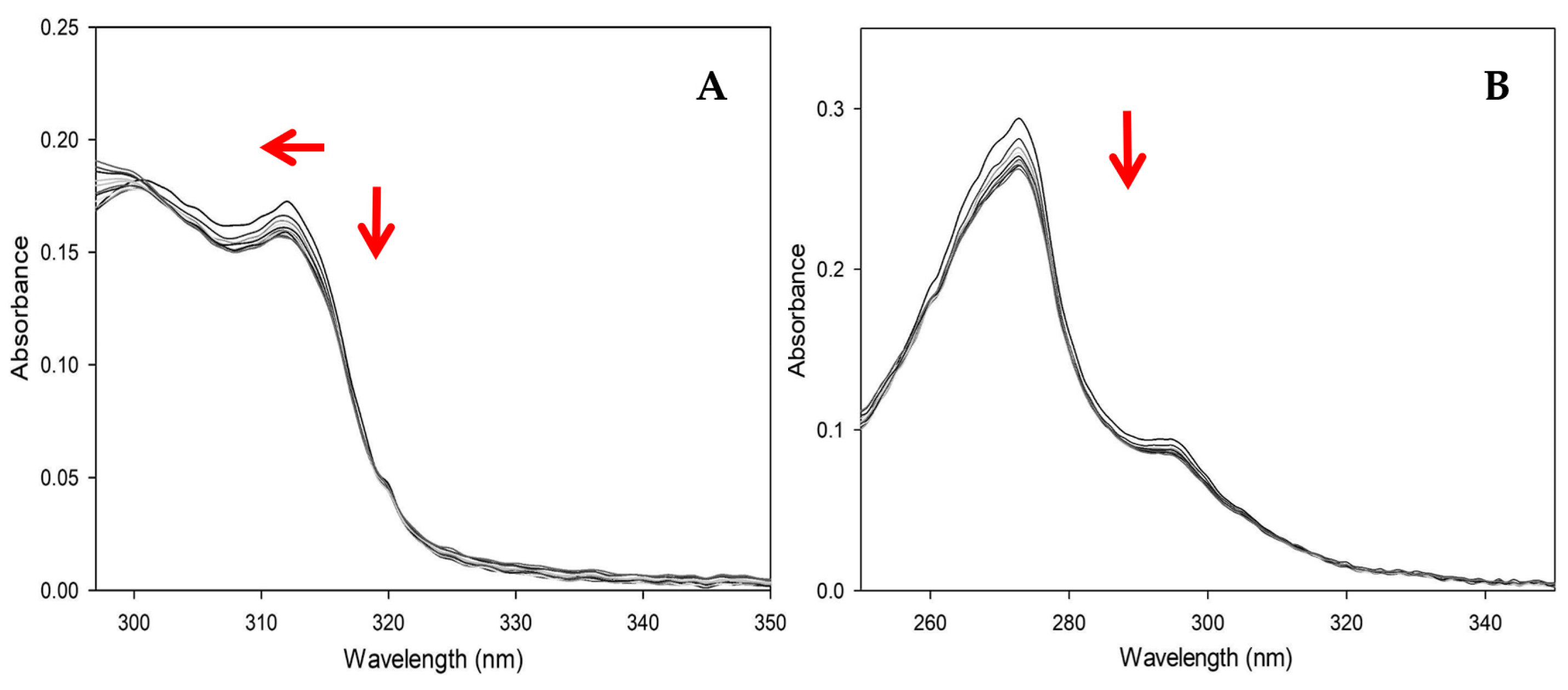
3.4.1. UV-Vis Experiments with CT-DNA
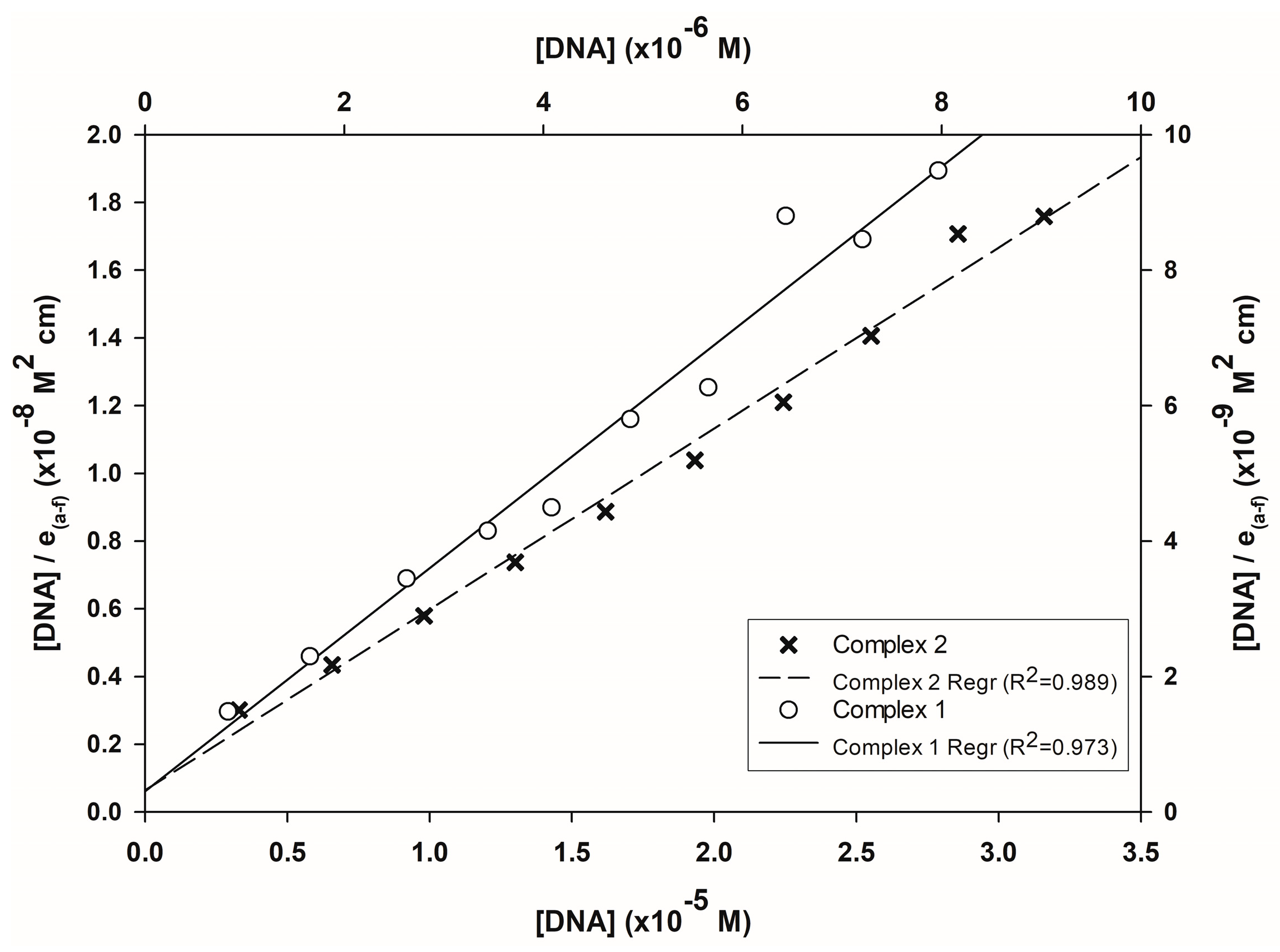
3.4.2. Fluorometric Experiments with CT-DNA
3.4.3. Interaction with pUC19
3.4.4. Cytotoxic Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goodsell, D.S. The Molecular Perspective: Cisplatin. Oncologist 2006, 11, 316–317. [Google Scholar] [CrossRef] [PubMed]
- Alderden, R.A.; Hall, M.D.; Hambley, T.W.; Kauffman, G.B. Chemistry for Everyone: The Discovery and Development of Cisplatin Products of Chemistry. J. Chem. Educ. 2006, 8, 728–734. [Google Scholar] [CrossRef]
- Muggia, F.; Leone, R.; Bonetti, A. Platinum and Other Heavy Metal Coordinating Compounds in Cancer Chemotherapy: Overview of Verona ISPCC XI. In Proceedings of the XI International Symposium of Platinum Coordination Compounds in Cancer Chemotherapy, Verona, Italy, 11–14 October 2012; pp. 416–421. [Google Scholar]
- Kennedy, I.C.S.; Fitzharris, B.M.; Coils, B.M.; Atkinson, C.H. Carboplatin Is Ototoxic. Cancer Chemother. Pharmacol. 1990, 26, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Crawford, J.; Dale, D.C.; Lyman, G.H. Chemotherapy-Induced Neutropenia: Risks, Consequences, and New Directions for Its Management. Cancer 2004, 100, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Meistrich, M.L. Effects of Chemotherapy and Radiotherapy on Spermatogenesis in Humans. Fertil. Steril. 2013, 100, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Love, R.R.; Leventhal, H.; Easterling, D.V.; Nerenz, D.R. Side Effects and Emotional Distress during Cancer Chemotherapy. Cancer 1989, 63, 604–612. [Google Scholar] [CrossRef]
- Ward, R.A.; Fawell, S.; Floc’H, N.; Flemington, V.; McKerrecher, D.; Smith, P.D. Challenges and Opportunities in Cancer Drug Resistance. Chem. Rev. 2021, 121, 3297–3351. [Google Scholar] [CrossRef] [PubMed]
- Kenny, R.G.; Marmion, C.J. Toward Multi-Targeted Platinum and Ruthenium Drugs—A New Paradigm in Cancer Drug Treatment Regimens? Chem. Rev. 2019, 119, 1058–1137. [Google Scholar] [CrossRef] [PubMed]
- Anthony, E.J.; Bolitho, E.M.; Bridgewater, H.E.; Carter, O.W.L.; Donnelly, J.M.; Imberti, C.; Lant, E.C.; Lermyte, F.; Needham, R.J.; Palau, M.; et al. Metallodrugs Are Unique: Opportunities and Challenges of Discovery and Development. Chem. Sci. 2020, 11, 12888–12917. [Google Scholar] [CrossRef]
- Mjos, K.D.; Orvig, C. Metallodrugs in Medicinal Inorganic Chemistry. Chem. Rev. 2014, 114, 4540–4563. [Google Scholar] [CrossRef]
- Masuri, S.; Vaňhara, P.; Cabiddu, M.G.; Moráň, L.; Havel, J.; Cadoni, E.; Pivetta, T. Copper (II) Phenanthroline-Based Complexes as Potential Anticancer Drugs: A Walkthrough on the Mechanisms of Action. Molecules 2022, 27, 49. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Copper Complexes as Anticancer Agents Targeting Topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef] [PubMed]
- Marzano, C.; Pellei, M.; Tisato, F.; Santini, C. Anti-Cancer Agents in Medicinal Chemistry. Anticancer Agents Med. Chem. 2009, 9, 185–211. [Google Scholar] [CrossRef] [PubMed]
- Farkas, E.; Sóvágó, I. Metal Complexes of Amino Acids and Peptides. In Amino Acids, Peptides and Proteins; Royal Society of Chemistry Books: Cambridge, UK, 2007; Volume 36, pp. 287–345. [Google Scholar] [CrossRef]
- Rimando, A.M.; Perkins-Veazie, P.M. Determination of Citrulline in Watermelon Rind. J. Chromatogr. A 2005, 1078, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Curis, E.; Crenn, P.; Cynober, L. Citrulline and the Gut. Curr. Opin. Clin. Nutr. Metab. Care 2007, 10, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Aguayo, E.; Martínez-Sánchez, A.; Fernández-Lobato, B.; Alacid, F. L-Citrulline: A Non-Essential Amino Acid with Important Roles in Human Health. Appl. Sci. 2021, 11, 3293. [Google Scholar] [CrossRef]
- Alghamdi, M.; Alasmari, D.; Assiri, A.; Mattar, E.; Aljaddawi, A.A.; Alattas, S.G.; Redwan, E.M. An Overview of the Intrinsic Role of Citrullination in Autoimmune Disorders. J. Immunol. Res. 2019, 2019, 7592851. [Google Scholar] [CrossRef] [PubMed]
- Maric, S.; Restin, T.; Muff, J.L.; Camargo, S.M.; Guglielmetti, L.C.; Holland-cunz, S.G.; Crenn, P.; Vuille-dit-bille, R.N. Citrulline, Biomarker of Enterocyte Functional Mass and Dietary Supplement. Metabolism, Transport, and Current Evidence for Clinical Use. Nutrients 2021, 13, 2794. [Google Scholar] [CrossRef] [PubMed]
- Eren, C.Y.; Gurer, H.G.; Gursoy, O.O.; Sezer, C.V. Antitumor Effects of L-Citrulline on Hela Cervical Cancer Cell Lines. Anticancer Agents Med. Chem. 2022, 22, 3157–3162. [Google Scholar] [CrossRef]
- Fragkos, K.C.; Forbes, A. Was Citrulline First a Laxative Substance?: The Truth about Modern Citrulline and Its Isolation. Nihon Ishigaku Zasshi 2011, 57, 275–292. [Google Scholar]
- Wada, M. On the Occurrence of a New Amino Acid in Watermelon, Citrullus Vulgaris, Schrad. Bull. Agric. Chem. Soc. Jpn. 1930, 6, 32–34. [Google Scholar] [CrossRef]
- Ramírez-Contreras, D.; García-García, A.; Mendoza, A.; Serrano-de la Rosa, L.E.; Sánchez-Gaytán, B.L.; Melendez, F.J.; Castro, M.E.; González-Vergara, E. D,L-Citrullinato-Bipyridine Copper Complex: Experimental and Theoretical Characterization. Crystals 2023, 13, 1391. [Google Scholar] [CrossRef]
- Mascaliovas, B.Z.; Bergamini, F.R.G.; Cuin, A.; Corbi, P.P. Synthesis and Crystal Structure of a Palladium(II) Complex with the Amino Acid L-Citrulline. Powder Diffr. 2015, 30, 357–361. [Google Scholar] [CrossRef]
- Trikha, K.C.; Nair, B.C.; Singh, R.P. Complexation of Bivalent Metal Ions with Amino Acids. I. L-Citrulline Complexes. Indian. J. Chem. 1968, 6, 532. [Google Scholar]
- Clarke, E.R.; Martell, A.E. Metal Chelates of Arginine and Related Ligands. J. Inorg. Nucl. Chem. 1970, 32, 911–926. [Google Scholar] [CrossRef]
- Yamauchi, O.; Sakurai, T.; Nakahara, A. Histidine-Containing Ternary Amino Acid-Copper(II) Complexes. Syntheses and Properties. J. Am. Chem. Soc. 1979, 101, 4164–4172. [Google Scholar] [CrossRef]
- Ganadu, M.L.; Leoni, V.; Crisponi, G.; Nurchi, V. An Investigation on the Interaction between Palladium(II) and L-Citrulline by 1H and 13C NMR Spectroscopy and Potentiometry. Polyhedron 1991, 10, 333–336. [Google Scholar] [CrossRef]
- Singh, M.; Shankar, V.; Singh, D.; Krishna, V. Chelation and Stabilization Properties of Citrulline and Uracil with Hg(II) as a Heavy Metal Ion in Solution. Chem. Sci. Trans. 2017, 6, 646–652. [Google Scholar] [CrossRef]
- Singh, M.; Sinha, S.; Krishna, V. Computed Distribution of Quaternary Complexes of Cu(II), Zn(II) Co(II) and Ni(II) with Citrulline and Tryphtophan as Primary Ligand and Thymine as Secondary Ligand. Proc. Natl. Acad. Sci. India Sect. A Phys. Sci. 2021, 91, 1–7. [Google Scholar] [CrossRef]
- Tisato, F.; Marzano, C.; Porchia, M.; Pellei, M.; Santini, C. Copper in Diseases and Treatments, and Copper-Based Anticancer Strategies. Med. Res. Rev. 2010, 30, 708–749. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Proschak, E.; Stark, H.; Merk, D. Polypharmacology by Design: A Medicinal Chemist’s Perspective on Multitargeting Compounds. J. Med. Chem. 2019, 62, 420–444. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Azuara, L.; Bastian, G.; Bravo-Gómez, M.E.; Cañas, R.C.; Flores-Alamo, M.; Fuentes, I.; Mejia, C.; García-Ramos, J.C.; Serrano, A. Abstract CT408: Phase I Study of One Mixed Chelates Copper (II) Compound, Casiopeína CasIIIia with Antitumor Activity and Its Mechanism of Action. Cancer Res. 2014, 74 (Suppl. S19), CT408. [Google Scholar] [CrossRef]
- Aguilar-Jiménez, Z.; Espinoza-Guillén, A.; Resendiz-Acevedo, K.; Fuentes-Noriega, I.; Mejía, C.; Ruiz-Azuara, L. The Importance of Being Casiopeina as Polypharmacologycal Profile (Mixed Chelate–Copper (II) Complexes and Their In Vitro and In Vivo Activities). Inorganics 2023, 11, 394. [Google Scholar] [CrossRef]
- Da Costa Ferreira, A.M.; Hureau, C.; Facchin, G. Bioinorganic Chemistry of Copper: From Biochemistry to Pharmacology. Inorganics 2024, 12, 97. [Google Scholar] [CrossRef]
- Ramírez-Contreras, D.; García-García, A.; Sánchez-Gaytán, B.L.; Serrano-de la Rosa, L.E.; Melendez, F.J.; Choquesillo-Lazarte, D.; Rodríguez-Diéguez, A.; Castro, M.E.; González-Vergara, E. Bis-Citrullinato Copper(II) Complex: Synthesis, Crystal Structure, and Non-Covalent Interactions. Crystals 2022, 12, 1386. [Google Scholar] [CrossRef]
- American Type Cell Culture. Animal Cell Culture Guide. Available online: https://www.atcc.org/resources/culture-guides/animal-cell-culture-guide (accessed on 28 February 2023).
- CrysAlis, C.C.D. CRYSALIS RED in XCALIBUR PX and KUMA KM-4-CCD Software; Oxford Diffraction Ltd.: Oxfordshire, UK, 2009. [Google Scholar]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. C Struct. Chem. 2015, 71 Pt 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Hohenberg, P.; Kohn, W. Inhomogeneous Electron Gas. Phys. Rev. 1964, 136, B864–B871. [Google Scholar] [CrossRef]
- Adamo, C.; Jacquemin, D. The Calculations of Excited-State Properties with Time-Dependent Density Functional Theory. Chem. Soc. Rev. 2013, 42, 845–856. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab Initio Effective Core Potentials for Molecular Calculations. Potentials for the Transition Metal Atoms Sc to Hg. J. Chem. Phys. 1985, 82. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R. Gaussian 16; Revision, B.01; Gaussian Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Dennington, R.D.; Keith, T.A.; Millam, J.M. Gauss View, Version 6.0.16; Semi-Chem Inc.: Shawnee Mission, UK, 2016. [Google Scholar]
- Keith, T.A. TK Gristmill Software; Version 19.02.13; AIMAll: Overland Park, KS, USA, 2019. [Google Scholar]
- Morris, G.M.; Ruth, H.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Software News and Updates AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Drew, H.R.; Wing, R.M.; Takano, T.; Broka, C.; Tanaka, S.; Itakura, K.; Dickerson, R.E. Structure of a B-DNA Dodecamer. Conformation and Dynamics. Proc. Natl. Acad. Sci. USA 1981, 78, 2179–2183. [Google Scholar] [CrossRef]
- Lipscomb, L.A.; Peek, M.E.; Zhou, F.X.; Bertrand, J.A.; VanDerveer, D.; Williams, L.D. Diversity of Water Ring Size at DNA Interfaces: Hydration and Dynamics of DNA-Anthracycline Complexes. Biochemistry 1994, 33, 3649–3659. [Google Scholar] [CrossRef]
- Corona-Motolinia, N.D.; Martínez-Valencia, B.; Noriega, L.; Sánchez-Gaytán, B.L.; Mendoza, A.; Meléndez-Bustamante, F.J.; Castro, M.E.; González-Vergara, E. Ternary Copper Complex of L-Glutamine and Phenanthroline as Counterions of Cyclo-Tetravanadate Anion: Experimental–Theoretical Characterization and Potential Antineoplastic Activity. Metals 2021, 11, 1541. [Google Scholar] [CrossRef]
- Vazquez-Rodriguez, S.; Ramírez-Contreras, D.; Noriega, L.; García-García, A.; Sánchez-Gaytán, B.L.; Melendez, F.J.; Castro, M.E.; de Azevedo, W.F.; González-Vergara, E. Interaction of Copper Potential Metallodrugs with TMPRSS2: A Comparative Study of Docking Tools and Its Implications on COVID-19. Front. Chem. 2023, 11, 1128859. [Google Scholar] [CrossRef]
- Discovery Studio. Dassault Systèmes BIOVIA; Dassault Systèmes: Vélizy-Villacoublay, France, 2016. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, S.A.; Thomas, C.A.; Doty, P. A Further Examination of the Molecular Weight and Size of Desoxypentose Nucleic Acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H.; Meehan, T. Polycyclic Aromatic Hydrocarbons Physically Intercalate into Duplex Regions of Denatured DNA. Biochemistry 1987, 26, 6369–6392. [Google Scholar] [CrossRef] [PubMed]
- Geall, A.J.; Blagbrough, I.S. Rapid and Sensitive Ethidium Bromide Fluorescence Quenching Assay of Polyamine Conjugate-DNA Interactions for the Analysis of Lipoplex Formation in Gene Therapy. J. Pharm. Biomed. Anal. 2000, 22, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Le Pecq, J.-B.; Paoletti, C. A New Fluorometric Method for RNA and DNA Determination. Anal. Biochem. 1966, 17, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Keizer, J. Nonlinear Fluorescence Quenching and the Origin of Positive Curvature in Stern-Volmer Plots. J. Am. Chem. Soc. 1983, 105, 1494–1498. [Google Scholar] [CrossRef]
- Balagurumoorthy, P.; Adelstein, S.J.; Kassis, A.I. Method to Eliminate Linear DNA from Mixture Containing Nicked Circular, Supercoiled, and Linear Plasmid DNA. Anal. Biochem. 2008, 381, 172–174. [Google Scholar] [CrossRef] [PubMed]
- Pyle, A.M.; Rehmann, J.P.; Meshoyrer, R.; Kumar, C.V.; Turro, N.J.; Barton, J.K. Mixed-Ligand Complexes of Ruthenium: Factors Governing Binding to DNA. J. Am. Chem. Soc. 1989, 111, 3051–3058. [Google Scholar] [CrossRef]
- Roehm, N.W.; Rodgers, G.H.; Hatfield, S.M.; Glasebrook, A.L. An Improved Colorimetric Assay for Cell Proliferation and Viability Utilizing the Tetrazolium Salt XTT. J. Immunol. Methods 1991, 142, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Halcrow, M.A. Jahn–Teller Distortions in Transition Metal Compounds, and Their Importance in Functional Molecular and Inorganic Materials. Chem. Soc. Rev. 2013, 42, 1784–1795. [Google Scholar] [CrossRef] [PubMed]
- Corona-Motolinia, N.D.; Martínez-Valencia, B.; Noriega, L.; Sánchez-Gaytán, B.L.; Méndez-Rojas, M.Á.; Melendez, F.J.; Castro, M.E.; González-Vergara, E. Synthesis, Crystal Structure, and Computational Methods of Vanadium and Copper Compounds as Potential Drugs for Cancer Treatment. Molecules 2020, 25, 4679. [Google Scholar] [CrossRef]
- Leite, S.M.G.; Lima, L.M.P.; Gama, S.; Mendes, F.; Orio, M.; Bento, I.; Paulo, A.; Delgado, R.; Iranzo, O. Copper(II) Complexes of Phenanthroline and Histidine Containing Ligands: Synthesis, Characterization and Evaluation of Their DNA Cleavage and Cytotoxic Activity. Inorg. Chem. 2016, 55, 11801–11814. [Google Scholar] [CrossRef]
- Figueroa-DePaz, Y.; Resendiz-Acevedo, K.; Dávila-Manzanilla, S.G.; García-Ramos, J.C.; Ortiz-Frade, L.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA, a Target of Mixed Chelate Copper(II) Compounds (Casiopeinas®) Studied by Electrophoresis, UV–Vis and Circular Dichroism Techniques. J. Inorg. Biochem. 2022, 231, 111772. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, B.J.M.L.; Brandão, P.; Meireles, M.; Martel, F.; Correia-Branco, A.; Fernandes, D.M.; Santos, T.M.; Félix, V. Synthesis, Structural Characterization, Cytotoxic Properties and DNA Binding of a Dinuclear Copper(II) Complex. J. Inorg. Biochem. 2016, 161, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sun, Q.; Li, J.L.; Jiang, L.; Gu, W.; Liu, X.; Tian, J.L.; Yan, S.P. Two Water-Soluble Copper(II) Complexes: Synthesis, Characterization, DNA Cleavage, Protein Binding Activities and in Vitro Anticancer Activity Studies. J. Inorg. Biochem. 2014, 137, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Lin, D.; Kokot, S. Synchronous Fluorescence, UV-Visible Spectrophotometric, and Voltammetric Studies of the Competitive Interaction of Bis(1,10-Phenanthroline)Copper(II) Complex and Neutral Red with DNA. Anal. Biochem. 2006, 352, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Li, G.Y.; Guan, R.L.; Ji, L.N.; Chao, H. DNA Condensation Induced by Metal Complexes. Coord. Chem. Rev. 2014, 281, 100–113. [Google Scholar] [CrossRef]
- Rajalakshmi, S.; Kiran, M.S.; Nair, B.U. DNA Condensation by Copper(II) Complexes and Their Anti-Proliferative Effect on Cancerous and Normal Fibroblast Cells. Eur. J. Med. Chem. 2014, 80, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Bloomfield, V.A. DNA Condensation. Curr. Opin. Struct. Biol. 1996, 6, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Srdić-Rajić, T.; Zec, M.; Todorović, T.; Anelković, K.; Radulović, S. Non-Substituted N-Heteroaromatic Selenosemicarbazone Metal Complexes Induce Apoptosis in Cancer Cells via Activation of Mitochondrial Pathway. Eur. J. Med. Chem. 2011, 46, 3734–3747. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.L.; Shen, W.Y.; Chen, Z.F.; Zhao, L.F.; Qin, Q.P.; Yu, Y.C.; Liang, H. Oxoaporphine Metal Complexes (CoII, NiII, ZnII) with High Antitumor Activity by Inducing Mitochondria- Mediated Apoptosis and S-Phase Arrest in HepG2. Sci. Rep. 2017, 7, 46056. [Google Scholar] [CrossRef]
- Masuda, H.; Odani, A.; Yamauchi, O. Structural Evidence for the Intramolecular Charge-Transfer Interaction Involving an Indole Ring in Ternary Copper(II) Complexes with L-Tryptophan and Aromatic Diamines. Inorganica Chim. Acta 1991, 180, 73. [Google Scholar] [CrossRef]
- Inci, D.; Aydin, R.; Sevgi, T.; Zorlu, Y.; Demirkan, E. Synthesis, Crystal Structure, Stability Studies, DNA/Albumin Interactions, and Antimicrobial Activities of Two Cu(II) Complexes with Amino Acids and 5-Nitro-1,10-Phenanthroline. J. Coord. Chem. 2017, 70, 512–543. [Google Scholar] [CrossRef]
- Baskaran, S.; Murali Krishnan, M.; Arumugham, M.N. Synthesis, Crystal Structure, DNA Binding, Cleavage and Cytotxicity, Antimicrobial Activity of New Copper(II) Complex with L-Ornithine and 1,10-Phenanthroline. Inorg. Nano-Metal. Chem. 2017, 47, 269–277. [Google Scholar] [CrossRef]
- Man-Yu, L.; Ya-Jing, S.; Lan-Zhi, W.; Hui-Hua, S. Syntheses, Crystal Structures and Electrochemical Properties of Two Chiral Cu Complexes Based on D (−)/L (+)-4-Hydroxyphenylglycine. Chin. J. Inorg. Chem. 2019, 35, 1065–1075. [Google Scholar] [CrossRef]
- Jiang, X.; Lin, L.; Zhu, Y.F.; Xia, H. Auxiliary Ligand Effects on the Structural Dimensionality of Copper(II) Coordination Complexes: From 3D Networks to 1D Chains. Transit. Metal. Chem. 2011, 36, 901–906. [Google Scholar] [CrossRef]
- Wang, C.; Yang, G.; Mao, C.; Liang, Y.; Li, S.; Zheng, Y.; Li, H.; Jiang, Y. Synthesis, Crystal Structure, and Magnetic Properties of Cu(II) Coordination Polymer with N-3-Pyridine Sulfonyl Amino Acid and 1,10-Phen. Synth. React. Inorg. Met. Org. Nano-Metal. Chem. 2015, 45, 993–996. [Google Scholar] [CrossRef]
- Patra, A.K.; Nethaji, M.; Chakravarty, A.R. Synthesis, Crystal Structure, DNA Binding and Photo-Induced DNA Cleavage Activity of (S-Methyl-l-Cysteine)Copper(II) Complexes of Heterocyclic Bases. J. Inorg. Biochem. 2007, 101, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Baggio, R.F.; Calvo, R.; Brondino, C.; Garland, M.T.; Atria, A.M.; Spodine, E. A Novel Structure of (L-Aspartato)(1,10-Phenanthroline)Copper(II) Hydrate. Acta Crystallogr. C 1995, 51, 382–385. [Google Scholar] [CrossRef]
- Batalha, P.N.; Mocanu, T.; Calancea, S.; Vaz, M.G.F.; Andruh, M. Zinc(II) and Copper(II) Complexes Constructed from New Bis(1H-1,2,3-Triazole-4-Carboxylate)-Based Ligands. J. Mol. Struct. 2022, 1259, 132703. [Google Scholar] [CrossRef]
- Wojciechowska, A.; Rojek, T.; Misiaszek, T.; Gągor, A.; Rytlewski, P. The Supramolecular Hybrid Inorganic–Organic L-Argininato-Based Copper(II) Materials—Preparation, Structural, Spectroscopic and Thermal Properties. Inorganica Chim. Acta 2023, 557, 121698. [Google Scholar] [CrossRef]
- Antolini, L.; Marcotrigiano, I.G.; Menabue, L.; C Pellacani, I.G.; Salad, I.M.; SOLAIa, M. Coordination Behavior of L-Glutamic Acid: Spectroscopic and Structural Properties of (L-Glutamato) (Imidazole)Copper(II), (L-Glutamato) (2,2′-Bipyridine)Copper(II), and Aqua (L-Glutamato) ( 1,LO-Phenanthroline) Copper (11) Trihydrate Complexes. Inorg. Chem. 1985, 24, 3621–3626. [Google Scholar] [CrossRef]
- Liu, G.F.; Li, L.L.; Ren, Y.S.; Li, H.X.; Ren, Z.G.; Zhang, Y.; Lang, J.P. Two 1D Coordination Polymeric Isomers with the Same Chemical Formula [Cu(2,2′-Bipy)(Htda)]n: Synthesis, Crystal Structures and Electrochemical Properties. Inorg. Chem. Commun. 2009, 12, 563–565. [Google Scholar] [CrossRef]
- Zhou, X.; Yang, C.; Le, X.; Chen, S.; Liu, J.; Huang, Z. Synthesis and Crystal Structures of One-Dimensional ClO4- Bridged Coordination Polymers: {[Cu(Bpy)2(μ2-ClO4)]·ClO4}n and {[Cu2(L-arg)2(Bpy)2(μ2-ClO4)2]·2ClO4·4H2O}n. J. Coord. Chem. 2004, 57, 401–409. [Google Scholar] [CrossRef]
- Seko, H.; Tsuge, K.; Igashira-Kamiyama, A.; Kawamoto, T.; Konno, T. Autoxidation of Thiol-Containing Amino Acid to Its Disulfide Derivative That Links Two Copper(II) Centers: The Important Role of Auxiliary Ligand. Chem. Commun. 2010, 46, 1962–1964. [Google Scholar] [CrossRef] [PubMed]
- Sugimori, T.; Masuda, H.; Ohata, N.; Koiwai, K.; Odani, A.; Yamauchi, O. Structural Dependence of Aromatic Ring Stacking and Related Weak Interactions in Ternary Amino Acid-Copper(II) Complexes and Its Biological Implication. Inorg. Chem. 1997, 36, 576–583. [Google Scholar] [CrossRef]
- Walsh, D.P.; Clérac, R.; Hearns, N.G.R.; Kruger, P.E.; Schmitt, W. Modulating Topologies and Magnetic Properties of Coordination Polymers Using 2,2′-Bipyridine and 5-Aminodiacetic Isophthalic Acid as Ligands. CrystEngComm 2009, 11, 1666–1673. [Google Scholar] [CrossRef]
- Sánchez-Lara, E.; García-García, A.; González-Vergara, E.; Cepeda, J.; Rodríguez-Diéguez, A. Magneto-Structural Correlations of Cyclo-Tetravanadates Functionalized with Mixed-Ligand Copper(II) Complexes. New J. Chem. 2021, 45, 5081–5092. [Google Scholar] [CrossRef]
- Biswas, C.; Drew, M.G.B.; Estrader, M.; Ghosh, A. Copper(II) Complexes of Mono-Anionic Glutamate: Anionic Influence in the Variations of Molecular and Supramolecular Structures. Dalton Trans. 2009, 25, 5015–5022. [Google Scholar] [CrossRef]
- Patra, A.K.; Roy, S.; Chakravarty, A.R. Synthesis, Crystal Structures, DNA Binding and Cleavage Activity of l-Glutamine Copper(II) Complexes of Heterocyclic Bases. Inorganica Chim. Acta 2009, 362, 1591–1599. [Google Scholar] [CrossRef]
- Liu, H.K.; Sadler, P.J. Metal Complexes as DNA Intercalators. Acc. Chem. Res. 2011, 44, 349–359. [Google Scholar] [CrossRef]
- Sigman, D.S.; Graham, D.R.; D’aurora, V.; Stern, A.M. Oxygen-Dependent Cleavage of DNA by the 1,10-Phenanthroline. Cuprous Complex. Inhibition of Escherichia coli DNA Polymerase I. J. Biol. Chem. 1979, 254, 12269–12272. [Google Scholar] [CrossRef]
- McGivern, T.J.P.; Afsharpour, S.; Marmion, C.J. Copper Complexes as Artificial DNA Metallonucleases: From Sigman’s Reagent to next Generation Anti-Cancer Agent? Inorganica Chim. Acta 2018, 472, 12–39. [Google Scholar] [CrossRef]
- Tsang, S.Y.; Tam, S.C.; Bremner, I.; Burkitt, M.J. Copper-1,10-Phenanthroline Induces Internucleosomal DNA Fragmentation in HepG2 Cells, Resulting from Direct Oxidation by the Hydroxyl Radical. Biochem. J. 1996, 317, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Hultberg, B.; Andersson, A.; Isaksson, A. Alterations of Thiol Metabolism in Human Cell Lines Induced by Low Amounts of Copper, Mercury or Cadmium Ions. Toxicology 1998, 126, 203–212. [Google Scholar] [CrossRef]
- McCann, M.; Geraghty, M.; Devereux, M.; O’shea, D.; Mason, J.; O’sullivan, L. Iinsights into the Mode of Action of the Anti-Candida Activity of 1,10-Phenanthroline and Its Metal Chelates. Metal-Based Drugs 2000, 7, 185–193. [Google Scholar] [CrossRef]
- De Vizcaya-Ruiz, A.; Rivero-Müller, A.; Ruiz-Ramirez, L.; Howarth, J.A.; Dobrota, M. Hematotoxicity Response in Rats by the Novel Copper-Based Anticancer Agent: Casiopeina II. Toxicology 2003, 194, 103–113. [Google Scholar] [CrossRef]
- Byrnes, R.W.; Antholine, W.E.; Petering, D.H. Oxidation-Reduction Reactions in Ehrlich Cells Treated with Copper-Neocuproine. Free Radic. Biol. Med. 1992, 13, 469–478. [Google Scholar] [CrossRef]
- Galindo-Murillo, R.; García-Ramos, J.C.; Ruiz-Azuara, L.; Cheatham, T.E.; Cortés-Guzmán, F. Intercalation Processes of Copper Complexes in DNA. Nucleic Acids Res. 2015, 43, 5364–5376. [Google Scholar] [CrossRef] [PubMed]
- Kowol, C.R.; Heffeter, P.; Miklos, W.; Gille, L.; Trondl, R.; Cappellacci, L.; Berger, W.; Keppler, B.K. Mechanisms Underlying Reductant-Induced Reactive Oxygen Species Formation by Anticancer Copper(II) Compounds. J. Biolog Inorg. Chem. 2012, 17, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Zehra, S.; Tabassum, S.; Arjmand, F. Biochemical Pathways of Copper Complexes: Progress over the Past 5 Years. Drug Discov. Today 2021, 26, 1086–1096. [Google Scholar] [CrossRef]
- Tabti, R.; Tounsi, N.; Gaiddon, C.; Bentouhami, E.; Desaubry, L. Progress in Copper Complexes as Anticancer Agents. Med. Chem. 2017, 7, 875–879. [Google Scholar] [CrossRef]
- Figueroa-DePaz, Y.; Pérez-Villanueva, J.; Soria-Arteche, O.; Martínez-Otero, D.; Gómez-Vidales, V.; Ortiz-Frade, L.; Ruiz-Azuara, L. Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and Structure–Activity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands. Molecules 2022, 27, 3504. [Google Scholar] [CrossRef]
- Godínez-Loyola, Y.; Gracia-Mora, J.; Rojas-Montoya, I.D.; Hernández-Ayala, L.F.; Reina, M.; Ortiz-Frade, L.A.; Rascón-Valenzuela, L.A.; Robles-Zepeda, R.E.; Gómez-Vidales, V.; Bernad-Bernad, M.J.; et al. Casiopeinas® Third Generation, with Indomethacin: Synthesis, Characterization, DFT Studies, Antiproliferative Activity, and Nanoencapsulation. RSC Adv. 2022, 12, 21662–21673. [Google Scholar] [CrossRef] [PubMed]
- Patra, A.K.; Bhowmick, T.; Roy, S.; Ramakumar, S.; Chakravarty, A.R. Copper(II) Complexes of L-Arginine as Netropsin Mimics Showing DNA Cleavage Activity in Red Light. Inorg. Chem. 2009, 48, 2932–2943. [Google Scholar] [CrossRef] [PubMed]
- Chetana, P.R.; Rao, R.; Saha, S.; Policegoudra, R.S.; Vijayan, P.; Aradhya, M.S. Oxidative DNA Cleavage, Cytotoxicity and Antimicrobial Studies of l-Ornithine Copper (II) Complexes. Polyhedron 2012, 48, 43–50. [Google Scholar] [CrossRef]
- Kiraz, S.; İnci, D.; Aydın, R.; Vatan, Ö.; Zorlu, Y.; Cavaş, T. Antiproliferative Activity of Copper(II) Glutamine Complexes with N,N-Donor Ligands: Synthesis, Characterization, Potentiometric Studies and DNA/BSA Interactions. J. Mol. Struct. 2019, 1194, 245–255. [Google Scholar] [CrossRef]
- Bravo-Gómez, M.E.; García-Ramos, J.C.; Gracia-Mora, I.; Ruiz-Azuara, L. Antiproliferative Activity and QSAR Study of Copper(II) Mixed Chelate [Cu(N-N)(Acetylacetonato)]NO3 and [Cu(N-N)(Glycinato)]NO3 Complexes, (Casiopeínas®). J. Inorg. Biochem. 2009, 103, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, M.A.; Sewell, G.J.; Rowland, C.G.; Riley, C.M.; Schowen, R.L. The Degradation of Carboplatin in Aqueous Solutions Containing Chloride or Other Selected Nucleophiles. Int. J. Pharm. 1991, 69, 197–210. [Google Scholar] [CrossRef]
- Ciancetta, A.; Coletti, C.; Marrone, A.; Re, N. Activation of Carboplatin by Chloride Ions: A Theoretical Investigation. Theor. Chem. Acc. 2011, 129, 757–769. [Google Scholar] [CrossRef]
- Pessoa, J.C.; Correia, I. Misinterpretations in Evaluating Interactions of Vanadium Complexes with Proteins and Other Biological Targets. Inorganics 2021, 9, 17. [Google Scholar] [CrossRef]
- Correia, I.; Borovic, S.; Cavaco, I.; Matos, C.P.; Roy, S.; Santos, H.M.; Fernandes, L.; Capelo, J.L.; Ruiz-Azuara, L.; Pessoa, J.C. Evaluation of the Binding of Four Anti-Tumor Casiopeínas® to Human Serum Albumin. J. Inorg. Biochem. 2017, 175, 284–297. [Google Scholar] [CrossRef]
- Ugone, V.; Pisanu, F.; Sanna, D.; Garribba, E. Interaction of the Potent Antitumoral Compounds Casiopeinas® with Blood Serum and Cellular Bioligands. J. Inorg. Biochem. 2021, 224, 111566. [Google Scholar] [CrossRef]
- Nunes, P.; Correia, I.; Marques, F.; Matos, A.P.; Dos Santos, M.M.C.; Azevedo, C.G.; Capelo, J.L.; Santos, H.M.; Gama, S.; Pinheiro, T.; et al. Copper Complexes with 1,10-Phenanthroline Derivatives: Underlying Factors Affecting Their Cytotoxicity. Inorg. Chem. 2020, 59, 9116–9134. [Google Scholar] [CrossRef]
- Türkel, N.; Şahın, Ç. Stability of binary and ternary copper (II) complexes with 1,10-phenanthroline, 2,2′-bipyridyl and some α-amino acids in aqueous medium. Chem. Pharm. Bull. 2009, 57, 694–699. [Google Scholar] [CrossRef]

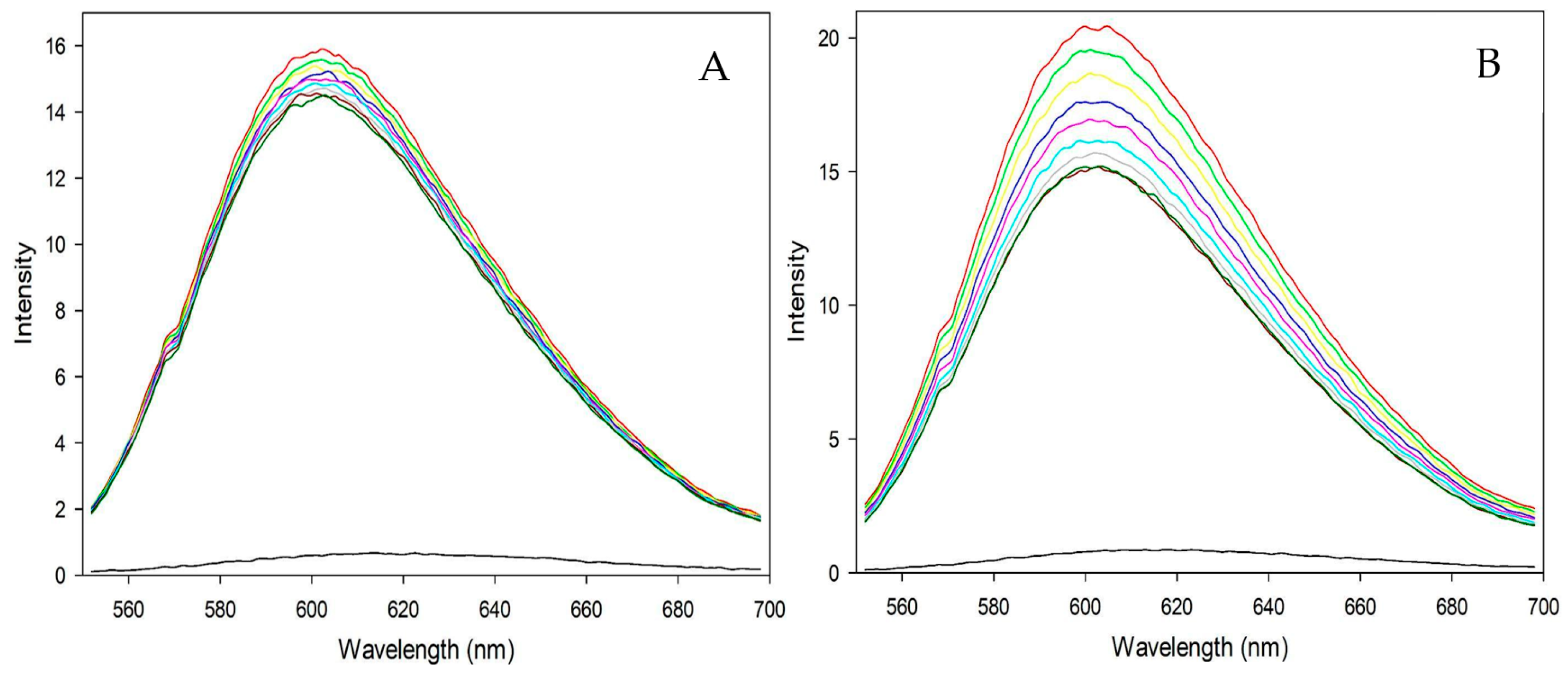
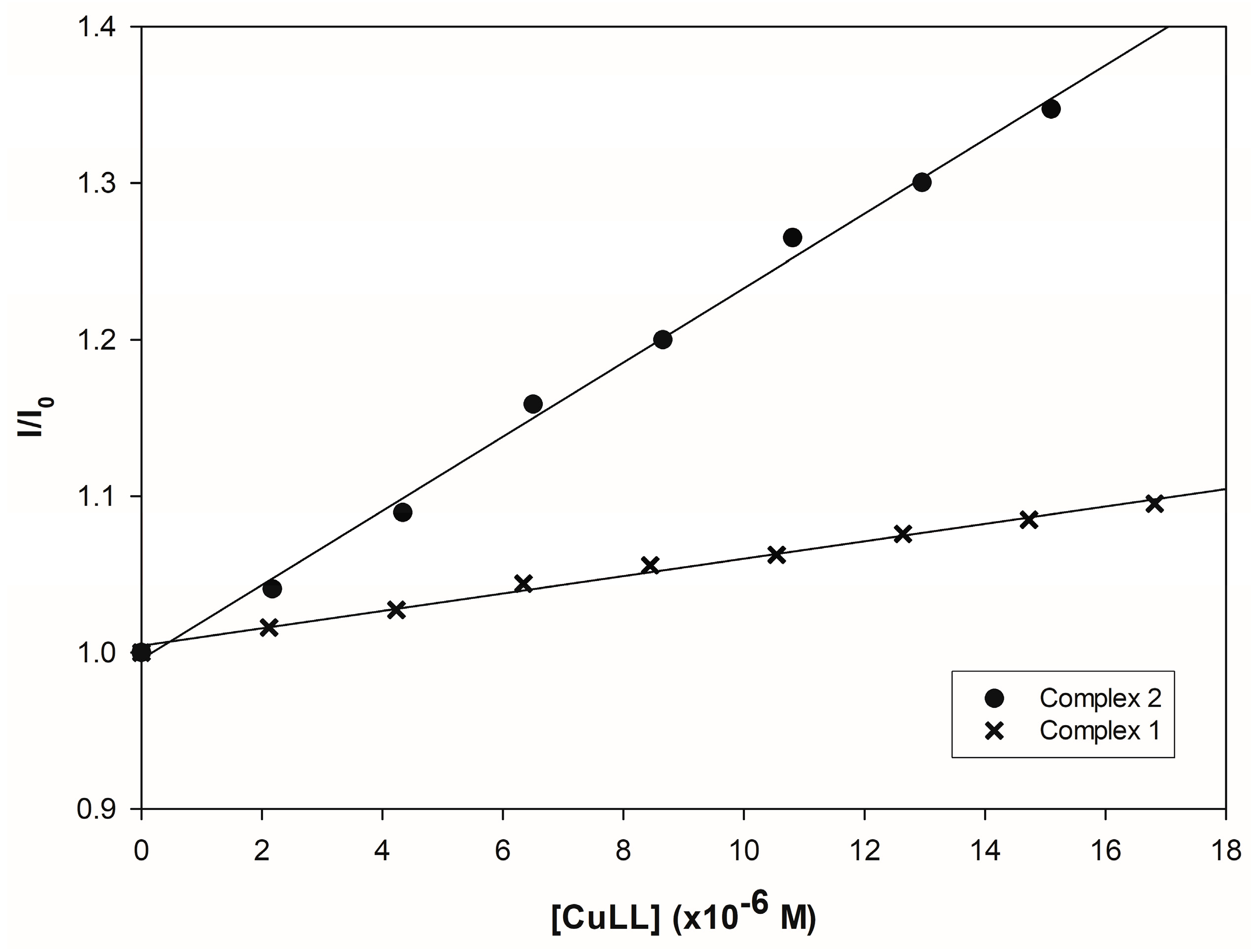
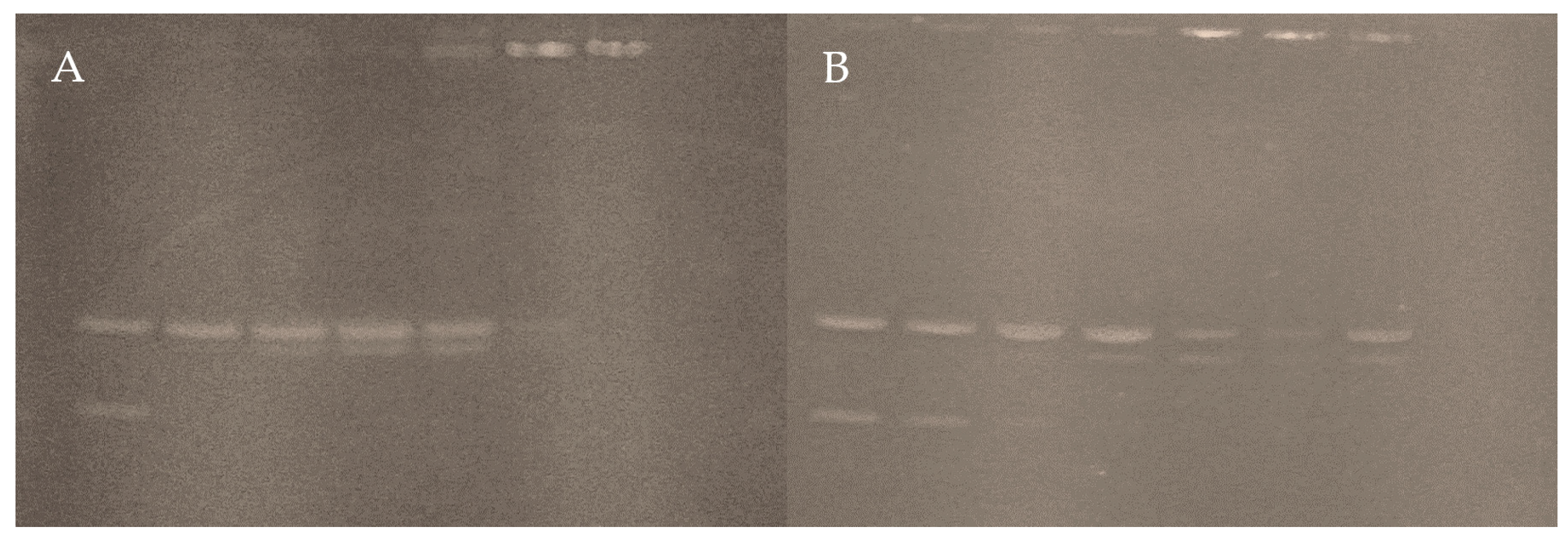
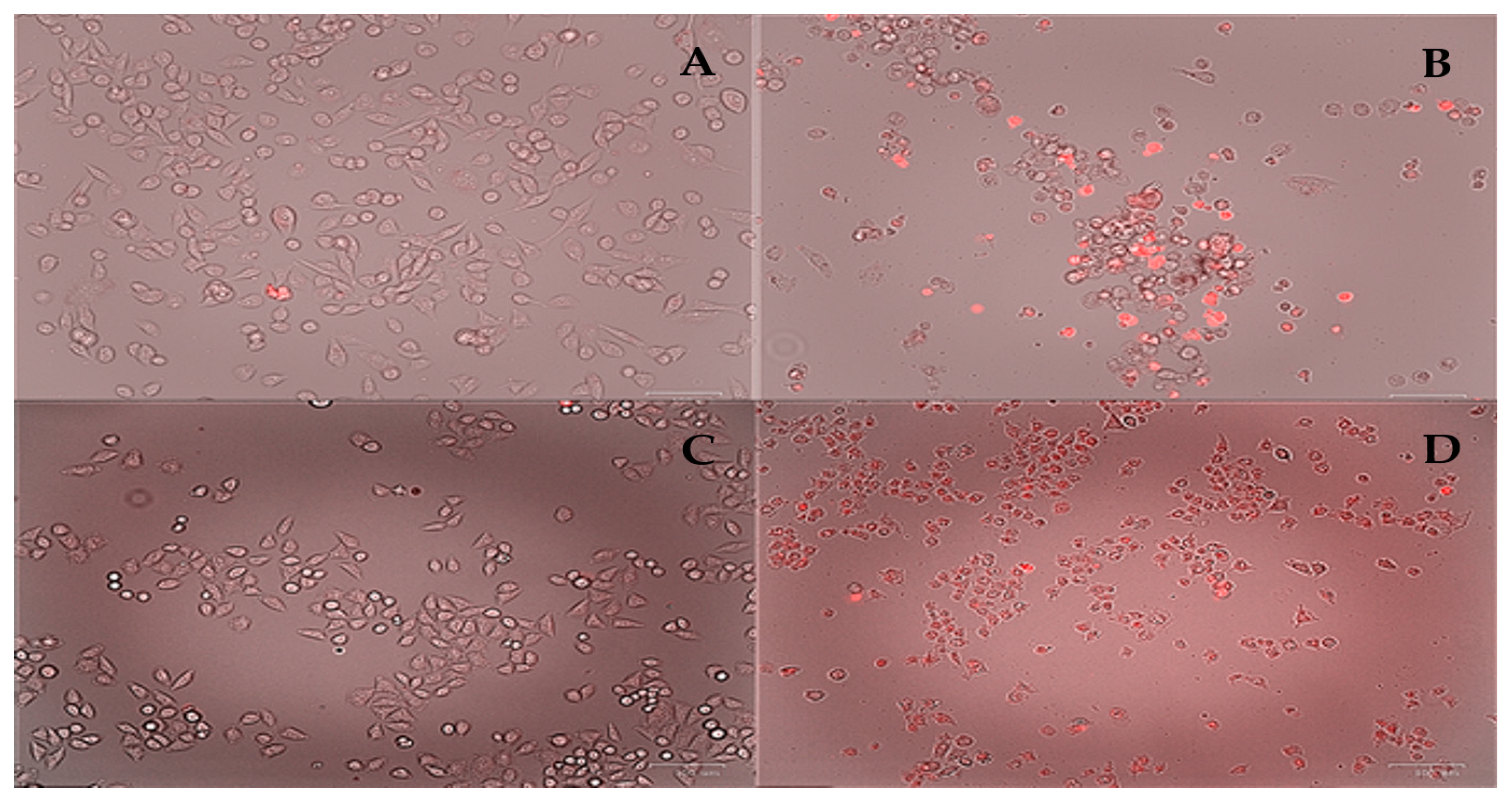
| Complex 1 | Complex 2 | |
|---|---|---|
| Empirical formula | C16H20CuN6O6 | C18H20CuN6O6 |
| CCDC | 2341818 | 2341817 |
| Formula weight (g/mol) | 455.92 | 479.94 |
| Temperature (K) | 293(2) | 293(2) |
| Crystal system | Monoclinic | Triclinic |
| Space group | P21 | P1 |
| a (Å) | 5.5321(2) | 9.2213(3) |
| b (Å) | 9.1050(3) | 9.4194(3) |
| c (Å) | 18.6006(6) | 12.3091(4) |
| a (°) | 90 | 93.660(3) |
| β (°) | 92.816(3) | 93.753(3) |
| γ (°) | 90 | 110.190(3) |
| Volume (Å3) | 935.78(6) | 997.04(6) |
| Z | 2 | 2 |
| ρcalc (g/cm3) | 1.618 | 1.599 |
| μ (mm−1) | 1.216 | 1.146 |
| Radiation | MoKα (λ = 0.71073) | MoKα (λ = 0.71073) |
| Index ranges | −8 ≤ h ≤ 8, −14 ≤ k ≤ 14, −29 ≤ l ≤ 30 | −14 ≤ h ≤ 14, −14 ≤ k ≤ 14, −18 ≤ l ≤ 18 |
| Largest diff. peak/hole (e Å−3) | 0.34/−0.47 | 0.42/−0.43 |
| Rint | 0.0496 | 0.0482 |
| GoF on F2 | 1.044 | 1.004 |
| Final R indexes [I > 2σ(I)] | R1 = 0.0366, wR2 = 0.0652 | R1 = 0.0461, wR2 = 0.0807 |
| Final R indexes [all data] | R1 = 0.0647, wR2 = 0.0756 | R1 = 0.0834, wR2 = 0.0934 |
| D-H···A | D-H Distance | H···A Distance | D···A Distance | Angle |
|---|---|---|---|---|
| N24-H24B···O26 | 0.89 | 2.289 | 3.103(3) | 151.9 |
| N24 i-H24A···O14 | 0.89 | 2.347 | 3.236(3) | 177.6 |
| N20 ii-H20···O23 | 0.86 | 2.158 | 2.947(3) | 152.3 |
| N22 ii-H22A···O23 | 0.83(3) | 2.37(3) | 3.113(3) | 149.7(3) |
| N22 iii-H22B···O23 | 0.88(4) | 2.32(4) | 3.161(3) | 158.2(3) |
| D-H···A | D-H Distance | H···A Distance | D···A Distance | Angle |
|---|---|---|---|---|
| N3-H3A···O7 | 0.89 | 2.26 | 3.004(5) | 141.5 |
| N3-H3B···O2 i | 0.89 | 2.41 | 3.100(4) | 134.7 |
| N4-H4···O12 i | 0.86 | 2.22 | 3.021(5) | 154.7 |
| N5-H5A···O2 ii | 0.86 | 2.08 | 2.916(5) | 164.3 |
| N5-H5B···O12 i | 0.86 | 2.38 | 3.152(6) | 149.5 |
| N8-H8A···O5 | 0.89 | 2.09 | 2.963(4) | 165.6 |
| N8-H8B···O10 | 0.89 | 2.18 | 3.013(4) | 155.5 |
| N9-H9A···O9 | 0.86 | 2.12 | 2.957(5) | 163.3 |
| N10-H10A···O5 iii | 0.86 | 2.15 | 2.990(5) | 164.4 |
| N10-H10B···O9 | 0.86 | 2.42 | 3.174(6) | 147.3 |
| Complex 1′ | |||
|---|---|---|---|
| BCP | ρ (r) | ∇2ρ (r) | EH···Y |
| Cu1···O1 | 0.0747 | 0.4718 | 38.00 |
| Cu1···N1 | 0.0760 | 0.3964 | 35.23 |
| Cu1···N2 | 0.0812 | 0.4601 | 41.04 |
| Cu1···N3 | 0.0753 | 0.4257 | 36.68 |
| Cu1···O3 | 0.0377 | 0.2307 | 13.99 |
| H2···O1 | 0.0124 | 0.0598 | 2.73 |
| Complex 2′ | |||
| BCP | ρ (r) | ∇2ρ (r) | EH···Y |
| Cu1···O1 | 0.0747 | 0.4711 | 37.90 |
| Cu1···N1 | 0.0771 | 0.3997 | 35.85 |
| Cu1···N2 | 0.0793 | 0.4500 | 39.69 |
| Cu1···N3 | 0.0713 | 0.4035 | 33.85 |
| Cu1···O3 | 0.0375 | 0.2289 | 13.87 |
| H2···O1 | 0.0097 | 0.0472 | 1.98 |
| Compound | Binding Energy (Kcal/mol) | |
|---|---|---|
| DNA/1BNA | DNA/151D | |
| Complex 1′ | −9.3 | −7.7 |
| Complex 2′ | −9.8 | −9.1 |
| Complex | Cancer Cell Line | |||
|---|---|---|---|---|
| HeLa | MCF-7 | MDA MB 231 | HCT-15 | |
| Complex 1 | >10 | 11.77 ± 0.61 | ||
| Complex 2 | 2.53 ± 0.04 | 4.67 ± 0.59 | 6.69 ± 0.60 | 5.20 ± 0.09 |
| Complex | Secondary Ligand | Coordinating Moiety | Crystallization Solvent | Ref. |
|---|---|---|---|---|
| [Cu(L-Trp) (1,10-phen)](ClO4)(H2O) | L-Trp | Trp α-CO (μ-O) | Water/EtOH | [78] |
| [Cu(Asn) (1,10-nphen)](ClO4) | L-Asn | Asn δ-CO (μ-O) | Water/MeOH | [79] |
| [Cu(Orn)(1,10-phen)](NO3)2(H2O) | L-Orn | CI (NO3) (μ-O) | Water/EtOH | [80] |
| [Cu(D/L-dhpg)(1,10-phen) (NO3)]x(H2O) | D/L-dhpg | CI (NO3) (μ-O) | Water/EtOH (5:3) | [81] |
| [Cu2(H2ttha)(1,10-phen)2] | (H2ttha)−4 | -NAc2 (μ4-N, O) | Water/EtOH (4:1) | [82] |
| [Cu(3-pysaa)(1,10-phen)]2(H2O) | 3-pysaa | Pyr (μ-N) | Water/EtOH (2:3) | [83] |
| [Cu(SMe-L-Cys)(dppz)(H2O)](NO3) | SMe-L-Cys | Cys α-CO (μ-O) | Water/MeOH (1:2) | [84] |
| [Cu(L-Asp)(1,10-phen)](H2O) | L-Asp | Asp δ-CO (μ-O) | Water/EtOH (1:1) | [85] |
| [Cu(sptc)(1,10-phen)] | (sptc)−2 | TC (μ2-N, O) | MeCN/MeOH (5:4) | [86] |
| Complex | Secondary Ligand | Coordinating Moiety | Crystallization Solvent | Ref. |
|---|---|---|---|---|
| [Cu(L-Glu)(2,2′-bipy)] | L-Glu | Glu ε-COO (μ-O) | Water/MeOH (3:1) | [88] |
| [Cu(L-Arg)(2,2′-bipy)(NO3)](NO3) | L-Arg | CI (NO3) (μ-O) | Water/MeOH (2:1) | [87] |
| [Cu(Hdta)(2,2′-bipy)] | Htda−2 | TA (μ-N) | MeCN | [89] |
| [Cu2(L-Arg)2(2,2′-bipy)2(ClO4)2]2(ClO4)4(H2O) | L-Arg | CI (ClO4) (μ2-O) | Water/EtOH | [90] |
| [Cu4(L-Cys-Cys)(2,2′-bipy)4]4(ClO4)1.5(H2O) | L-Cys-Cys | Cys α-COO(μ-O) | MeOH | [91] |
| [Cu(L-NH2Phe)(2,2′bipy)](NO3)(H2O) | L-NH2-Phe | -NH2 (μ-N) | Water/MeOH | [92] |
| [Cu(H2adip)2(2,2′-Bipy)] | adip−2 | MeCOO (μ-O) | Water | [93] |
| [Cu(D,L-Lys)(2,2′-bipy)]2(V4O10)7(H2O) | D, L-Lys | CI (V4O10) (μ-O) | Water | [94] |
| [Cu(L-Glu)(2,2′-bipy)(H2O)] [Cu(L-Glu)(2,2′-bipy)(ClO4)](ClO4)2(H2O) | L-Glu | CI (ClO4) (μ-O) | MeOH | [95] |
| [Cu(L-Gln)(2,2′-bipy)(H2O)] [Cu(L-Gln)(2,2′-bipy)](SO4)4(H2O) | L-Gln | L-Gln α-COO (μ-O) | Water/MeOH | [96] |
| Compound | Kb (M−1) | Reference |
|---|---|---|
| [Cu(L-Citr)(1,10-phen)(H2O)]NO3 (Complex 2′) | 8.32 × 105 | This work |
| [Cu(L-Citr)(2,2′-bipy)(H2O]NO3 (Complex 1′) | 3.76 × 104 | |
| [Cu(2,2′ -bipy)(gly)(H2O)]NO3 (CasVII-gly) | 3.23 × 105 | [69] |
| [Cu(4,4-dimethyl-2,2′-bipy)(acac)(H2O)]NO3 (CasIII-ia) | 3.31 × 105 | |
| [Cu(1,10-phen)(gly)(H2O)]NO3 (CasVII-gly) | 7.15 × 105 | |
| [Cu(phen)(acac)(H2O)]NO3 (CasIII-Ba) | 7.60 × 105 | |
| [Cu(4,7-dimethyl-1,10-phen)(acac)(H2O)]NO3 (CasIII-Ea) | 7.90 × 105 | |
| [Cu(5,6-dimethyl-1,10-phen)(gly)(H2O)]NO3 (CasVI-gly) | 7.89 × 105 | |
| [Cu(L-Arg)(2,2′-bipy)(Cl)](Cl) | 1.70 × 103 | [112] |
| [Cu(L-Orn)(1,10-phen)(Cl)](Cl) | 2.70 × 104 | [113] |
| [Cu(L-Asn)(1,10-nphen)](ClO4) | 2.43 × 103 | [79] |
| [Cu(L-Gln)(1,10-nphen)(H2O)](ClO4) | 1.05 × 104 | |
| [Cu(L-Gln)(1,10-phen)(H2O)](NO3)(H2O) | 3.62 × 103 | [113] |
| [Cu(L-Gln)(dmphen)(H2O](ClO4) | 7.33 × 103 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Contreras, D.; Vázquez-Rodríguez, S.; García-García, A.; Noriega, L.; Mendoza, A.; Sánchez-Gaytán, B.L.; Meléndez, F.J.; Castro, M.E.; Cárdenas-García, M.; González-Vergara, E. L-Citrullinato-Bipyridine and L-Citrullinato-Phenanthroline Mixed Copper Complexes: Synthesis, Characterization and Potential Anticancer Activity. Pharmaceutics 2024, 16, 747. https://doi.org/10.3390/pharmaceutics16060747
Ramírez-Contreras D, Vázquez-Rodríguez S, García-García A, Noriega L, Mendoza A, Sánchez-Gaytán BL, Meléndez FJ, Castro ME, Cárdenas-García M, González-Vergara E. L-Citrullinato-Bipyridine and L-Citrullinato-Phenanthroline Mixed Copper Complexes: Synthesis, Characterization and Potential Anticancer Activity. Pharmaceutics. 2024; 16(6):747. https://doi.org/10.3390/pharmaceutics16060747
Chicago/Turabian StyleRamírez-Contreras, Diego, Sergio Vázquez-Rodríguez, Amalia García-García, Lisset Noriega, Angel Mendoza, Brenda L. Sánchez-Gaytán, Francisco J. Meléndez, María Eugenia Castro, Maura Cárdenas-García, and Enrique González-Vergara. 2024. "L-Citrullinato-Bipyridine and L-Citrullinato-Phenanthroline Mixed Copper Complexes: Synthesis, Characterization and Potential Anticancer Activity" Pharmaceutics 16, no. 6: 747. https://doi.org/10.3390/pharmaceutics16060747
APA StyleRamírez-Contreras, D., Vázquez-Rodríguez, S., García-García, A., Noriega, L., Mendoza, A., Sánchez-Gaytán, B. L., Meléndez, F. J., Castro, M. E., Cárdenas-García, M., & González-Vergara, E. (2024). L-Citrullinato-Bipyridine and L-Citrullinato-Phenanthroline Mixed Copper Complexes: Synthesis, Characterization and Potential Anticancer Activity. Pharmaceutics, 16(6), 747. https://doi.org/10.3390/pharmaceutics16060747





