In Vitro Studies to Evaluate the Intestinal Permeation of an Ursodeoxycholic Acid-Conjugated Oligonucleotide for Duchenne Muscular Dystrophy Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
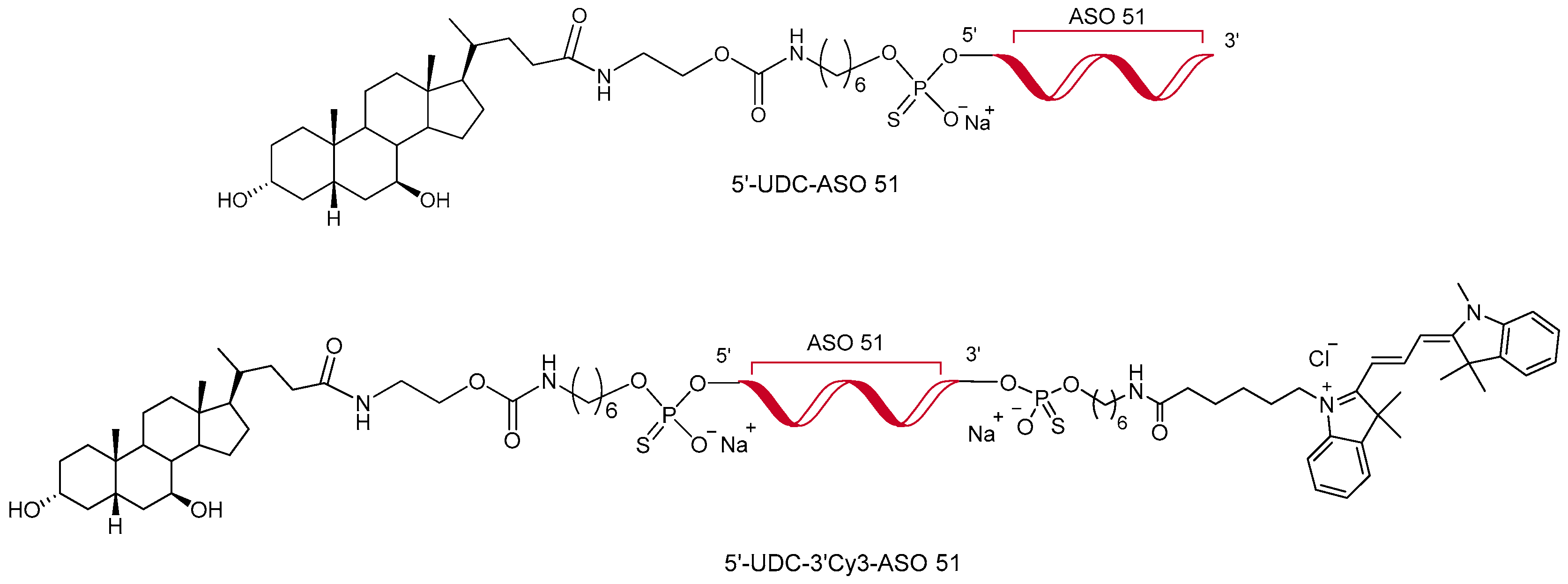
2.2. Synthesis of Antisense Oligonucleotides
2.3. HPLC Analysis
2.4. IEC-6 Cells’ Culture and Differentiation to Polarized Monolayers
2.5. MTT Assay for Evaluation of ASO 51, 5′-UDC-ASO 51 and 5′-UDC-3′Cy3-ASO 51 Toxicity on IEC-6 Cells
2.6. Permeation Studies across Intestinal Cell Monolayers
2.7. Exosome Isolation and Characterization
2.8. Exosome Loading
2.9. Lipophilic Membrane Dye Labeling
2.10. RNA Extraction, Retrotranscritpion, RT–PCR and Exon Skipping Quantification
2.11. Non-Contact Co-Culture of DMD Myotubes and Intestinal IEC-6 Cells
2.12. Statistical Analyses
2.13. Microscopy and Image Elaboration
3. Results and Discussion
3.1. IEC-6 Cell Viability
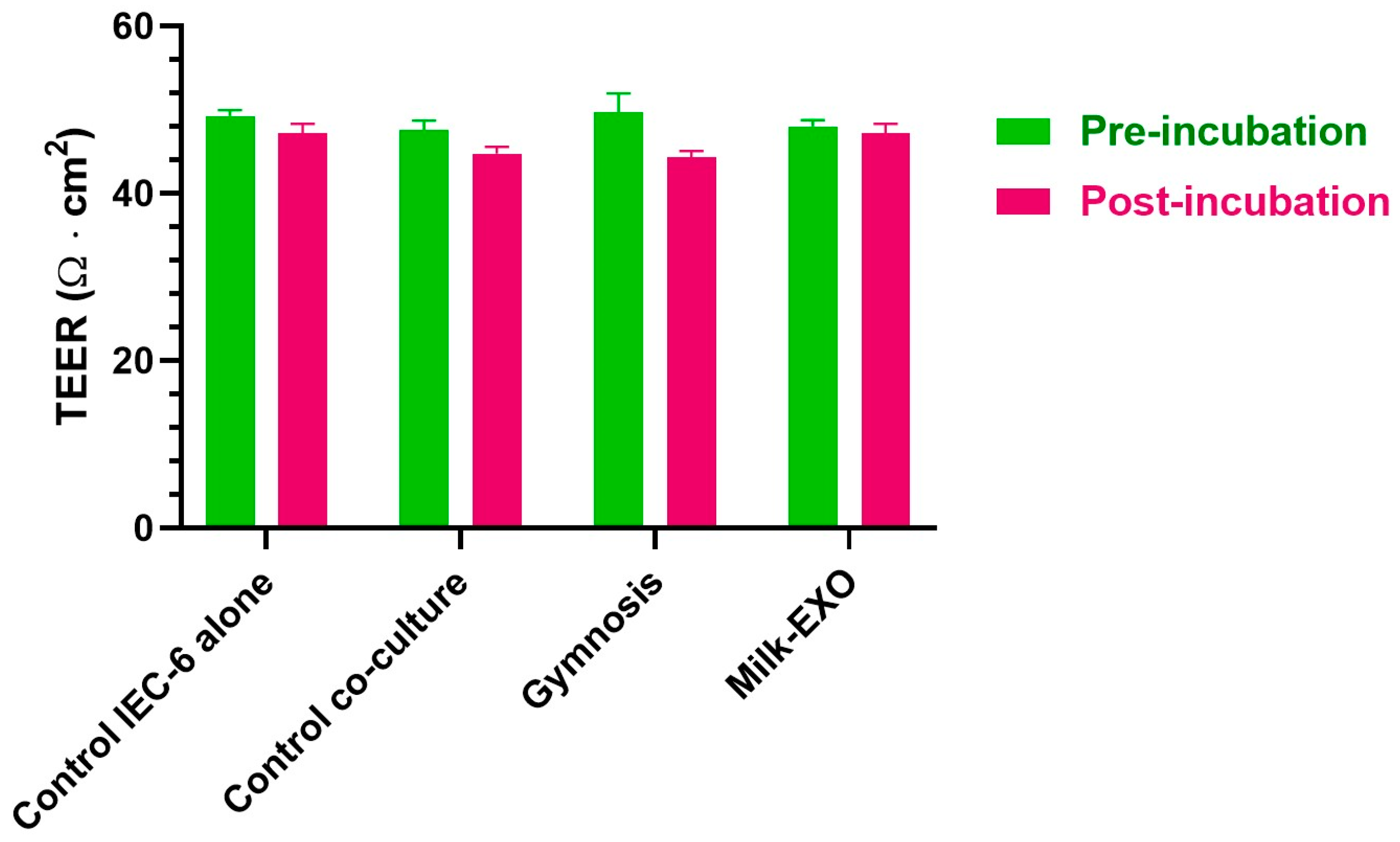
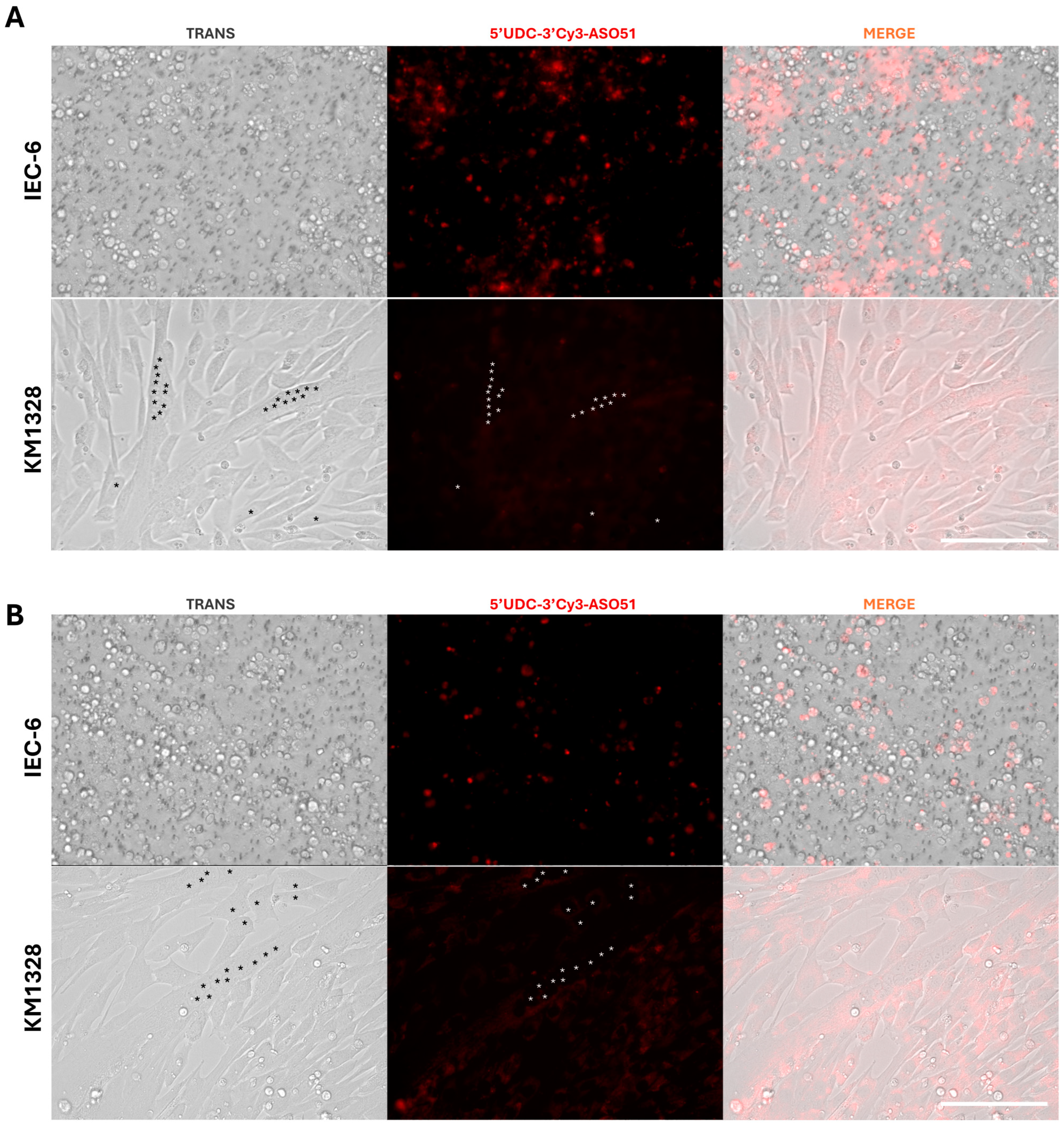
3.2. Permeation Studies across Intestinal Monolayers
3.3. Exosome Isolation and Characterization
3.4. Exosomes Are Efficiently Loaded with Both 5′-UDC-3′Cy3-ASO 51 and 3′Cy3-ASO 51
3.5. EXO–ASO Complexes Efficiently Fuse to Myotubes, Distributing ASOs in the Cytoplasms, but Not in the Nucleus
3.6. MusEXO-ASO Complexes Induce Higher Levels of Exon Skipping in Myotubes Compared to MilkEXO-ASO Complexes and Gymnotic Delivery
3.7. Non-Contact Co-Culture of IEC-6 and DMD Muscle Cells: 5′-UDC-3′Cy3-ASO 51 after Crossing the IEC-6 Monolayers Can Induce Exon Skipping in Muscle Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duan, D.; Goemans, N.; Takeda, S.; Mercuri, E.; Aartsma-Rus, A. Duchenne Muscular Dystrophy. Nat. Rev. Dis. Primers 2021, 7, 13. [Google Scholar] [CrossRef] [PubMed]
- Fox, H.; Millington, L.; Mahabeer, I.; van Ruiten, H. Duchenne Muscular Dystrophy. BMJ 2020, 368, l7012. [Google Scholar] [CrossRef] [PubMed]
- Vinjamuri, B.P.; Pan, J.; Peng, P. A Review on Commercial Oligonucleotide Drug Products. J. Pharm. Sci. 2024, 113, 1749–1768. [Google Scholar] [CrossRef]
- Bylo, M.; Farewell, R.; Coppenrath, V.A.; Yogaratnam, D. A Review of Deflazacort for Patients with Duchenne Muscular Dystrophy. Ann. Pharmacother. 2020, 54, 788–794. [Google Scholar] [CrossRef]
- FDA Approves Nonsteroidal Treatment for Duchenne Muscular Dystrophy. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-nonsteroidal-treatment-duchenne-muscular-dystrophy (accessed on 27 June 2024).
- Arechavala-Gomeza, V.; Anthony, K.; Morgan, J.; Muntoni, F. Antisense Oligonucleotide-Mediated Exon Skipping for Duchenne Muscular Dystrophy: Progress and Challenges. Curr. Gene Ther. 2012, 12, 152–160. [Google Scholar] [CrossRef]
- Patterson, G.; Conner, H.; Groneman, M.; Blavo, C.; Parmar, M.S. Duchenne Muscular Dystrophy: Current Treatment and Emerging Exon Skipping and Gene Therapy Approach. Eur. J. Pharmacol. 2023, 947, 175675. [Google Scholar] [CrossRef]
- Łoboda, A.; Dulak, J. Muscle and Cardiac Therapeutic Strategies for Duchenne Muscular Dystrophy: Past, Present, and Future. Pharmacol. Rep. 2020, 72, 1227–1263. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Shen, L.; Zhang, Z.; Xie, X. Therapeutic Strategies for Duchenne Muscular Dystrophy: An Update. Genes 2020, 11, 837. [Google Scholar] [CrossRef] [PubMed]
- Verhaart, I.E.C.; Aartsma-Rus, A. Therapeutic Developments for Duchenne Muscular Dystrophy. Nat. Rev. Neurol. 2019, 15, 373–386. [Google Scholar] [CrossRef]
- Aartsma-Rus, A.; Fokkema, I.; Verschuuren, J.; Ginjaar, I.; van Deutekom, J.; van Ommen, G.-J.; den Dunnen, J.T. Theoretic Applicability of Antisense-Mediated Exon Skipping for Duchenne Muscular Dystrophy Mutations. Hum. Mutat. 2009, 30, 293–299. [Google Scholar] [CrossRef]
- Le, B.T.; Chen, S.; Veedu, R.N. Evaluation of Chemically Modified Nucleic Acid Analogues for Splice Switching Application. ACS Omega 2023, 8, 48650–48661. [Google Scholar] [CrossRef] [PubMed]
- Engelbeen, S.; O’Reilly, D.; Van De Vijver, D.; Verhaart, I.; van Putten, M.; Hariharan, V.; Hassler, M.; Khvorova, A.; Damha, M.J.; Aartsma-Rus, A. Challenges of Assessing Exon 53 Skipping of the Human DMD Transcript with Locked Nucleic Acid-Modified Antisense Oligonucleotides in a Mouse Model for Duchenne Muscular Dystrophy. Nucleic Acid Ther. 2023, 33, 348–360. [Google Scholar] [CrossRef]
- Kandasamy, P.; McClorey, G.; Shimizu, M.; Kothari, N.; Alam, R.; Iwamoto, N.; Kumarasamy, J.; Bommineni, G.R.; Bezigian, A.; Chivatakarn, O.; et al. Control of Backbone Chemistry and Chirality Boost Oligonucleotide Splice Switching Activity. Nucleic Acids Res. 2022, 50, 5443–5466. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Clemens, P.R.; Hoffman, E.P. Exon-Skipping in Duchenne Muscular Dystrophy. J. Neuromuscul. Dis. 2021, 8, S343–S358. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, C.; Desviat, L.R.; Smedsrød, B.; Piétri-Rouxel, F.; Denti, M.A.; Disterer, P.; Lorain, S.; Nogales-Gadea, G.; Sardone, V.; Anwar, R.; et al. Delivery Is Key: Lessons Learnt from Developing Splice-switching Antisense Therapies. EMBO Mol. Med. 2017, 9, 545–557. [Google Scholar] [CrossRef]
- Juliano, R.L. The Delivery of Therapeutic Oligonucleotides. Nucleic Acids Res. 2016, 44, 6518–6548. [Google Scholar] [CrossRef] [PubMed]
- Bost, J.P.; Barriga, H.; Holme, M.N.; Gallud, A.; Maugeri, M.; Gupta, D.; Lehto, T.; Valadi, H.; Esbjörner, E.K.; Stevens, M.M.; et al. Delivery of Oligonucleotide Therapeutics: Chemical Modifications, Lipid Nanoparticles, and Extracellular Vesicles. ACS Nano 2021, 15, 13993–14021. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Benizri, S.; Gissot, A.; Martin, A.; Vialet, B.; Grinstaff, M.W.; Barthélémy, P. Bioconjugated Oligonucleotides: Recent Developments and Therapeutic Applications. Bioconjug. Chem. 2019, 30, 366–383. [Google Scholar] [CrossRef]
- Østergaard, M.E.; Jackson, M.; Low, A.; Chappell, A.E.; Lee, R.G.; Peralta, R.Q.; Yu, J.; Kinberger, G.A.; Dan, A.; Carty, R.; et al. Conjugation of Hydrophobic Moieties Enhances Potency of Antisense Oligonucleotides in the Muscle of Rodents and Non-Human Primates. Nucleic Acids Res. 2019, 47, 6045–6058. [Google Scholar] [CrossRef]
- Seth, P.P.; Tanowitz, M.; Bennett, C.F. Selective Tissue Targeting of Synthetic Nucleic Acid Drugs. J. Clin. Investig. 2019, 129, 915–925. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Allen, N.; Prakash, T.P.; Liang, X.; Crooke, S.T. Lipid Conjugates Enhance Endosomal Release of Antisense Oligonucleotides into Cells. Nucleic Acid Ther. 2019, 29, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, E.; Bovolenta, M.; Preti, L.; Capobianco, M.L.; Mamchaoui, K.; Bertoldo, M.; Perrone, D. Synthesis and Exon-Skipping Properties of a 3′-Ursodeoxycholic Acid-Conjugated Oligonucleotide Targeting DMD Pre-MRNA: Pre-Synthetic versus Post-Synthetic Approach. Molecules 2021, 26, 7662. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, E.; Cortesi, R.; Preti, L.; Rimessi, P.; Sguizzato, M.; Bovolenta, M.; Perrone, D. Antisense Oligonucleotides Conjugated with Lipophilic Compounds: Synthesis and In Vitro Evaluation of Exon Skipping in Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2022, 23, 4270. [Google Scholar] [CrossRef] [PubMed]
- Goemans, N.M.; Tulinius, M.; van den Hauwe, M.; Kroksmark, A.-K.; Buyse, G.; Wilson, R.J.; van Deutekom, J.C.; de Kimpe, S.J.; Lourbakos, A.; Campion, G. Long-Term Efficacy, Safety, and Pharmacokinetics of Drisapersen in Duchenne Muscular Dystrophy: Results from an Open-Label Extension Study. PLoS ONE 2016, 11, e0161955. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Morena, C.; Cavero-Redondo, I.; Álvarez-Bueno, C.; Mesas, A.E.; Pozuelo-Carrascosa, D.; Martínez-Vizcaíno, V. Restorative Treatments of Dystrophin Expression in Duchenne Muscular Dystrophy: A Systematic Review. Ann. Clin. Transl. Neurol. 2020, 7, 1738–1752. [Google Scholar] [CrossRef] [PubMed]
- van Deutekom, J.C.; Janson, A.A.; Ginjaar, I.B.; Frankhuizen, W.S.; Aartsma-Rus, A.; Bremmer-Bout, M.; den Dunnen, J.T.; Koop, K.; van der Kooi, A.J.; Goemans, N.M.; et al. Local Dystrophin Restoration with Antisense Oligonucleotide PRO051. N. Engl. J. Med. 2007, 357, 2677–2686. [Google Scholar] [CrossRef] [PubMed]
- Quaroni, A.; Wands, J.; Trelstad, R.L.; Isselbacher, K.J. Epithelioid Cell Cultures from Rat Small Intestine. Characterization by Morphologic and Immunologic Criteria. J. Cell. Biol. 1979, 80, 248–265. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.; Costa, J.; Sarmento, B.; Araújo, F. Cell-Based in Vitro Models for Intestinal Permeability Studies. In Concepts and Models for Drug Permeability Studies; Elsevier: Amsterdam, The Netherlands, 2016; pp. 57–81. [Google Scholar]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2022, 9, 811971. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Bi, J.; Huang, J.; Tang, Y.; Du, S.; Li, P. Exosome: A Review of Its Classification, Isolation Techniques, Storage, Diagnostic and Targeted Therapy Applications. Int. J. Nanomed. 2020, 15, 6917–6934. [Google Scholar] [CrossRef]
- Huda, M.N.; Nafiujjaman, M.; Deaguero, I.G.; Okonkwo, J.; Hill, M.L.; Kim, T.; Nurunnabi, M. Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 2021, 7, 2106–2149. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.I.; Park, J.; Zhu, Y.; Wang, X.; Han, Y.; Zhang, D. Recent Advances in Extracellular Vesicles for Therapeutic Cargo Delivery. Exp. Mol. Med. 2024, 56, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shen, Y.; Kim, I.; Weintraub, N.L.; Hamrick, M.; Tang, Y. Extracellular Vesicles for Muscle Atrophy Treatment. In Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 2023; pp. 119–126. [Google Scholar]
- Zeng, H.; Guo, S.; Ren, X.; Wu, Z.; Liu, S.; Yao, X. Current Strategies for Exosome Cargo Loading and Targeting Delivery. Cells 2023, 12, 1416. [Google Scholar] [CrossRef] [PubMed]
- Aminzadeh, M.A.; Rogers, R.G.; Fournier, M.; Tobin, R.E.; Guan, X.; Childers, M.K.; Andres, A.M.; Taylor, D.J.; Ibrahim, A.; Ding, X.; et al. Exosome-Mediated Benefits of Cell Therapy in Mouse and Human Models of Duchenne Muscular Dystrophy. Stem Cell Rep. 2018, 10, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Gartz, M.; Lin, C.-W.; Sussman, M.A.; Lawlor, M.W.; Strande, J.L. Duchenne Muscular Dystrophy (DMD) Cardiomyocyte-Secreted Exosomes Promote the Pathogenesis of DMD-Associated Cardiomyopathy. Dis. Models Mech. 2020, 13, dmm045559. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Dong, X.; Gao, X.; Ran, N.; Geng, M.; Zuo, B.; Wu, Y.; Li, W.; Yan, H.; Han, G.; et al. Exosome-Mediated Improvement in Membrane Integrity and Muscle Function in Dystrophic Mice. Mol. Ther. 2021, 29, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Pandurangan, M.; Hwang, I. Application of Cell Co-Culture System to Study Fat and Muscle Cells. Appl. Microbiol. Biotechnol. 2014, 98, 7359–7364. [Google Scholar] [CrossRef] [PubMed]
- Artursson, P.; Karlsson, J. Correlation between Oral Drug Absorption in Humans and Apparent Drug Permeability Coefficients in Human Intestinal Epithelial (Caco-2) Cells. Biochem. Biophys. Res. Commun. 1991, 175, 880–885. [Google Scholar] [CrossRef]
- Pal, D.; Udata, C.; Mitra, A.K. Transport of Cosalane—A Highly Lipophilic Novel Anti-HIV Agent—Across Caco-2 Cell Monolayers. J. Pharm. Sci. 2000, 89, 826–833. [Google Scholar] [CrossRef]
- Raje, S.; Cao, J.; Newman, A.H.; Gao, H.; Eddington, N.D. Evaluation of the Blood-Brain Barrier Transport, Population Pharmacokinetics, and Brain Distribution of Benztropine Analogs and Cocaine Using In Vitro and In Vivo Techniques. J. Pharmacol. Exp. Ther. 2003, 307, 801–808. [Google Scholar] [CrossRef]
- Yee, S. In Vitro Permeability Across Caco-2 Cells (Colonic) Can Predict In Vivo (Small Intestinal) Absorption in Man—Fact or Myth. Pharm. Res. 1997, 14, 763–766. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, L.; Ouandaogo, Z.G.; Anakor, E.; Connolly, O.; Butler Browne, G.; Laine, J.; Duddy, W.; Duguez, S. Optimized Method for Extraction of Exosomes from Human Primary Muscle Cells. Skelet. Muscle 2020, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Didiot, M.-C.; Haraszti, R.A.; Aronin, N.; Khvorova, A. Loading of Extracellular Vesicles with Hydrophobically Modified SiRNAs. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 199–214. [Google Scholar]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Zhang, L.; Li, W.; Liu, X.; Xiao, J.; Chen, G.; Li, N. Natural CAC Chemopreventive Agents from Ilex Rotunda Thunb. J. Nat. Med. 2019, 73, 456–467. [Google Scholar] [CrossRef]
- Oliveira, P.V.; Aguiar, G.P.S.; Siebel, A.M.; Müller, L.G.; Lerin, L.A.; Botti, G.; Bianchi, A.; Bernardi, T.; Gentili, V.; Rizzo, R.; et al. Synthesis of Naringenin-Betaine Cocrystal by Gas Antisolvent Technique and Cell Models for in Vitro Permeation Studies. J. Drug Deliv. Sci. Technol. 2024, 96, 105671. [Google Scholar] [CrossRef]
- Hansen, M.S.; Gadegaard, I.S.E.; Arnspang, E.C.; Blans, K.; Nejsum, L.N.; Rasmussen, J.T. Specific and Non-Invasive Fluorescent Labelling of Extracellular Vesicles for Evaluation of Intracellular Processing by Intestinal Epithelial Cells. Biomedicines 2020, 8, 211. [Google Scholar] [CrossRef]
- Rothen-Rutishauser, B.; Braun, A.; Günthert, M.; Wunderli-Allenspach, H. Formation of Multilayers in the Caco-2 Cell Culture Model: A Confocal Laser Scanning Microscopy Study. Pharm. Res. 2000, 17, 460–465. [Google Scholar] [CrossRef]
- Gildea, J.J.; Roberts, D.A.; Bush, Z. Protective Effects of Lignite Extract Supplement on Intestinal Barrier Function in Glyphosate-Mediated Tight Junction Injury. J. Clin. Nutr. Diet. 2017, 3, 1–6. [Google Scholar] [CrossRef]
- Dalpiaz, A.; Paganetto, G.; Pavan, B.; Fogagnolo, M.; Medici, A.; Beggiato, S.; Perrone, D. Zidovudine and Ursodeoxycholic Acid Conjugation: Design of a New Prodrug Potentially Able to Bypass the Active Efflux Transport Systems of the Central Nervous System. Mol. Pharm. 2012, 9, 957–968. [Google Scholar] [CrossRef]
- Fischer, H.; Gottschlich, R.; Seelig, A. Blood-Brain Barrier Permeation: Molecular Parameters Governing Passive Diffusion. J. Membr. Biol. 1998, 165, 201–211. [Google Scholar] [CrossRef]
- Pavan, B.; Dalpiaz, A. Prodrugs and Endogenous Transporters: Are They Suitable Tools for Drug Targeting into the Central Nervous System? Curr. Pharm. Des. 2011, 17, 3560–3576. [Google Scholar] [CrossRef] [PubMed]
- Riccardi, C.; Musumeci, D.; Russo Krauss, I.; Piccolo, M.; Irace, C.; Paduano, L.; Montesarchio, D. Exploring the Conformational Behaviour and Aggregation Properties of Lipid-Conjugated AS1411 Aptamers. Int. J. Biol. Macromol. 2018, 118, 1384–1399. [Google Scholar] [CrossRef] [PubMed]
- Farrell, T.L.; Poquet, L.; Dew, T.P.; Barber, S.; Williamson, G. Predicting Phenolic Acid Absorption in Caco-2 Cells: A Theoretical Permeability Model and Mechanistic Study. Drug Metab. Dispos. 2012, 40, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Pavan, B.; Dalpiaz, A.; Marani, L.; Beggiato, S.; Ferraro, L.; Canistro, D.; Paolini, M.; Vivarelli, F.; Valerii, M.C.; Comparone, A.; et al. Geraniol Pharmacokinetics, Bioavailability and Its Multiple Effects on the Liver Antioxidant and Xenobiotic-Metabolizing Enzymes. Front. Pharmacol. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Dalpiaz, A.; Fogagnolo, M.; Ferraro, L.; Capuzzo, A.; Pavan, B.; Rassu, G.; Salis, A.; Giunchedi, P.; Gavini, E. Nasal Chitosan Microparticles Target a Zidovudine Prodrug to Brain HIV Sanctuaries. Antivir. Res. 2015, 123, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Dalpiaz, A.; Fogagnolo, M.; Ferraro, L.; Beggiato, S.; Hanuskova, M.; Maretti, E.; Sacchetti, F.; Leo, E.; Pavan, B. Bile Salt-Coating Modulates the Macrophage Uptake of Nanocores Constituted by a Zidovudine Prodrug and Enhances Its Nose-to-Brain Delivery. Eur. J. Pharm. Biopharm. 2019, 144, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, Biodistribution and Cell Uptake of Antisense Oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Ejazi, S.A.; Louisthelmy, R.; Maisel, K. Mechanisms of Nanoparticle Transport across Intestinal Tissue: An Oral Delivery Perspective. ACS Nano 2023, 17, 13044–13061. [Google Scholar] [CrossRef] [PubMed]
- Casper, J.; Schenk, S.H.; Parhizkar, E.; Detampel, P.; Dehshahri, A.; Huwyler, J. Polyethylenimine (PEI) in Gene Therapy: Current Status and Clinical Applications. J. Control. Release 2023, 362, 667–691. [Google Scholar] [CrossRef]
- Poukalov, K.K.; Valero, M.C.; Muscato, D.R.; Adams, L.M.; Chun, H.; Lee, Y.I.; Andrade, N.S.; Zeier, Z.; Sweeney, H.L.; Wang, E.T. Myospreader Improves Gene Editing in Skeletal Muscle by Myonuclear Propagation. Proc. Natl. Acad. Sci. USA 2024, 121, e2321438121. [Google Scholar] [CrossRef]
- Chen, H.; Wang, L.; Zeng, X.; Schwarz, H.; Nanda, H.S.; Peng, X.; Zhou, Y. Exosomes, a New Star for Targeted Delivery. Front. Cell Dev. Biol. 2021, 9, 751079. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Harris-Hooker, S.; Kumar, R.; Sanford, G. Co-Culture of Retinal and Endothelial Cells Results in the Modulation of Genes Critical to Retinal Neovascularization. Vasc. Cell 2011, 3, 27, Corrected in Vasc. Cell 2012, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Wolf, T.; Baier, S.R.; Zempleni, J. The Intestinal Transport of Bovine Milk Exosomes Is Mediated by Endocytosis in Human Colon Carcinoma Caco-2 Cells and Rat Small Intestinal IEC-6 Cells. J. Nutr. 2015, 145, 2201–2206. [Google Scholar] [CrossRef] [PubMed]
- Shaughnessey, E.M.; Kann, S.H.; Azizgolshani, H.; Black, L.D.; Charest, J.L.; Vedula, E.M. Evaluation of Rapid Transepithelial Electrical Resistance (TEER) Measurement as a Metric of Kidney Toxicity in a High-Throughput Microfluidic Culture System. Sci. Rep. 2022, 12, 13182. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Hu, Y.; Xu, S.; Liu, F.; Gao, Y. Exosomal MicroRNAs as Potential Biomarkers and Therapeutic Agents for Acute Ischemic Stroke: New Expectations. Front. Neurol. 2022, 12, 747380. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf–Bensussan, N.; Heyman, M. Intestinal Epithelial Cells Secrete Exosome–Like Vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, W.; Masuno, T.; Yokota, H.; Takizawa, T. Expression Profiles and Circulation Dynamics of Rat Mesenteric Lymph MicroRNAs. Mol. Med. Rep. 2017, 15, 1989–1996. [Google Scholar] [CrossRef] [PubMed]
- Orellana, E.A.; Abdelaal, A.M.; Rangasamy, L.; Tenneti, S.; Myoung, S.; Low, P.S.; Kasinski, A.L. Enhancing MicroRNA Activity through Increased Endosomal Release Mediated by Nigericin. Mol. Ther. Nucleic Acids 2019, 16, 505–518. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faiella, M.; Botti, G.; Dalpiaz, A.; Gnudi, L.; Goyenvalle, A.; Pavan, B.; Perrone, D.; Bovolenta, M.; Marchesi, E. In Vitro Studies to Evaluate the Intestinal Permeation of an Ursodeoxycholic Acid-Conjugated Oligonucleotide for Duchenne Muscular Dystrophy Treatment. Pharmaceutics 2024, 16, 1023. https://doi.org/10.3390/pharmaceutics16081023
Faiella M, Botti G, Dalpiaz A, Gnudi L, Goyenvalle A, Pavan B, Perrone D, Bovolenta M, Marchesi E. In Vitro Studies to Evaluate the Intestinal Permeation of an Ursodeoxycholic Acid-Conjugated Oligonucleotide for Duchenne Muscular Dystrophy Treatment. Pharmaceutics. 2024; 16(8):1023. https://doi.org/10.3390/pharmaceutics16081023
Chicago/Turabian StyleFaiella, Marika, Giada Botti, Alessandro Dalpiaz, Lorenzo Gnudi, Aurélie Goyenvalle, Barbara Pavan, Daniela Perrone, Matteo Bovolenta, and Elena Marchesi. 2024. "In Vitro Studies to Evaluate the Intestinal Permeation of an Ursodeoxycholic Acid-Conjugated Oligonucleotide for Duchenne Muscular Dystrophy Treatment" Pharmaceutics 16, no. 8: 1023. https://doi.org/10.3390/pharmaceutics16081023
APA StyleFaiella, M., Botti, G., Dalpiaz, A., Gnudi, L., Goyenvalle, A., Pavan, B., Perrone, D., Bovolenta, M., & Marchesi, E. (2024). In Vitro Studies to Evaluate the Intestinal Permeation of an Ursodeoxycholic Acid-Conjugated Oligonucleotide for Duchenne Muscular Dystrophy Treatment. Pharmaceutics, 16(8), 1023. https://doi.org/10.3390/pharmaceutics16081023












