The Use of Some Species of Bacteria and Algae in the Bioremediation of Pollution Caused by Hydrocarbons and Some Heavy Metals in Al Asfar Lake Water
Abstract
1. Introduction
2. Methodology
2.1. Collection of Algal Samples
2.2. Isolation, Purification, and Identification of Algal Samples
2.3. Morphological Examination and Establishment of Monocultures
2.4. Isolation and Purification of Bacterial Strains
2.5. Detection of Heavy Metals by Chemical Analysis of Samples
2.6. Chemical Element Analysis after Incubation with Algal Consortium
2.7. Molecular Identification
2.8. Polyaromatic Hydrocarbon Analysis
3. Results
3.1. Microscopic Examination of Predominant Microbial Consortium Members
3.2. Bioremediation of Water Samples from Al Asfar Lake
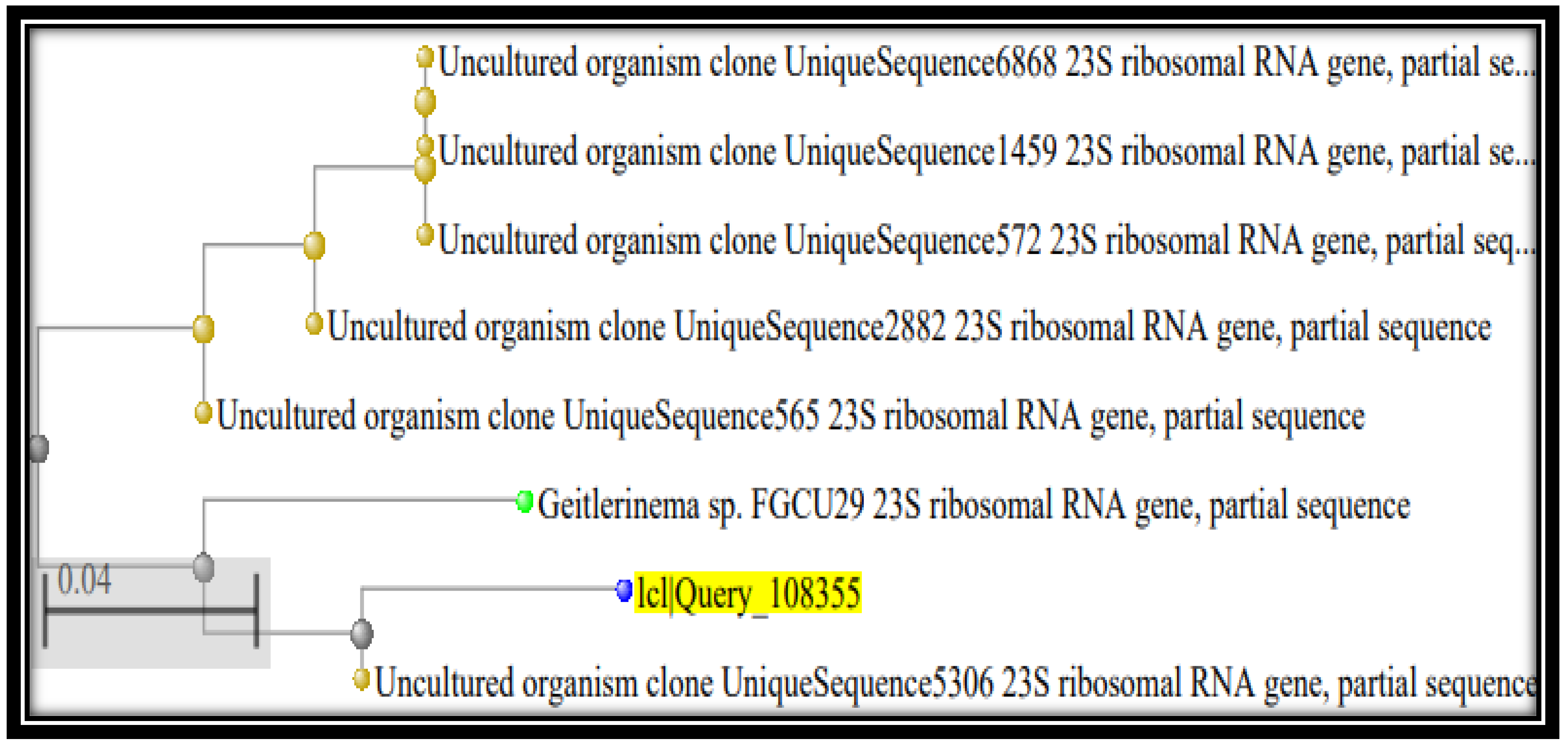
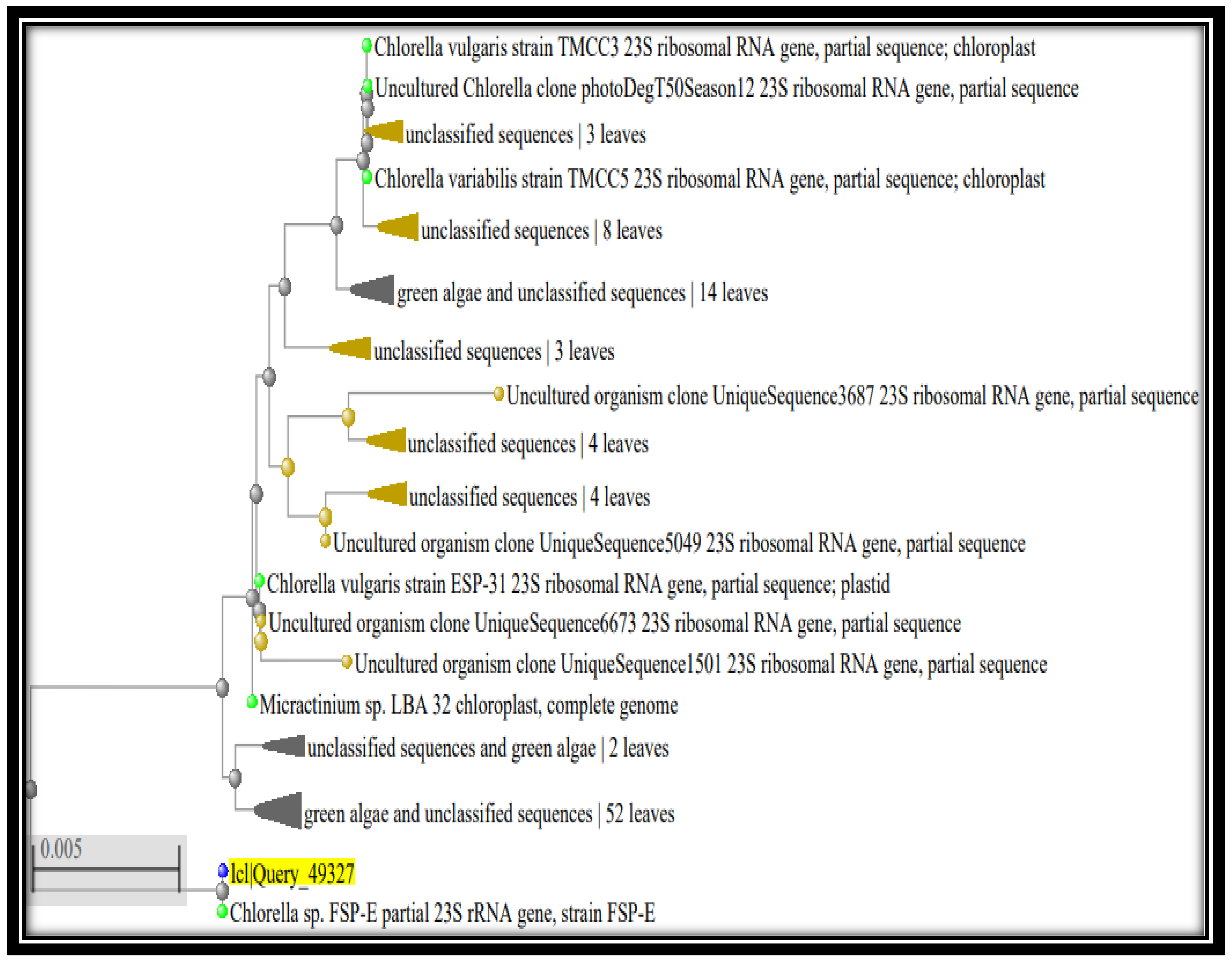
3.3. Characterization of Taxonomic Identity of Most Efficient Bio-Remediating Algal Strains Using Molecular Analysis and Phylogenetic Inference
3.4. Polyaromatic Hydrocarbon Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akl, F.M.A.; Ahmed, S.I.; El-Sheekh, M.M.; Makhlof, M.E.M. Bioremediation of n-alkanes, polycyclic aromatic hydrocarbons, and heavy metals from wastewater using seaweeds. Environ. Sci. Pollut. Res. 2023, 30, 104814–104832. [Google Scholar] [CrossRef]
- Sharma, R.K.; Agrawal, M.; Marshall, F. Heavy metal contamination of soil and vegetables in suburban areas of Varanasi, India. Ecotoxicol. Environ. Saf. 2007, 66, 258–266. [Google Scholar] [CrossRef] [PubMed]
- El Mahmoudi, A.; Massoud, M.; Al-Dakheel, Y.; Hussein, A. Studies of Al Asfar and Al Uyoun Evaporation Lakes Water Quality and the Potential of Its Reuse in Agriculture Activities, Al Hassa Area, KSA. J. King Abdulaziz Univ. -Meteorol. Environ. Arid. Land Agric. Sci. 2011, 22, 67–85. [Google Scholar] [CrossRef]
- El-Sheekh, M.M.; Mahmoud, Y.A.-G. Technological Approach of Bioremediation Using Microbial Tools: Bacteria, Fungi, and Algae. In Handbook of Research on Inventive Bioremediation Techniques; IGI Global: Hershey, PA, USA, 2017; Available online: https://www.igi-global.com/chapter/technological-approach-of-bioremediation-using-microbial-tools/176461 (accessed on 25 June 2024).
- Nagm El Deen, F. Master’s Thesis, Mansoura University, Mansoura, Egypt, 2013.
- Spain, O.; Plöhn, M.; Funk, C. The cell wall of green microalgae and its role in heavy metal removal. Physiol. Plant. 2021, 173, 526–535. [Google Scholar] [CrossRef]
- Groudeva, V.I.; Groudev, S.N.; Doycheva, A.S. Bioremediation of waters contaminated with crude oil and toxic heavy metals. Int. J. Miner. Process. 2001, 62, 293–299. [Google Scholar] [CrossRef]
- Hassan, Z.U.; Ali, S.; Rizwan, M.; Ibrahim, M.; Nafees, M.; Waseem, M. Role of Bioremediation Agents (Bacteria, Fungi, and Algae) in Alleviating Heavy Metal Toxicity. In Probiotics in Agroecosystem; Springer: Berlin/Heidelberg, Germany, 2017; pp. 517–537. [Google Scholar] [CrossRef]
- Mondal, M.; Halder, G.; Oinam, G.; Indrama, T.; Tiwari, O.N. Chapter 17—Bioremediation of Organic and Inorganic Pollutants Using Microalgae. In New and Future Developments in Microbial Biotechnology and Bioengineering; Gupta, V.K., Pandey, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 223–235. ISBN 9780444635044. [Google Scholar] [CrossRef]
- Raklami, A.; Meddich, A.; Oufdou, K.; Baslam, M. Plants—Microorganisms-Based Bioremediation for Heavy Metal Cleanup: Recent Developments, Phytoremediation Techniques, Regulation Mechanisms, and Molecular Responses. Int. J. Mol. Sci. 2022, 23, 5031. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, A.R.; Presting, G.G. Universal primers amplify a 23s rdna plastid marker in eukaryotic algae and cyanobacteria. J. Phycol. 2007, 43, 605–608. [Google Scholar] [CrossRef]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Pérez, S.; Guillamón, M.; Barceló, D. Quantitative analysis of polycyclic aromatic hydrocarbons in sewage sludge from wastewater treatment plants. J. Chromatogr. A 2001, 938, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Picó, Y.; Alvarez-Ruiz, R.; Alfarhan, A.H.; El-Sheikh, M.A.; Alshahrani, H.O.; Barceló, D. Pharmaceuticals, pesticides, personal care products and microplastics contamination assessment of Al-Hassa irrigation network (Saudi Arabia) and its shallow lakes. Sci. Total Environ. 2020, 701, 135021. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Verma, T.; Gaur, R. Detoxification of hexavalent chromium by an indigenous facultative anaerobic Bacillus cereus strain isolated from tannery effluent. Afr. J. Biotechnol. 2013, 12, 1091–1103. [Google Scholar]
- Cornu, J.Y.; Huguenot, D.; Jézéquel, K.; Lollier, M.; Lebeau, T. Bioremediation of copper-contaminated soils by bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–9. [Google Scholar]
- Danouche, M.; El Ghachtouli, N.; El Arroussi, H. Phycoremediation mechanisms of heavy metals using living green microalgae: Physicochemical and molecular approaches for enhancing selectivity and removal capacity. Heliyon 2021, 7, e07609. [Google Scholar] [CrossRef] [PubMed]
- Al-Amin, A.; Parvin, F.; Chakraborty, J.; Kim, Y.I. Cyanobacteria mediated heavy metal removal: A review on mechanism, biosynthesis, and removal capability. Environ. Technol. Rev. 2021, 10, 44–57. [Google Scholar] [CrossRef]
- Ince, M.; Ince, O.K. Introductory Chapter: Sources, Health Impact, and Environment Effect of Hydrocarbons. In Hydrocarbon Pollution and its Effect on the Environment; IntechOpen: London, UK, 2019; p. 1. [Google Scholar]
- Srogi, K. Monitoring of environmental exposure to polycyclic aromatic hydrocarbons: A review. Environ. Chem. Lett. 2007, 5, 169–195. [Google Scholar] [CrossRef] [PubMed]
- Das, N.; Chandran, P. Microbial degradation of petroleum hydrocarbon contaminants: An overview. Biotechnol. Res. Int. 2011, 2011, 941810. [Google Scholar] [CrossRef] [PubMed]
- Essa, E.; Abu El-Hassan, G.; Farag, S. Biochemical Composition, Toxicity and Bioactivities of the Essential Oil extracted from Coffea arabica L. husks against the Cotton Leafworm, Spodoptera littoralis (Boisduval) (Lepidoptera: Noctudiae). Egypt. Acad. J. Biol. Sci. A Entomol. 2022, 15, 37–49. [Google Scholar] [CrossRef]
- Elumalai, P.; Parthipan, P.; Huang, M.; Muthukumar, B.; Cheng, L.; Govarthanan, M.; Rajasekar, A. Enhanced biodegradation of hydrophobic organic pollutants by the bacterial consortium: Impact of enzymes and biosurfactants. Environ. Pollut. 2021, 289, 117956. [Google Scholar] [CrossRef] [PubMed]
| Component | Stock Solution (g·L−1 dH2O) | Quantity Used/L |
|---|---|---|
| Fe citrate solution | – | 1 mL |
| Citric acid | 6 | 1 mL |
| Ferric ammonium citrate | 6 | 1 mL |
| NaNO3 | – | 1.5 g |
| K2HPO4·3H3O | 40 | 1 mL |
| MgSO4·7H2O | 75 | 1 mL |
| CaCl2·2H2O | 36 | 1 mL |
| Na2CO3 | 20 | 1 mL |
| Na2EDTA·H2O | 1.0 | 1 mL |
| Trace metal solution | – | 1 mL |
| Trace element solution composition | ||
| Component | Stock Solution (g·L−1 dH2O) | Quantity Used/L |
| H3BO3 | – | 2.860 g |
| MnCl2·4H2O | – | 1.810 g |
| ZnSO4·7H2O | – | 0.22 g |
| CuSO4·5H2O | 79.0 | 1 mL |
| Na2MoO4·2H2O | – | 0.391 g |
| Co(NO3)2·6H2O | 49.4 | 1 mL |
| Bacterial Pathogen Strain (Code) | Colony Color | Size of Colony (Diameter) | Shape | Texture | Elevation |
|---|---|---|---|---|---|
| A1 | Orange | 2 mL | Round | Shiny, smooth | Flat |
| A2 | Yellowish | 1 mL | Round | Shiny, smooth | Flat |
| B | Light, orange | 2 mL | Round | Shiny, smooth | Flat |
| C1 | White | 2.5 mL | Round | Shiny, smooth | Flat |
| C2 | White | 1.5 mL | Round | Shiny, smooth | Flat |
| Chemical Element | Concentration Average | Concentration RSD | Concentration SD |
|---|---|---|---|
| Mn | 1.678 | 35.1% | 0.6 ppm |
| Co | 0.994 | 0.5% | 0.0 ppm |
| Cu | 0.769 | 92.7% | 0.7 ppm |
| Zn | 1.059 | 0.3% | 0.0 ppm |
| Cd | 1.037 | 0.3% | 0.0 ppm |
| Pb | 1.060 | 1.2% | 0.0 ppm |
| As | 0.753 | 0.6% | 0.0 ppm |
| Fe | 0.887 | 0.2% | 0.0 ppm |
| Ni | 1.032 | 0.3% | 0.0 ppm |
| Cr | 0.588 | 259.5% | 1.5 ppm |
| B | 1.0090 | 0.3% | 0.0 ppm |
| Sample | (C1) 10 mL Culture | (C1) 15 mL Culture | (C2) 10 mL Culture | (C2) 15 mL Culture | (F) 10 mL Culture | (F) 15 mL Culture |
|---|---|---|---|---|---|---|
| Mn | 0.511 ppm | 0.220 ppm | 0.126 ppm | 0.081 ppm | 0.362 ppm | −0.668 ppm |
| Co | 0.004 ppm | 0.002 ppm | 0.002 ppm | 0.001 ppm | 0.002 ppm | 0.003 ppm |
| Cu | 0.105 ppm | −0.068 ppm | 0.282 ppm | −0.096 ppm | 0.149 ppm | 0391 ppm |
| Zn | 0.029 ppm | 0.000 ppm | 0.030 ppm | 0.036 ppm | 0.082 ppm | 0.100 ppm |
| Cd | 0.002 ppm | 0.017 ppm | 0.000 ppm | 0.000 ppm | 0.000 ppm | 0.000 ppm |
| Pb | 0.018 ppm | 0.008 ppm | 0.019 ppm | 0.020 ppm | 0.027 ppm | 0.018 ppm |
| As | 0.011 ppm | 0.008 ppm | 0.006 ppm | 0.008 ppm | 0.007 ppm | 0.005 ppm |
| Fe | 0.060 ppm | 0.053 ppm | 0.058 ppm | 0.062 ppm | 0.027 ppm | 0.037 ppm |
| Ni | 0.004 ppm | 0.0 03 ppm | 0.002 ppm | 0.004 ppm | 0.003 ppm | 0.001 ppm |
| Cr | −0.103 ppm | −1.116 ppm | −1.642 ppm- | 0.172 ppm | −0.400 ppm | −2.307 ppm |
| B | 1.427 ppm | 1.544 ppm | 1.426 ppm | 1.676 ppm | 1.737 ppm | 1.419 ppm |
| Compound Name | % | Retention Time (Rt), min |
|---|---|---|
| Phenol, 2,4-bis(1,1-dimethylethyl) | 0.94 | 7.597 |
| Cycloheptasiloxane, tetradecamethyl | 3.62 | 7.801 |
| Cyclooctasiloxane, hexadecamethyl | 7.69 | 9.2 |
| Cyclononasiloxane, octadecamethyl | 8.15 | 10.365 |
| Cyclodecasiloxane, eicosamethyl | 7.38 | 11.423 |
| Cyclooctasiloxane, hexadecamethyl | 8.7 | 12.635 |
| Cyclononasiloxane, octadecamethyl | 11.72 | 13.723 |
| Tetracosamethyl-cyclododecasiloxane | 13.56 | 14.759 |
| Tetracosamethyl-cyclododecasiloxane | 17.48 | 16.067 |
| Tetracosamethyl-cyclododecasiloxane | 20.76 | 17.836 |
| Compound Name | % | Retention Time (Rt), min |
|---|---|---|
| Phenol, 2,6-bis(1,1-dimethylethyl) | 1.42 | 7.593 |
| 7-(1,5-Dimethylhexyl)-10,13-dimethyl2,3,4,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthren3-ol | 72.14 | 14.341 |
| Cholest-5-ene, 3.beta.-chloro | 26.44 | 14.711 |
| Compound Name | % | Retention Time (Rt), min |
|---|---|---|
| Phenol, 2,4-bis(1,1-dimethylethyl) | 10.44 | 7.595 |
| Cyclooctasiloxane, hexadecamethyl | 5.11 | 9.197 |
| Cyclononasiloxane, octadecamethyl | 7.4 | 10.362 |
| Cyclodecasiloxane, eicosamethyl | 6.49 | 11.421 |
| Cyclooctasiloxane, hexadecamethyl- | 8.26 | 12.633 |
| Tetracosamethyl-cyclododecasiloxane | 11.44 | 13.721 |
| Tetracosamethyl-cyclododecasiloxane | 13.89 | 14.755 |
| Tetracosamethyl-cyclododecasiloxane | 17.53 | 16.064 |
| Tetracosamethyl-cyclododecasiloxane | 19.44 | 17.833 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altammar, F.; El Semary, N.; Aldayel, M. The Use of Some Species of Bacteria and Algae in the Bioremediation of Pollution Caused by Hydrocarbons and Some Heavy Metals in Al Asfar Lake Water. Sustainability 2024, 16, 7896. https://doi.org/10.3390/su16187896
Altammar F, El Semary N, Aldayel M. The Use of Some Species of Bacteria and Algae in the Bioremediation of Pollution Caused by Hydrocarbons and Some Heavy Metals in Al Asfar Lake Water. Sustainability. 2024; 16(18):7896. https://doi.org/10.3390/su16187896
Chicago/Turabian StyleAltammar, Fatimah, Nermin El Semary, and Munirah Aldayel. 2024. "The Use of Some Species of Bacteria and Algae in the Bioremediation of Pollution Caused by Hydrocarbons and Some Heavy Metals in Al Asfar Lake Water" Sustainability 16, no. 18: 7896. https://doi.org/10.3390/su16187896
APA StyleAltammar, F., El Semary, N., & Aldayel, M. (2024). The Use of Some Species of Bacteria and Algae in the Bioremediation of Pollution Caused by Hydrocarbons and Some Heavy Metals in Al Asfar Lake Water. Sustainability, 16(18), 7896. https://doi.org/10.3390/su16187896






