Effects of GLP-1 Analogues and Agonists on the Gut Microbiota: A Systematic Review
Abstract
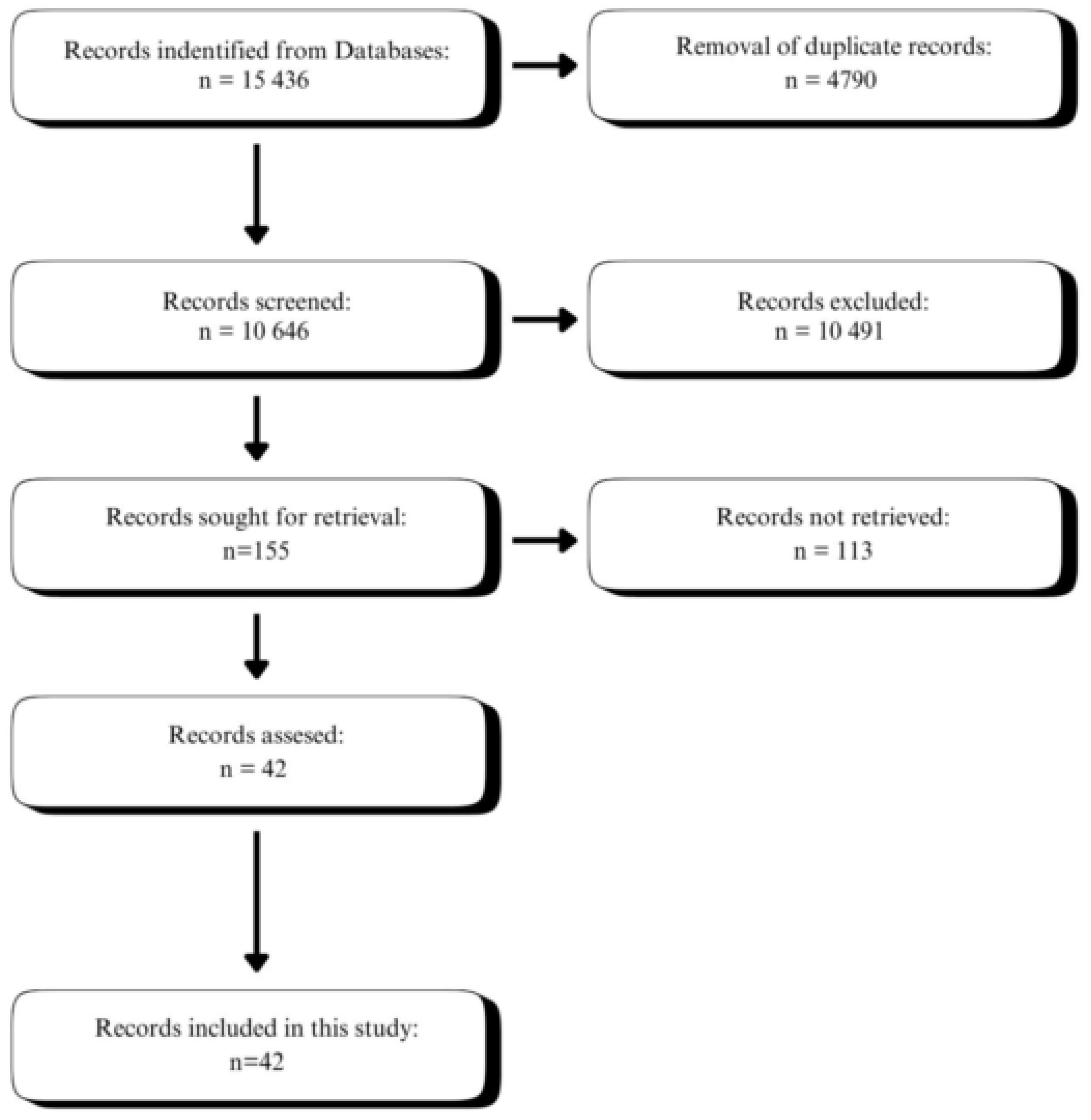
:1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Study Selection Criteria
2.3. Study Selection
2.4. Data Collection Process
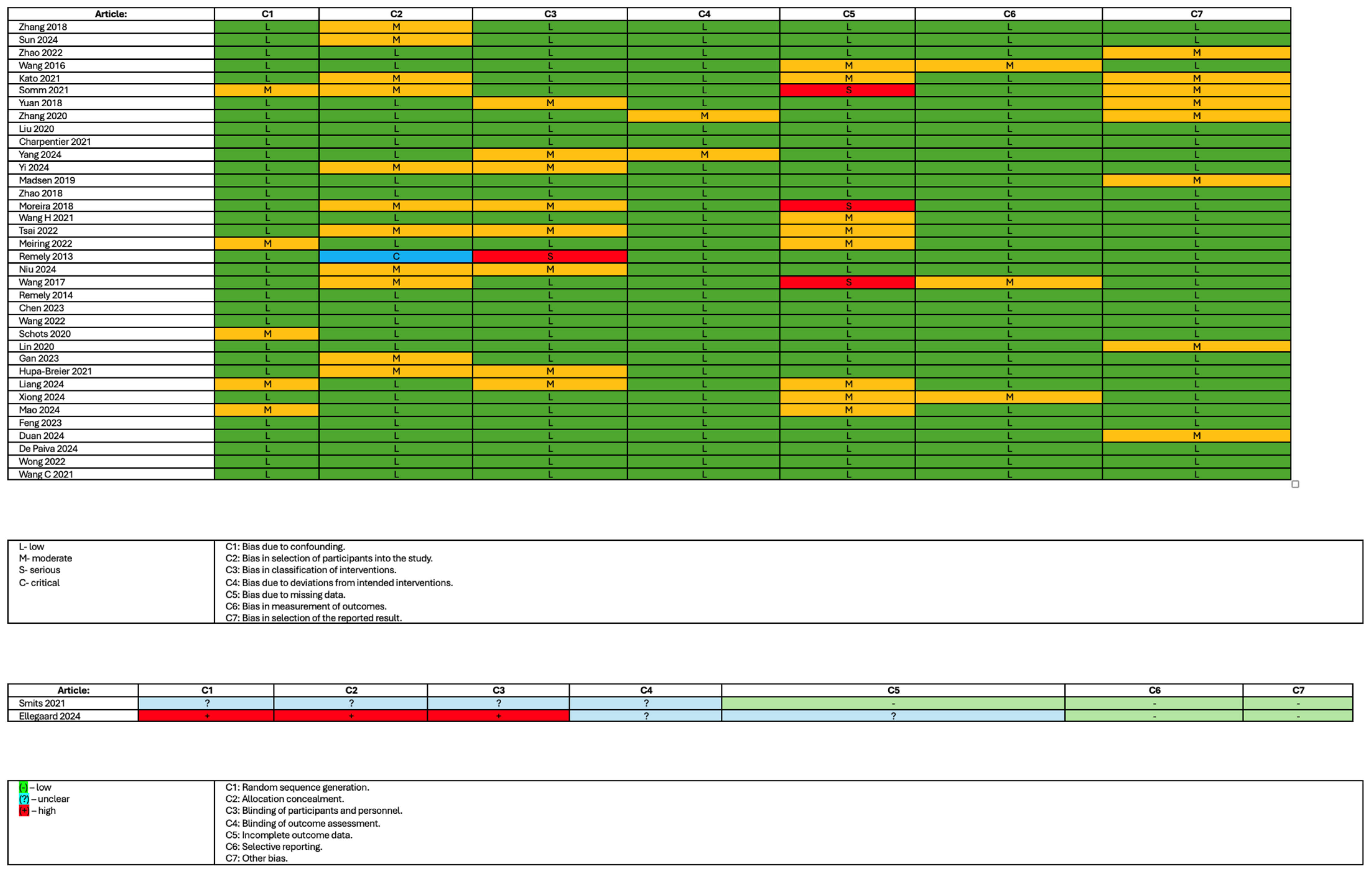
2.5. Study Risk of Bias Assessment
2.6. Assessment Outcomes
3. Results
3.1. Reviewed Studies
3.2. Methods of Analysis for Gut Microbiota Composition in Studies
3.3. The Risk of Bias
3.4. Changes in Bacterial Composition Resulting from GLP-1 Analogue Treatment
3.4.1. Changes in Bacterial Genera and Species Induced by Liraglutide Treatment in the Animal Models
3.4.2. Changes in Bacterial Genera and Species Induced by Liraglutide Treatment in Humans
3.4.3. Changes in Bacterial Genera and Species Induced by Exenatide or Exendin-4 in the Animal Model
3.4.4. Changes in Bacterial Genera and Species Induced by Exenatide in Humans
3.4.5. Changes in Bacterial Genera and Species Induced by Dulaglutide Treatment in an Animal Model
3.4.6. Changes in Bacterial Genera and Species Induced by Dulaglutide Treatment in Humans
3.4.7. Changes in Bacterial Genera and Species Induced by Semaglutide Treatment in the Animal Model
3.5. Effects of GLP-1 Analogues on Gut Microbiota Diversity
3.5.1. Effects of Liraglutide Treatment on Gut Microbiota Diversity
3.5.2. Effects of Exenatide/Exentin-4 Treatment on Gut Microbiota Diversity
3.5.3. Effects of Dulaglutide Treatment on Gut Microbiota Diversity
3.5.4. Effects of Semaglutide Treatment on Gut Microbiota Diversity
3.6. Effects of GLP-1 Receptor Agonists on Metabolic Syndrome
4. Discussion
5. Limitations
6. Summary and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Li, S.; Vandvik, P.O.; Lytvyn, L.; Guyatt, G.H.; Palmer, S.C.; Rodriguez-Gutierrez, R.; Foroutan, F.; Agoritsas, T.; Siemieniuk, R.A.C.; Walsh, M.; et al. SGLT-2 inhibitors or GLP-1 receptor agonists for adults with type 2 diabetes: A clinical practice guideline. BMJ 2021, 373, n1091, Erratum in BMJ 2022, 377, o1080. https://doi.org/10.1136/bmj.o1080. [Google Scholar] [CrossRef] [PubMed]
- ElSayed, N.A.; Aleppo, G.; Aroda, V.R.; Bannuru, R.R.; Brown, F.M.; Bruemmer, D.; Collins, B.S.; Cusi, K.; Das, S.R.; Gibbons, C.H.; et al. Introduction and Methodology: Standards of Care in Diabetes-2023. Diabetes Care 2023, 46 (Suppl. 1), S1–S4. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- International Hypoglycaemia Study Group. Hypoglycaemia, cardiovascular disease, and mortality in diabetes: Epidemiology, pathogenesis, and management. Lancet Diabetes Endocrinol. 2019, 7, 385–396, Erratum in Lancet Diabetes Endocrinol. 2019, 7, e18. https://doi.org/10.1016/S2213-8587(19)30139-1. [Google Scholar] [CrossRef] [PubMed]
- SAXENDA® (Liraglutide) Injection 3 mg Medication Guide. Available online: https://www.novo-pi.com/saxenda.pdf#guide (accessed on 5 April 2025).
- Wegovy® (Semaglutide) Injection 2.4 mg Medication Guide. Available online: https://www.novo-pi.com/wegovy.pdf#guide (accessed on 5 April 2025).
- Xu, D.; Nair, A.; Sigston, C.; Ho, C.; Li, J.; Yang, D.; Liao, X.; Chen, W.; Kuang, M.; Li, Y.; et al. Potential Roles of Glucagon-Like Peptide 1 Receptor Agonists (GLP-1 RAs) in Nondiabetic Populations. Cardiovasc. Ther. 2022, 2022, 6820377. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Morris, A.P.; Voight, B.F.; Teslovich, T.M.; Ferreira, T.; Segrè, A.V.; Steinthorsdottir, V.; Strawbridge, R.J.; Khan, H.; Grallert, H.; Mahajan, A.; et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012, 44, 981–990. [Google Scholar]
- Dietrich, S.; Jacobs, S.; Zheng, J.S.; Meidtner, K.; Schwingshackl, L.; Schulze, M.B. Gene-lifestyle interaction on risk of type 2 diabetes: A systematic review. Obes. Rev. 2019, 20, 1557–1571. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mehta, A.; Singh, S.; Saeed, A.; Mahtta, D.; Bittner, V.A.; Sperling, L.S.; Virani, S.S. Pathophysiological Mechanisms Underlying Excess Risk for Diabetes and Cardiovascular Disease in South Asians: The Perfect Storm. Curr. Diabetes Rev. 2021, 17, e070320183447. [Google Scholar] [CrossRef] [PubMed]
- Gurung, M.; Li, Z.; You, H.; Rodrigues, R.; Jump, D.B.; Morgun, A.; Shulzhenko, N. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine 2020, 51, 102590. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Petakh, P.; Kamyshna, I.; Kamyshnyi, A. Effects of metformin on the gut microbiota: A systematic review. Mol. Metab. 2023, 77, 101805. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sterne, J.A.; Hernán, M.A.; Reeves, B.C.; Savović, J.; Berkman, N.D.; Viswanathan, M.; Henry, D.; Altman, D.G.; Ansari, M.T.; Boutron, I.; et al. ROBINS-I: A tool for assessing risk of bias in non-randomised studies of interventions. BMJ 2016, 355, i4919. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X.; Zheng, J.; Li, M.; Yu, M.; Ping, F.; Wang, T.; Wang, X. Featured article: Structure moderation of gut microbiota in liraglutide-treated diabetic male rats. Exp. Biol. Med. 2017, 243, 34–44. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sun, H.; Shu, J.; Tang, J.; Li, Y.; Qiu, J.; Ding, Z.; Xuan, B.; Chen, M.; Gan, C.; Lin, J.; et al. GLP-1 receptor agonists alleviate colonic inflammation by modulating intestinal microbiota and the function of group 3 innate lymphoid cells. Immunology 2024, 172, 451–468. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Qiu, Y.; Zhang, P.; Wu, X.; Zhao, Z.; Deng, X.; Yang, L.; Wang, D.; Yuan, G. Gut microbiota mediates positive effects of liraglutide on dyslipidemia in mice fed a high-fat diet. Front. Nutr. 2022, 9, 1048693. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, L.; Li, P.; Tang, Z.; Yan, X.; Feng, B. Structural modulation of the gut microbiota and the relationship with body weight: Compared evaluation of liraglutide and saxagliptin treatment. Sci. Rep. 2016, 6, 33251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kato, S.; Sato, T.; Fujita, H.; Kawatani, M.; Yamada, Y. Effects of GLP-1 receptor agonist on changes in the gut bacterium and the underlying mechanisms. Sci. Rep. 2021, 11, 9167. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Somm, E.; Montandon, S.A.; Loizides-Mangold, U.; Gaïa, N.; Lazarevic, V.; De Vito, C.; Perroud, E.; Bochaton-Piallat, M.L.; Dibner, C.; Schrenzel, J.; et al. The GLP-1R agonist liraglutide limits hepatic lipotoxicity and inflammatory response in mice fed a methionine-choline deficient diet. Transl. Res. 2021, 227, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Ni, H.; Chen, X.; Feng, X.; Wu, Q.; Chen, J. Identification of therapeutic effect of glucagon-like peptide 1 in the treatment of STZ-induced diabetes mellitus in rats by restoring the balance of intestinal flora. J. Cell. Biochem. 2018, 119, 10067–10074. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Tao, J.; Gao, L.; Bi, Y.; Li, P.; Wang, H.; Zhu, D.; Feng, W. Liraglutide Attenuates Nonalcoholic Fatty Liver Disease by Modulating Gut Microbiota in Rats Administered a High-Fat Diet. BioMed Res. Int. 2020, 2020, 2947549. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, Q.; Cai, B.Y.; Zhu, L.X.; Xin, X.; Wang, X.; An, Z.M.; Li, S.; Hu, Y.Y.; Feng, Q. Liraglutide modulates gut microbiome and attenuates nonalcoholic fatty liver in db/db mice. Life Sci. 2020, 261, 118457. [Google Scholar] [CrossRef] [PubMed]
- Charpentier, J.; Briand, F.; Lelouvier, B.; Servant, F.; Azalbert, V.; Puel, A.; Christensen, J.E.; Waget, A.; Branchereau, M.; Garret, C.; et al. Liraglutide targets the gut microbiota and the intestinal immune system to regulate insulin secretion. Acta Diabetol. 2021, 58, 881–897. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Deng, L.; Feng, C.; Wen, J. Comparing the effects of empagliflozin and liraglutide on lipid metabolism and intestinal microflora in diabetic mice. PeerJ 2024, 12, e17055. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yi, B.; Su, K.; Cai, Y.L.; Chen, X.L.; Bao, Y.; Wen, Z.Y. Liraglutide ameliorates diabetic kidney disease by modulating gut microbiota and L-5-Oxoproline. Eur. J. Pharmacol. 2024, 983, 176905. [Google Scholar] [CrossRef] [PubMed]
- Madsen, M.S.A.; Holm, J.B.; Pallejà, A.; Wismann, P.; Fabricius, K.; Rigbolt, K.; Mikkelsen, M.; Sommer, M.; Jelsing, J.; Nielsen, H.B.; et al. Metabolic and gut microbiome changes following GLP-1 or dual GLP-1/GLP-2 receptor agonist treatment in diet-induced obese mice. Sci. Rep. 2019, 9, 15582. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, L.; Chen, Y.; Xia, F.; Abudukerimu, B.; Zhang, W.; Guo, Y.; Wang, N.; Lu, Y. A Glucagon-Like Peptide-1 Receptor Agonist Lowers Weight by Modulating the Structure of Gut Microbiota. Front. Endocrinol. 2018, 9, 233. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moreira, G.V.; Azevedo, F.F.; Ribeiro, L.M.; Santos, A.; Guadagnini, D.; Gama, P.; Liberti, E.A.; Saad, M.; Carvalho, C. Liraglutide modulates gut microbiota and reduces NAFLD in obese mice. J. Nutr. Biochem. 2018, 62, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Li, Y.; Luo, S.; Ye, J.; Lu, Z.; Li, X.; Lu, H. The HIF-2α/PPARα pathway is essential for liraglutide-alleviated, lipid-induced hepatic steatosis. Biomed. Pharmacother. 2021, 140, 111778. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.M.; Fluitman, K.S.; Herrema, H.; Davids, M.; Kramer, M.H.H.; Groen, A.K.; Belzer, C.; de Vos, W.M.; Cahen, D.L.; Nieuwdorp, M.; et al. Liraglutide and sitagliptin have no effect on intestinal microbiota composition: A 12-week randomized placebo-controlled trial in adults with type 2 diabetes. Diabetes Metab. 2021, 47, 101223. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.Y.; Lu, H.C.; Chou, Y.H.; Liu, P.Y.; Chen, H.Y.; Huang, M.C.; Lin, C.H.; Tsai, C.N. Gut Microbial Signatures for Glycemic Responses of GLP-1 Receptor Agonists in Type 2 Diabetic Patients: A Pilot Study. Front. Endocrinol. 2022, 12, 814770. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Meiring, S.; van Baar, A.C.G.; Sørensen, N.; Holleman, F.; Soeters, M.R.; Nieuwdorp, M.; Bergman, J.J.G.H.M. A Changed Gut Microbiota Diversity Is Associated With Metabolic Improvements After Duodenal Mucosal Resurfacing With Glucagon-Like-Peptide-1 Receptor Agonist in Type 2 Diabetes in a Pilot Study. Front. Clin. Diabetes Healthc. 2022, 3, 856661. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Remely, M.; Aumueller, E.; Merold, C.; Dworzak, S.; Hippe, B.; Zanner, J.; Pointner, A.; Brath, H.; Haslberger, A.G. Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene 2014, 537, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, A.M.; Kårhus, M.L.; Krych, L.; Sonne, D.P.; Forman, J.L.; Hansen, S.H.; Dragsted, L.O.; Nielsen, D.S.; Knop, F.K. Liraglutide and Colesevelam Change Serum and Fecal Bile Acid Levels in a Randomized Trial With Patients With Bile Acid Diarrhea. Clin. Transl. Gastroenterol. 2024, 15, e00772. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Niu, X.; Lu, P.; Huang, L.; Sun, Y.; Jin, M.; Liu, J.; Li, X. The effect of metformin combined with liraglutide on gut microbiota of Chinese patients with type 2 diabetes. Int. Microbiol. 2023, 27, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Saha, S.; Van Horn, S.; Thomas, E.; Traini, C.; Sathe, G.; Rajpal, D.K.; Brown, J.R. Gut microbiome differences between metformin- and liraglutide-treated T2DM subjects. Endocrinol. Diabetes Metab. 2017, 1, e00009. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Remely, M.; Hippe, B.; Zanner, J.; Aumueller, E.; Brath, H.; Haslberger, A.G. Gut Microbiota of Obese, Type 2 Diabetic Individuals is Enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after Weight Loss. Endocr. Metab. Immune Disord.-Drug Targets (Former. Curr. Drug Targets-Immune Endocr. Metab. Disord.) 2016, 16, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Shu, A.; Jiang, M.; Jiang, J.; Du, Q.; Chen, T.; Shaw, C.; Chai, W.; Chao, T.; Li, X.; et al. Exenatide improves hypogonadism and attenuates inflammation in diabetic mice by modulating gut microbiota. Int. Immunopharmacol. 2023, 120, 110339. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Chen, W.J.; Jin, R.; Xu, X.; Wei, J.; Huang, H.; Tang, Y.H.; Zou, C.W.; Chen, T.T. Engineered probiotics Clostridium butyricum-pMTL007-GLP-1 improves blood pressure via producing GLP-1 and modulating gut microbiota in spontaneous hypertension rat models. Microb. Biotechnol. 2022, 16, 799–812. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Schots, P.C.; Jansen, K.M.; Mrazek, J.; Pedersen, A.M.; Olsen, R.L.; Larsen, T.S. Obesity-induced alterations in the gut microbiome in female mice fed a high-fat diet are antagonized by dietary supplementation with a novel, wax ester-rich, marine oil. Nutr. Res. 2020, 83, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Dong, C.; Zhao, B.; Zhou, B.; Yang, L. Glucagon-like peptide-1 receptor agonist regulates fat browning by altering the gut microbiota and ceramide metabolism. Medcomm 2023, 4, e416. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gan, J.; Chen, J.; Ma, R.L.; Deng, Y.; Ding, X.S.; Zhu, S.Y.; Sun, A.J. Metagenomics study on taxonomic and functional change of gut microbiota in patients with obesity with PCOS treated with exenatide combination with metformin or metformin alone. Gynecol. Endocrinol. 2023, 39, 2219342. [Google Scholar] [CrossRef] [PubMed]
- Hupa-Breier, K.L.; Dywicki, J.; Hartleben, B.; Wellhöner, F.; Heidrich, B.; Taubert, R.; Mederacke, Y.E.; Lieber, M.; Iordanidis, K.; Manns, M.P.; et al. Dulaglutide Alone and in Combination with Empagliflozin Attenuate Inflammatory Pathways and Microbiome Dysbiosis in a Non-Diabetic Mouse Model of NASH. Biomedicines 2021, 9, 353. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liang, L.; Su, X.; Guan, Y.; Wu, B.; Zhang, X.; Nian, X. Correlation between intestinal flora and GLP-1 receptor agonist dulaglutide in type 2 diabetes mellitus treatment-A preliminary longitudinal study. iScience 2024, 27, 109784. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Xiong, C.; Wu, J.; Ma, Y.; Li, N.; Wang, X.; Li, Y.; Ding, X. Effects of Glucagon-Like Peptide-1 Receptor Agonists on Gut Microbiota in Dehydroepiandrosterone-Induced Polycystic Ovary Syndrome Mice: Compared Evaluation of Liraglutide and Semaglutide Intervention. Diabetes Metab. Syndr. Obes. Targets Ther. 2024, ume 17, 865–880. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mao, T.; Zhang, C.; Yang, S.; Bi, Y.; Li, M.; Yu, J. Semaglutide alters gut microbiota and improves NAFLD in db/db mice. Biochem. Biophys. Res. Commun. 2024, 710, 149882. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Teng, Z.; Yang, Y.; Liu, J.; Chen, S. Effects of semaglutide on gut microbiota, cognitive function and inflammation in obese mice. PeerJ 2024, 12, e17891. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Duan, X.; Zhang, L.; Liao, Y.; Lin, Z.; Guo, C.; Luo, S.; Wang, F.; Zou, Z.; Zeng, Z.; Chen, C.; et al. Semaglutide alleviates gut microbiota dysbiosis induced by a high-fat diet. Eur. J. Pharmacol. 2024, 969, 176440. [Google Scholar] [CrossRef] [PubMed]
- de Paiva, I.H.R.; da Silva, R.S.; Mendonça, I.P.; de Souza, J.R.B.; Peixoto, C.A. Semaglutide Attenuates Anxious and Depressive-Like Behaviors and Reverses the Cognitive Impairment in a Type 2 Diabetes Mellitus Mouse Model Via the Microbiota-Gut-Brain Axis. J. Neuroimmune Pharmacol. 2024, 19, 36. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Yusta, B.; Koehler, J.A.; Baggio, L.L.; McLean, B.A.; Matthews, D.; Seeley, R.J.; Drucker, D.J. Divergent roles for the gut intraepithelial lymphocyte GLP-1R in control of metabolism, microbiota, and T cell-induced inflammation. Cell Metab. 2022, 34, 1514–1531.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Hu, H.J.; Dong, Q.Q.; Huang, R.; Zhao, W.; Song, Y.J.; Li, Z.Y.; Wang, N.; Zhang, T.C.; Luo, X.G. Enhancing bile tolerance of Lactobacilli is involved in the hypolipidemic effects of liraglutide. Biosci. Biotechnol. Biochem. 2021, 85, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Asadifard, E.; Hokmabadi, M.; Hashemi, M.; Bereimipour, A. Linking gut microbiota dysbiosis to molecular pathways in Alzheimer’s disease. Brain Res. 2024, 1845, 149242. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Camargo Tavares, L.; Lee, H.C.; Steele, J.R.; Ribeiro, R.V.; Beale, A.L.; Yiallourou, S.; Carrington, M.J.; Kaye, D.M.; Head, G.A.; et al. Faecal metaproteomics analysis reveals a high cardiovascular risk profile across healthy individuals and heart failure patients. Gut Microbes 2024, 17, 2441356. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, F.; Li, Y.; Shi, Y.; Wang, X.; Chen, X.; Zheng, W.; Hsing, J.C.; Lu, Y.; Wu, Y.S.; et al. Distinct Gut Microbiota Profiles in Normal Weight Obesity and Their Association With Cardiometabolic Diseases: Results From Two Independent Cohort Studies. J. Cachexia Sarcopenia Muscle 2024, 16, e13644. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Misera, A.; Kaczmarczyk, M.; Łoniewski, I.; Liśkiewicz, P.; Podsiadło, K.; Misiak, B.; Skonieczna-Żydecka, K.; Samochowiec, J. Comparative analysis of gut microbiota in major depressive disorder and schizophrenia during hospitalisation—The case-control, post hoc study. Psychoneuroendocrinology 2024, 171, 107208. [Google Scholar] [CrossRef] [PubMed]
- Bellando-Randone, S.; Russo, E.; Di Gloria, L.; Lepri, G.; Baldi, S.; Fioretto, B.S.; Romano, E.; Ghezzi, G.; Bertorello, S.; El Aoufy, K.; et al. Gut microbiota in very early systemic sclerosis: The first case-control taxonomic and functional characterisation highlighting an altered butyric acid profile. RMD Open 2024, 10, e004647. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, Y.; Chen, Z.; Li, C.; Sun, T.; Luo, X.; Jiang, B.; Liu, M.; Wang, Q.; Li, T.; Cao, J.; et al. Associations between changes in the gut microbiota and liver cirrhosis: A systematic review and meta-analysis. BMC Gastroenterol. 2025, 25, 16. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, J.; Ou, G.; Wang, S.; Chen, Y.; Xu, L.; Deng, L.; Xu, H.; Chen, X. The predictive, preventive, and personalized medicine of depression: Gut microbiota and inflammation. EPMA J. 2024, 15, 587–598. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Adeshirlarijaney, A.; Gewirtz, A.T. Considering gut microbiota in treatment of type 2 diabetes mellitus. Gut Microbes 2020, 11, 253–264. [Google Scholar] [CrossRef]
- Cheng, Y.; Lin, S.; Cao, Z.; Yu, R.; Fan, Y.; Chen, J. The role of chronic low-grade inflammation in the development of sarcopenia: Advances in molecular mechanisms. Int. Immunopharmacol. 2025, 147, 114056. [Google Scholar] [CrossRef] [PubMed]
- Gofron, K.; Berezowski, A.; Gofron, M.; Borówka, M.; Dziedzic, M.; Kazimierczak, W.; Kwiatkowski, M.; Gofron, M.; Nowaczyk, Z.; Małgorzewicz, S. Akkermansia muciniphila—Impact on the cardiovascular risk, the intestine inflammation and obesity. Acta Biochim. Pol. 2024, 71, 13550. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.X.; Chang, S.Y.; Jin, Z.Y.; Li, D.; Zhu, J.; Luo, Z.B.; Han, S.Z.; Kang, J.D.; Quan, L.H. Lactobacillus reuteri-Enriched Eicosatrienoic Acid Regulates Glucose Homeostasis by Promoting GLP-1 Secretion to Protect Intestinal Barrier Integrity. J. Agric. Food Chem. 2024, 73, 393–408. [Google Scholar] [CrossRef] [PubMed]
- Bonab, S.F.; Tahmasebi, S.; Ghafouri-Fard, S.; Eslami, S. Preventive impact of probiotic supplements on heart injury and inflammatory indices in a rat model of myocardial infarction: Histopathological and gene expression evaluation. APMIS 2024, 133, e13479. [Google Scholar] [CrossRef] [PubMed]
- Chi, Y.Y.; Xiang, J.Y.; Li, H.M.; Shi, H.Y.; Ning, K.; Shi, C.; Xiang, H.; Xie, Q. Schisandra chinensis polysaccharide prevents alcohol-associated liver disease in mice by modulating the gut microbiota-tryptophan metabolism-AHR pathway axis. Int. J. Biol. Macromol. 2024, 282, 136843. [Google Scholar] [CrossRef] [PubMed]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
| No. | Article | Type of Study | Description of the Experiment and Study Groups/Description of the Study | Method of Analysis | Conclusion |
|---|---|---|---|---|---|
| 1 | Zhang 2018 [15] | Experimental | Diabetic male rats | 16S rRNA V3-V4 | In liraglutide-treated diabetic male rats, short-chain fatty acid (SCFA)-producing bacteria, including Bacteroides, Lachnospiraceae, and the probiotic Bifidobacterium, were selectively enhanced. Lactobacillus showed a negative correlation with fasting blood glucose. |
| 2 | Sun 2024 [16] | Experimental | Wild-type and Rag1−/−, Rag2−/− i, Rorcgfp/gfp mice | 16S rRNA V3-V4 | Liraglutide administration significantly alleviates the DSS-induced colitis symptoms and ameliorates histological damage. |
| 3 | Zhao 2022 [17] | Experimental | C57BL/6 mice | 16S rDNA V3-V4 | Correlation analysis showed that TC and LDL were linked to harmful bacteria and inversely related to beneficial ones. Liraglutide improves dyslipidemia in HFD-fed mice by modulating gut microbiota, especially Akkermansia, suggesting this as a key mechanism of its effect. |
| 4 | Wang 2016 [18] | Experimental | ApoE−/− mice on C57BL/6 background | 16s rDNA V1-V3 | The GLP-1 receptor agonist liraglutide modulates gut microbiota composition, promoting a lean-associated profile that aligns with its weight-loss effects. |
| 5 | Kato 2021 [19] | Experimental | Mice | 16S/18S rRNA | Increased levels of caseinolytic protease B, a component of Escherichia coli, and norepinephrine were observed in the cecum. |
| 6 | Somm 2021 [20] | Experimental | Mice fed a regular diet or a methionine-choline deficient (MCD) diet. | 16S rDNA V3-V4 | Liraglutide altered gut microbiota composition affected by the MCD diet, restoring normal Bacteroides levels and shifting Erysipelotrichaceae from Allobaculum to Turicibacter. |
| 7 | Yuan 2018 [21] | Experimental | STZ-induced Sprague-Dawley rats | 16S rRNA V3 | GLP-1, by restoring the intestinal flora dysbiosis, can moderate STZ-induced DM treatment. |
| 8 | Zhang 2020 [22] | Experimental | Male rats fed a high-fat diet. After 12 weeks, two rats | 16s rDNA V4-V5 | Liraglutide significantly modified the structure and composition of HFD-disrupted gut microbiota in comparison to rats on a normal diet. |
| 9 | Liu 2020 [23] | Experimental | Male db/db mice | 16S rRNA V3-V4 | Bacteria like Parabacteroides, Oscillibacter, and Prevotellaceae_UCG-001 produce anti-inflammatory SCFAs and influence T cell differentiation. In the LG group, bacteria such as Anaerotruncus and Lachnospiraceae were reduced and correlated with ALT and AST. Oscillibacter and Klebsiella correlated with ALT, while Desulfovibrio and Bacteroides correlated with AST. |
| 10 | Charpentier 2021 [24] | Experimental | Diet-induced dysmetabolic mice | 16S rRNA | Data showed that GLP-1 receptor agonists, like liraglutide, regulate insulin secretion by simultaneously modulating intestinal immunity and bacterial ecology. |
| 11 | Yang 2024 [25] | Experimental | Mouse diabetic model | 16S rRNA | Empagliflozin reduced the Firmicutes/Bacteroidota ratio, while liraglutide had no effect. Both decreased Helicobacter and increased Lactobacillus. Empagliflozin also raised Muribaculaceae, Muribaculum, Olsenella, and Odoribacter, whereas liraglutide increased Ruminococcus. |
| 12 | Yi 2024 [26] | Experimental | Diabetic kidney disease rats | 16S rRNA V3-V4 | The gut microbiota-metabolites-kidney axis, particularly Clostridium-5-OP-ELD, may serve as a potential target of Liraglutide in DKD. |
| 13 | Madsen 2019 [27] | Experimental | Diet-induced obese mice. | 16S rRNA V3-V4 | DIO mice showed similar gut bacterial changes after liraglutide and GUB09-145 treatment, with shifts in low-abundant species and metabolic pathways. |
| 14 | Zhao 2018 [28] | Experimental | Wistar and Goto–Kakizaki (GK) rats | 16S rDNA V3-V4 | Liraglutide prevents weight gain by altering gut microbiota, reducing diversity, increasing the Firmicutes/Bacteroidetes ratio, and promoting lean-associated microbial profiles in obesity and T2DM. |
| 15 | Moreira 2018 [29] | Experimental | C57BL/6J mice | 16S rRNA V3-V4 | Liraglutide modified gut microbiota by reducing Proteobacteria and increasing Akkermansia muciniphila, contributing to weight loss, microbiota balance, and NAFLD improvement. |
| 16 | Wang H 2021 [30] | Experimental | HIF-2α heterozygous knockout (HIF-2α+/−) and wild-type (WT) littermate mice | 16S rRNA V3-V4 | HIF-2α is essential for the effect of liraglutide in promoting the increased abundance of Akkermansia muciniphila. |
| 17 | Smits 2021 [31] | Randomized trial | Fifty-one adults (humans) with type 2 diabetes | 16S rRNA V3-V4 | Liraglutide and sitagliptin improve glucose metabolism, body weight, and bile acids as add-ons to metformin or sulphonylureas, independent of changes in intestinal microbiota. |
| 18 | Tsai 2022 [32] | Longitudinal study | Fifty-two adults | 16S rRNA | Bacteroides dorei, Lachnoclostridium sp., and Mitsuokella multacida were significant after adjusting for baseline glycohemoglobin and C-peptide concentrations, two clinical confounders. |
| 19 | Meiring 2022 [33] | spin-off study | Fecal samples from 16 patients were collected for Illumina shotgun sequencing at baseline and three months post-DMR. | shotgun sequencing | Gut microbiota diversity changes correlate with HbA1c, PDFF, and metabolic improvements after DMR and GLP-1 receptor agonist treatment in type 2 diabetes. |
| 20 | Remely 2013 [34] | Longitudinal study | Fourteen obese individuals (OC) with no established insulin resistance and twenty-four insulin-dependent type 2 diabetes (D) patients | 16S rRNA | Gut microbiota in obesity and type 2 diabetes may influence FFAR epigenetics, affecting satiety and hunger. Butyrate supports regenerative medicine by promoting epigenetic remodeling and gene expression. |
| 21 | Ellegaard 2024 [35] | Randomized trial | Fifty-two adults | 16S rRNA | Neither treatment resulted in changes to the composition of fecal microbiota. |
| 22 | Niu 2024 [36] | Longitudinal study | All 32 participants in the pre-diabetic/overweight control group (NCP) later transitioned to the untreated T2DM group (UNT) upon diagnosis. | 16S rRNA V4 | MET and MET + LRG treatment significantly altered gut microbiota from T2DM diagnosis, with MET + LRG showing distinct changes, suggesting LRG’s additive effect. |
| 23 | Wang 2017 [37] | Longitudinal study | 37 patients with T2DM | 16S rRNA V4 | The study suggests that altering the gut microbiome may play a key role in the mechanisms of action of metformin and liraglutide. |
| 24 | Remely 2014 [38] | Longitudinal study | T2DM patients | 16S rRNA | Gut microbiota composition varies between groups with GLP-1 analogue therapy and that with no therapy. |
| 25 | Chen 2023 [39] | Experimental | C57BL/6J mice | 16S rDNA V3-V4 | Exenatide reduced pathogenic bacteria, increased Akkermansia, improved testosterone, and lowered inflammation. Fecal bacteria from the Exe group reduced harmful microbes and protected against testicular damage in diabetic mice. |
| 26 | Wang 2022 [40] | Experimental | Hypertensive rats (SHR). SHR and Wistar rats. | 16S rRNA V3-V4 | The treatments corrected dysbiosis in SHR rats by reducing Porphyromonadaceae and increasing Lactobacillus. GLP-1 improved blood pressure and cardiomegaly by restoring the gut microbiome and reducing ventricular hypertrophy. |
| 27 | Schots 2020 [41] | Experimental | 57bl/6J mice | 16s rDNA V4-V5 | Exenatide prevented the HFD-induced increase in Lactococcus and caused a decrease in the abundance of Streptococcus compared to the HFD group. |
| 28 | Lin 2020 [42] | Experimental | Mice | 16S/18S rRNA | Lactobacillus reuteri increased in bacterial counts, while ceramide levels in mouse serum decreased, with a negative correlation between L. reuteri and ceramide. The decrease in ceramide was linked to overexpression of alkaline ceramidase 2 (Acer2). L. reuteri may work synergistically with GLP-1 RA for therapeutic benefits. |
| 29 | Gan 2023 [43] | Non-randomized controlled trial | Obese patients with polycystic ovary syndrome (PCOS) | Paired-end sequencing | Both groups had high levels of Firmicutes, Bacteroidetes, Uroviricota, Actinobacteria, and Proteobacteria. Post-treatment, probiotics like Phocaeicola and Anaerobutyricum increased. Clostridium, Fusobacterium, and Oxalobacter dominated in the post-MF group, while Lactococcus garvieae, Clostridium perfringens, and Coprococcus sp. AF16_5 were dominant in the post-COM group. |
| 30 | Hupa-Breier 2021 [44] | Experimental | C57BL/6HanJ Ztm. mice | real-time PCR | The study highlights dulaglutide’s significant anti-inflammatory effects, especially when combined with empagliflozin, by modulating pro-inflammatory immune responses and microbiome dysbiosis. |
| 31 | Liang 2024 [45] | Longitudinal study | Patients with newly diagnosed T2DM with no GLP-1RA history. | 16S rRNA | No significant changes in gut microbiota after 1 week of dulaglutide treatment in newly diagnosed T2DM patients. After 48 weeks, notable changes occurred, including a significant reduction in microbial abundance. |
| 32 | Xiong 2024 [46] | Experimental | Dehydroepiandrosterone (DHEA)-induced PCOS mice | shotgun sequencing | The greater efficacy in weight loss compared with liraglutide observed after semaglutide intervention was positively related with Helicobacter. |
| 33 | Mao 2024 [47] | Experimental | db/m and db/db mice | 16S rRNA | Significant changes in Alloprevotella, Alistipes, Ligilactobacillus, and Lactobacillus were observed between the db/db control group and the semaglutide treatment group. LDA scores showed increased Alloprevotella and Alistipes, while Ligilactobacillus and Lactobacillus decreased after semaglutide treatment. |
| 34 | Feng 2023 [48] | Experimental | C57BL/6J mice | 16S rRNA V3-V4 | Semaglutide may influence gut microbiota composition and structure, impacting cognitive function and inflammation. |
| 35 | Duan 2024 [49] | Experimental | Mice fed a high-fat diet | 16S rRNA | Semaglutide could alter gut microbiota dysbiosis, which may explain the anti-obesity effects of semaglutide. |
| 36 | De Paiva 2024 [50] | Experimental | C57BL/6 mice | 16S rRNA | Semaglutide may treat depression and anxiety by reducing hippocampal neuroinflammation and promoting neurogenesis through the insulin/GLP-1 pathway and gut microbiota modulation. |
| 37 | Wong 2022 [51] | Experimental | Rag2−/−;Il2rg−/− mice and Glp1rTcell−/− mice | 16S rRNA V3-V4 | The intestinal intraepithelial lymphocyte GLP-1R is required for actions of GLP-1 on gut microbiota and for selective restraint of local and systemic T cell-induced, but not lipopolysaccharide-induced, inflammation. |
| 38 | Wang C 2021 [52] | Experimental | Mice with a high-fat diet | 16S rRNA | Liraglutide increases Lactobacillaceae abundance in hyperlipidemic mice, enhances LAB bile tolerance by upregulating bile salt hydrolases, and liraglutide-sensitive LAB lysates directly downregulate HMGCR. |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Bacteroides | 5 | 3 |
| Bacteroidota | Alistipes | 2 | 0 |
| Bacteroidota | Parabacteroides | 2 | 1 |
| Bacteroidota | Butyricimonas | 2 | 0 |
| Bacteroidota | Prevotella_9 | 0 | 2 |
| Bacteroidota | Norank_F_Bacteroidales_S24-7_Group | 1 | 0 |
| Bacteroidota | Barnesiella | 1 | 0 |
| Bacillota | Ruminicoccus | 2 | 4 |
| Bacillota | Lactobacillus | 6 | 0 |
| Bacillota | Turicibacter | 2 | 1 |
| Bacillota | Staphylococcus | 0 | 1 |
| Bacillota | Faecalibaculum | 0 | 1 |
| Bacillota | Allobaculum | 4 | 1 |
| Bacillota | Oscillospira | 2 | 0 |
| Bacillota | Clostridium | 3 | 2 |
| Bacillota | Anaerotruncus | 0 | 2 |
| Bacillota | Anaerostipes | 1 | 0 |
| Bacillota | Blautia | 2 | 0 |
| Bacillota | Candidatus Arthromitus | 0 | 1 |
| Bacillota | Roseburia | 0 | 2 |
| Bacillota | Marvinbryantia | 0 | 1 |
| Bacillota | Flavonifractor | 1 | 1 |
| Bacillota | Lachnoclostridium | 1 | 0 |
| Bacillota | Cellulosilyticum | 1 | 0 |
| Bacillota | Christensenellaceae_R-7_group | 0 | 1 |
| Bacillota | Lachnospiraceae_UCG-010 | 1 | 1 |
| Bacillota | Peptoniphilus | 1 | 0 |
| Bacillota | Ruminoclostridium_6 | 0 | 2 |
| Bacillota | Romboutsia | 1 | 0 |
| Bacillota | Lachnospiraceae_NK4A136_group | 0 | 1 |
| Bacillota | Oscillibacter | 0 | 1 |
| Bacillota | Ruminiclostridium_9 | 0 | 1 |
| Bacillota | uncultured_f__Peptococcaceae | 0 | 1 |
| Bacillota | Sarcina | 1 | 0 |
| Bacillota | SMB53 | 1 | 0 |
| Bacillota | 02d06 | 1 | 0 |
| Verrucomicrobia | Akkermansia | 5 | 0 |
| Pseudomonadota | Helicobacter | 1 | 2 |
| Pseudomonadota | Desulfobacterota | 0 | 1 |
| Pseudomonadota | Sutterella | 1 | 0 |
| Pseudomonadota | Desulfovibrio | 2 | 1 |
| Pseudomonadota | Oxalobacter | 1 | 0 |
| Pseudomonadota | Sphingomonas | 1 | 0 |
| Pseudomonadota | Klebsiella | 0 | 1 |
| Pseudomonadota | Escherichia | 1 | 0 |
| Pseudomonadota | Shigella | 1 | 0 |
| Actinomycetota | Bifidobacteria | 2 | 0 |
| Actinomycetota | Enterorhabdus | 1 | 0 |
| Actinomycetota | Gardnerella | 1 | 0 |
| Actinomycetota | Johnsonella | 1 | 0 |
| Actinomycetota | Candidatus_Saccharimonas | 0 | 1 |
| Fusobacteria | Sneathia | 1 | 0 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Alistipes | 0 | 1 |
| Bacillota | Lactococcus | 1 | 0 |
| Bacillota | Blautia | 0 | 1 |
| Bacillota | Dialister | 0 | 1 |
| Bacillota | Megasphaera | 0 | 1 |
| Bacillota | unknown genus in the family Christensenellaceae | 1 | 0 |
| Verrucomicrobia | Akkermansia | 1 | 0 |
| Pseudomonadota | Sutterella | 0 | 1 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Odoribacter | 1 | 0 |
| Bacteroidota | Bacteroides | 0 | 1 |
| Bacteroidota | Barnesiella | 1 | 0 |
| Bacillota | Lactobacillus | 1 | 1 |
| Bacillota | Romboutsia | 0 | 1 |
| Bacillota | Streptococcus | 0 | 2 |
| Bacillota | Weissella | 0 | 1 |
| Bacillota | Marvinbryantia | 0 | 1 |
| Bacillota | Enterococcus | 0 | 1 |
| Bacillota | Lactococcus | 0 | 1 |
| Bacillota | Ruminococcus | 1 | 0 |
| Bacillota | Flavonifactor | 0 | 1 |
| Verrucomicrobia | Akkermansia | 1 | 0 |
| Actinomycetota | Enterorhabdus | 0 | 1 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Actinomycetota | Bifidobacterium | 1 | 0 |
| Bacteroidota | Prevotella | 1 | 0 |
| Bacillota | Oscillospira | 0 | 1 |
| Bacillota | Flavonifractor | 0 | 1 |
| Bacillota | Lactococcus | 0 | 1 |
| Bacillota | Anaerotignum | 0 | 1 |
| Bacillota | Coprococcus | 1 | 0 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Alistipes | 1 | 0 |
| Bacteroidota | Bacteroides | 1 | 0 |
| Bacteroidota | Barnesiella | 1 | 0 |
| Bacteroidota | Parabacteroides | 1 | 0 |
| Bacillota | Aerococcus | 1 | 0 |
| Bacillota | Clostridium sensu stricto | 1 | 0 |
| Bacillota | Clostridium XlVb | 1 | 0 |
| Bacillota | Coprobacillus | 0 | 1 |
| Bacillota | Enterococcus | 1 | 0 |
| Bacillota | Lachnospiracea_ incertae_sedis | 1 | 0 |
| Bacillota | Macrococcus | 0 | 1 |
| Bacillota | Oscillibacter | 1 | 0 |
| Bacillota | Pseudoflavonifractor | 1 | 0 |
| Bacillota | Roseburia | 0 | 1 |
| Bacillota | Ruminococcus | 1 | 0 |
| Bacillota | Streptococcus | 0 | 1 |
| Pseudomonadota | Escherichia/Shigella | 1 | 0 |
| Pseudomonadota | Parasutterella | 1 | 0 |
| Verrucomicrobia | Akkermansia | 1 | 0 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Bacteroides | 1 | 0 |
| Bacteroidota | Prevotella | 1 | 1 |
| Bacillota | Blautia | 0 | 1 |
| Bacillota | Lactobacillus | 1 | 0 |
| Bacillota | Ruminococcus | 0 | 1 |
| Actinomycetota | Bifidobacterium | 1 | 1 |
| Phylum | Genus | Increased | Decreased |
|---|---|---|---|
| Bacteroidota | Bacteriodes | 1 | 0 |
| Bacteroidota | Odoribacter | 0 | 1 |
| Bacteroidota | Muribaculaceae | 1 | 0 |
| Bacteroidota | Alloprevotella | 1 | 0 |
| Bacteroidota | Alistpes | 1 | 0 |
| Bacillota | Blautia | 1 | 0 |
| Bacillota | Dubosiella | 2 | 1 |
| Bacillota | Romboutsia | 0 | 2 |
| Bacillota | Enterococcus | 1 | 0 |
| Bacillota | Lactobacillus | 1 | 1 |
| Bacillota | Clostridia _UCG_014 | 1 | 0 |
| Bacillota | Ligilactobacillus | 0 | 1 |
| Bacillota | Allobaculum | 1 | 0 |
| Bacillota | Lachnospiraceae_NK4A136_group | 0 | 1 |
| Bacillota | unclassified_f_Oscillospiraceae | 0 | 1 |
| Pseudomonadota | Escherichia-Shigella | 1 | 0 |
| Pseudomonadota | Helicobacter | 1 | 0 |
| Actinomycetota | Coriobacteriaceae UCG-002 | 2 | 0 |
| Verrucomicrobia | Akkermansia | 2 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gofron, K.K.; Wasilewski, A.; Małgorzewicz, S. Effects of GLP-1 Analogues and Agonists on the Gut Microbiota: A Systematic Review. Nutrients 2025, 17, 1303. https://doi.org/10.3390/nu17081303
Gofron KK, Wasilewski A, Małgorzewicz S. Effects of GLP-1 Analogues and Agonists on the Gut Microbiota: A Systematic Review. Nutrients. 2025; 17(8):1303. https://doi.org/10.3390/nu17081303
Chicago/Turabian StyleGofron, Krzysztof Ksawery, Andrzej Wasilewski, and Sylwia Małgorzewicz. 2025. "Effects of GLP-1 Analogues and Agonists on the Gut Microbiota: A Systematic Review" Nutrients 17, no. 8: 1303. https://doi.org/10.3390/nu17081303
APA StyleGofron, K. K., Wasilewski, A., & Małgorzewicz, S. (2025). Effects of GLP-1 Analogues and Agonists on the Gut Microbiota: A Systematic Review. Nutrients, 17(8), 1303. https://doi.org/10.3390/nu17081303








