Imaging Collagen Alterations in STICs and High Grade Ovarian Cancers in the Fallopian Tubes by Second Harmonic Generation Microscopy
Abstract
:1. Introduction
2. Results
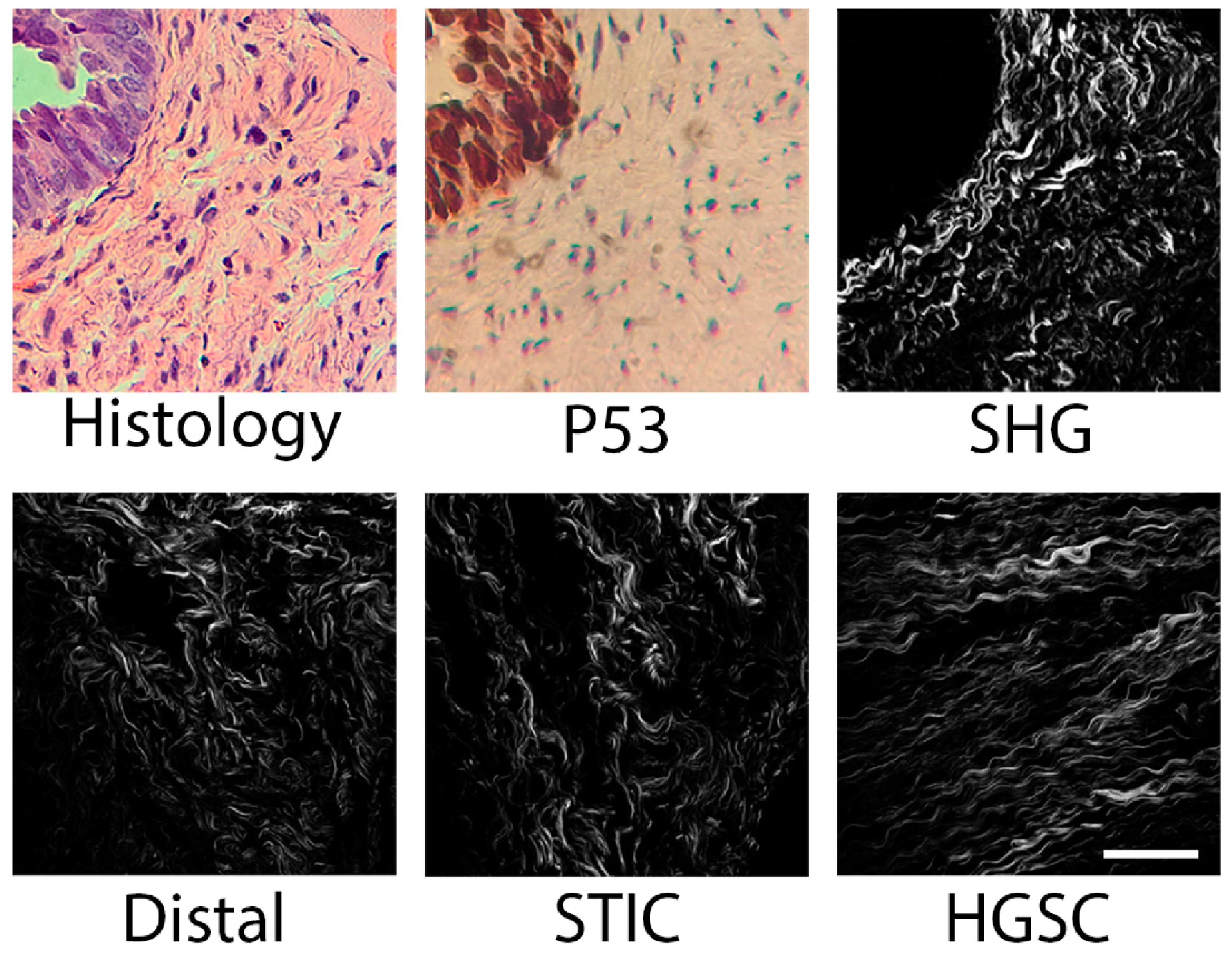
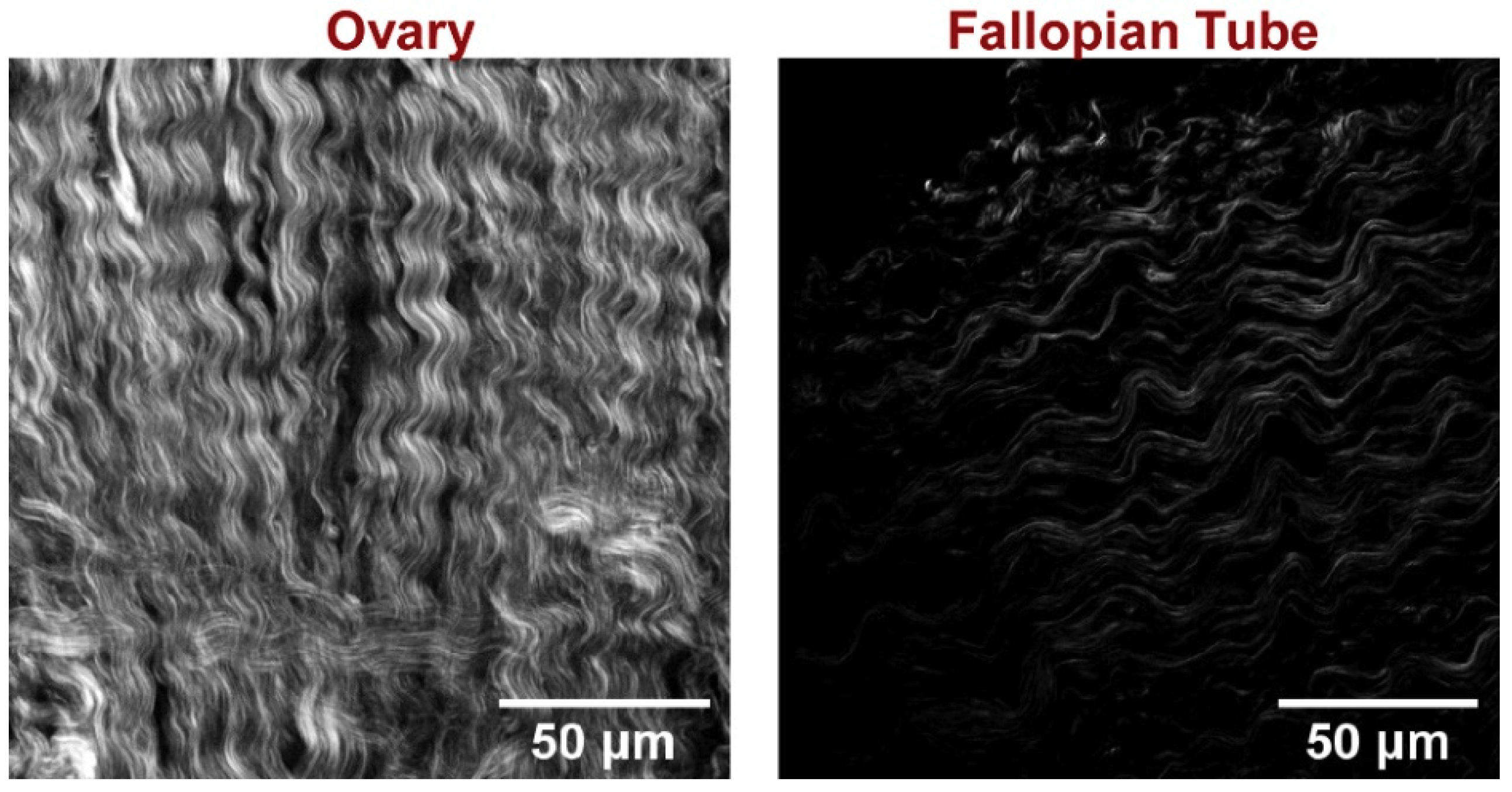
2.1. SHG Sampling
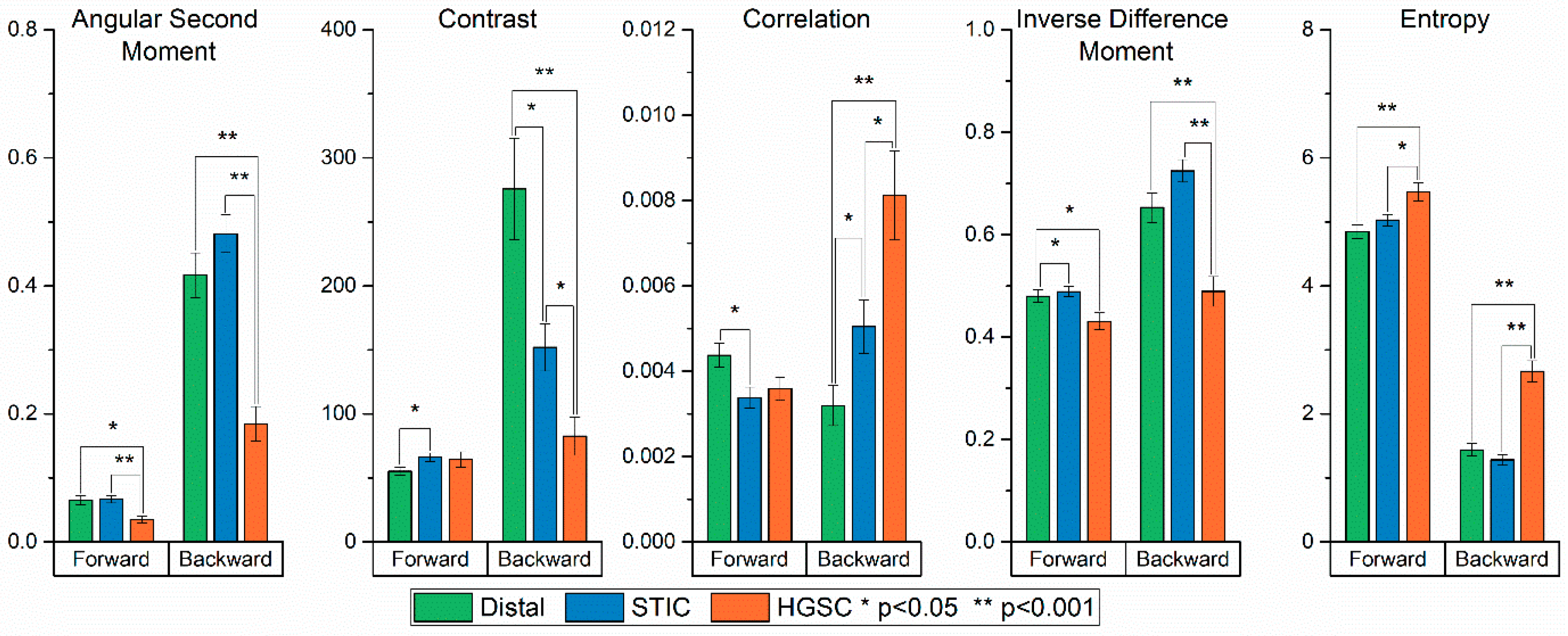
2.2. Gray Level Co-Occurrence Matrix Analysis
- (i)
- Angular Second Moment (ASM) and Energy measures the number of repeated pairs. The energy (square root of ASM) will be high if the occurrence of repeated pixel pairs is high.
- (ii)
- Inverse Difference Moment (IDM) is related to the smoothness or homogeneity across the image and will be high if the gray levels of the pixel pairs are similar.
- (iii)
- Contrast is a measure of the local contrast of an image and will be low if the gray levels of each pixel pair are similar.
- (iv)
- Entropy measures the randomness of a gray level distribution and will be high if the gray levels are distributed randomly throughout the image.
- (v)
- Correlation measures the linear dependency of gray levels on those of neighboring pixels.
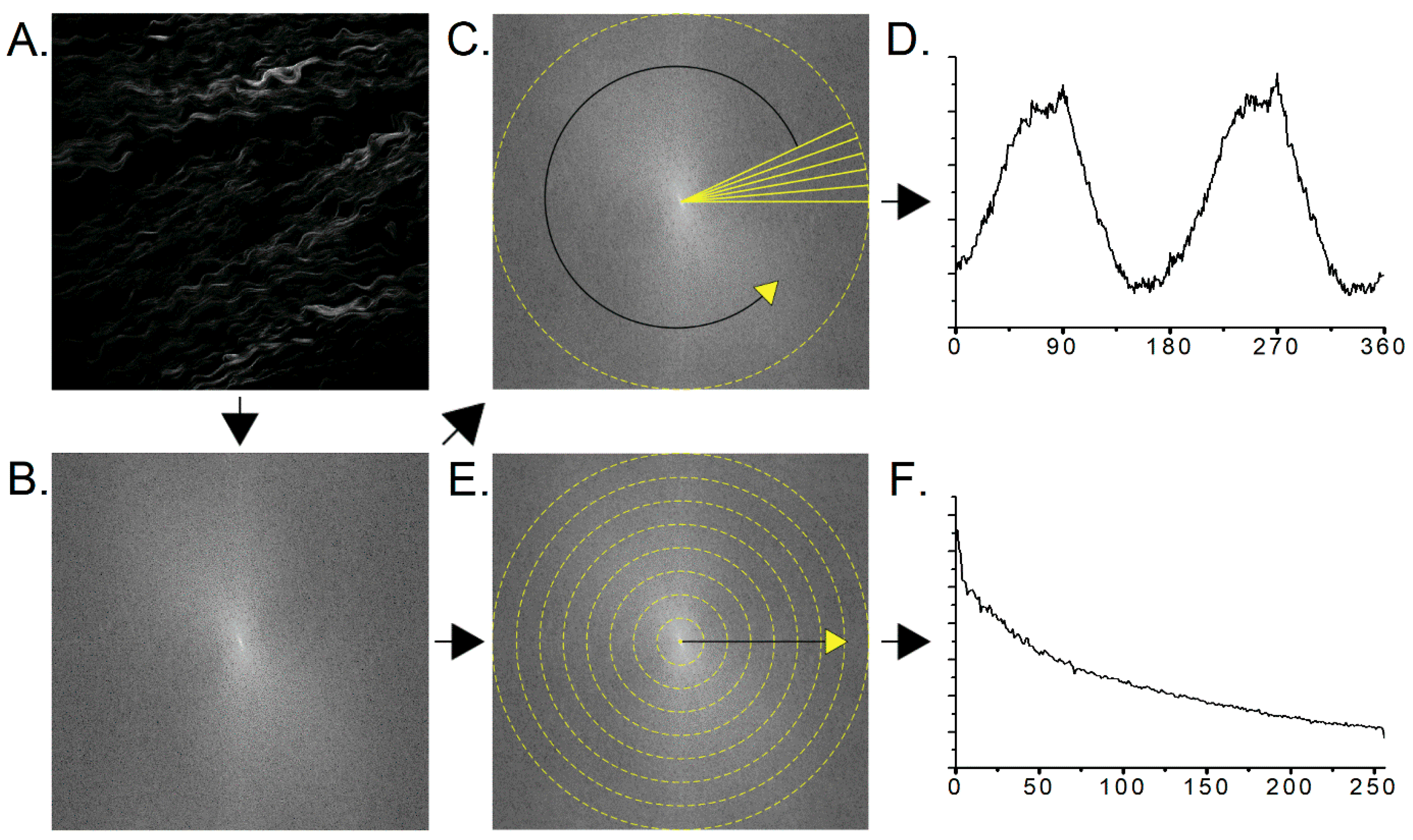
2.3. Two-Dimensional Fast Fourier Transform Analysis
2.4. CT-FIRE Analysis
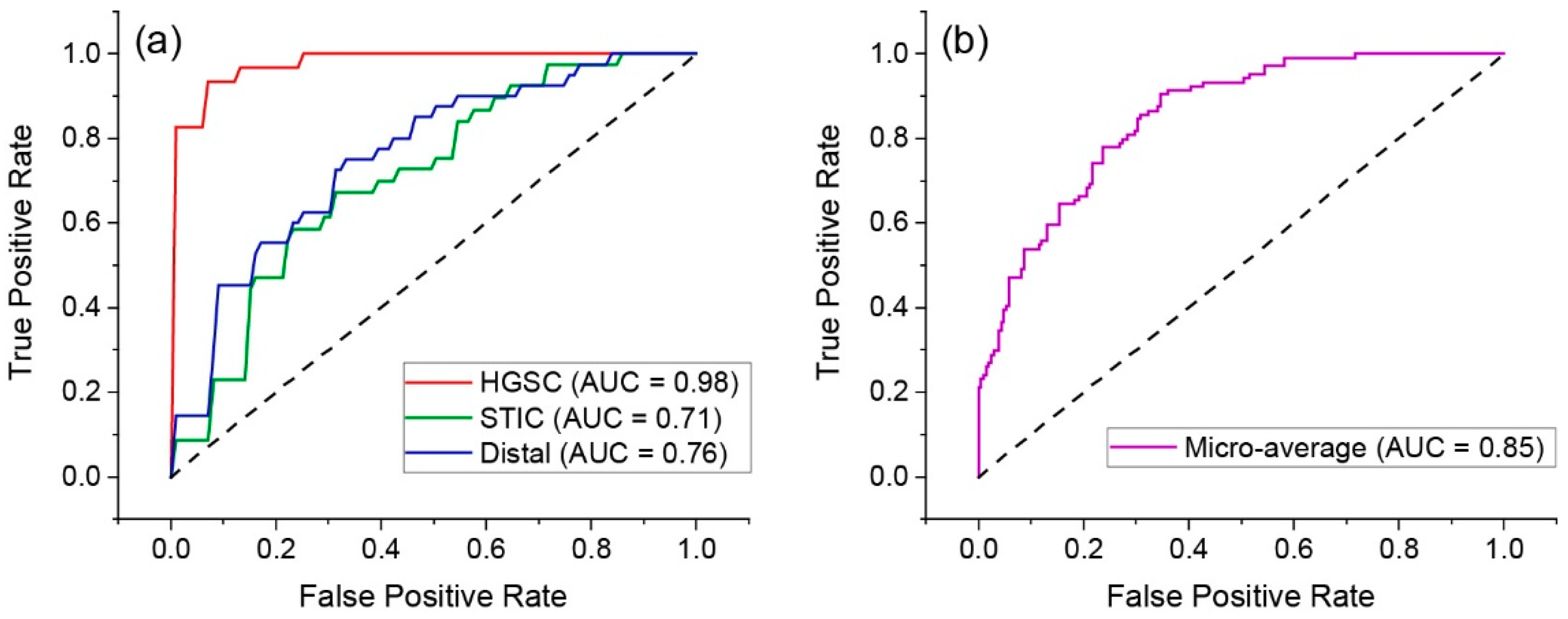
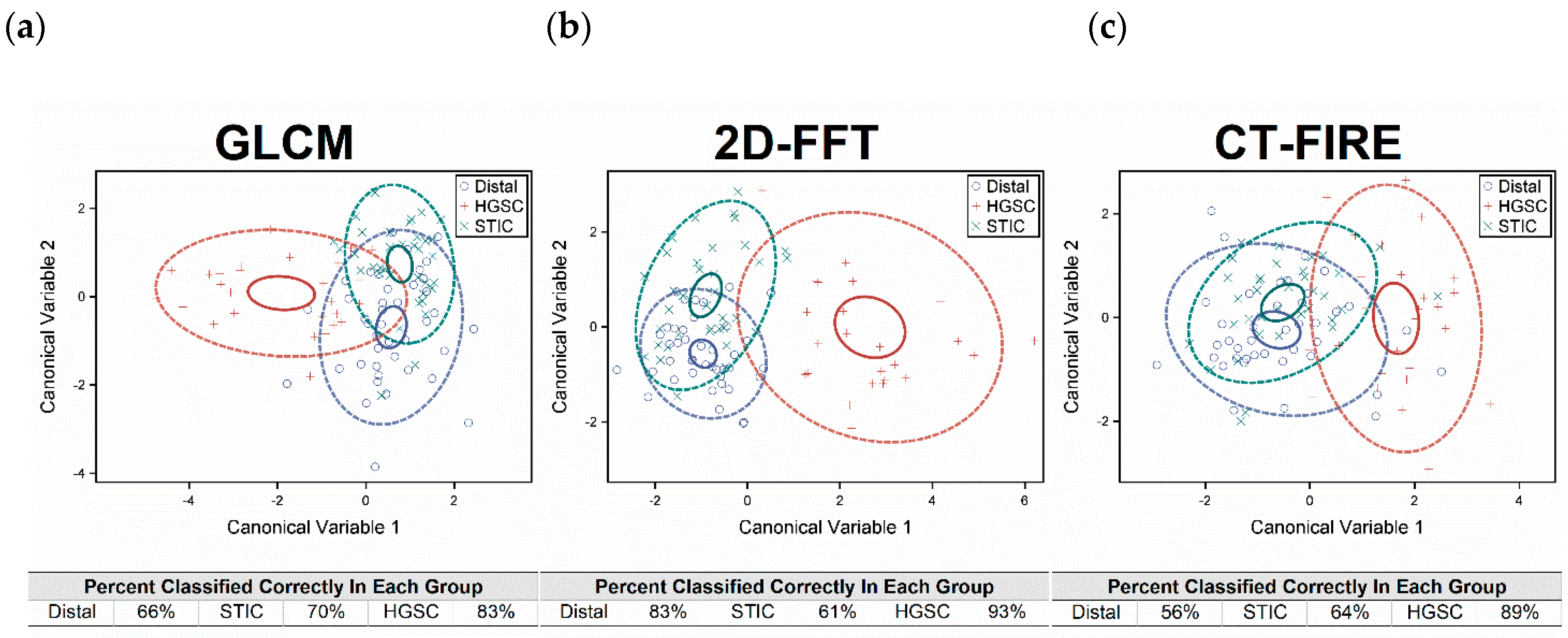
2.5. Classification by Linear Discriminate Analysis
3. Discussion
4. Methods
4.1. Fallopian Tube Tissues
4.2. SHG Microscopy
4.3. Image Analysis
4.4. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef] [PubMed]
- Karlan, B.Y. The status of ultrasound and color Doppler imaging for the early detection of ovarian carcinoma. Cancer Investig. 1997, 15, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Qayyum, A.; Coakley, F.V.; Westphalen, A.C.; Hricak, H.; Okuno, W.T.; Powell, B. Role of CT and MR imaging in predicting optimal cytoreduction of newly diagnosed primary epithelial ovarian cancer. Gynecol. Oncol. 2005, 96, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.E.; Giuntoli, R.L., 2nd; Pannu, H.K.; Schulick, R.D.; Fishman, E.K.; Wahl, R.L. Combined PET/CT for detecting recurrent ovarian cancer limited to retroperitoneal lymph nodes. Gynecol. Oncol. 2005, 99, 294–300. [Google Scholar] [CrossRef]
- Doubeni, C.A.; Doubeni, A.R.; Myers, A.E. Diagnosis and Management of Ovarian Cancer. Am Fam. Physician 2016, 93, 937–944. [Google Scholar]
- van Nagell, J.R., Jr.; DePriest, P.D.; Reedy, M.B.; Gallion, H.H.; Ueland, F.R.; Pavlik, E.J.; Kryscio, R.J. The efficacy of transvaginal sonographic screening in asymptomatic women at risk for ovarian cancer. Gynecol. Oncol. 2000, 77, 350–356. [Google Scholar] [CrossRef]
- Fishman, D.A.; Cohen, L.; Blank, S.V.; Shulman, L.; Singh, D.; Bozorgi, K.; Tamura, R.; Timor-Tritsch, I.; Schwartz, P.E. The role of ultrasound evaluation in the detection of early-stage epithelial ovarian cancer. Am. J. Obs. Gynecol. 2005, 192, 1214–1221, discussion 1221-1212. [Google Scholar] [CrossRef]
- Crum, C.P.; Drapkin, R.; Miron, A.; Ince, T.A.; Muto, M.; Kindelberger, D.W.; Lee, Y. The distal fallopian tube: A new model for pelvic serous carcinogenesis. Curr. Opin. Obs. Gynecol. 2007, 19, 3–9. [Google Scholar] [CrossRef]
- Folkins, A.K.; Jarboe, E.A.; Saleemuddin, A.; Lee, Y.; Callahan, M.J.; Drapkin, R.; Garber, J.E.; Muto, M.G.; Tworoger, S.; Crum, C.P. A candidate precursor to pelvic serous cancer (p53 signature) and its prevalence in ovaries and fallopian tubes from women with BRCA mutations. Gynecol. Oncol. 2008, 109, 168–173. [Google Scholar] [CrossRef]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1093. [Google Scholar] [CrossRef]
- Mehra, K.; Mehrad, M.; Ning, G.; Drapkin, R.; McKeon, F.D.; Xian, W.; Crum, C.P. STICS, SCOUTs and p53 signatures; a new language for pelvic serous carcinogenesis. Front Biosci. (Elite Ed) 2011, 3, 625–634. [Google Scholar] [PubMed]
- Ricciardelli, C.; Rodgers, R.J. Extracellular matrix of ovarian tumors. Semin. Reprod. Med. 2006, 24, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Campbell, K.R.; Tilbury, K.; Nadiarnykh, O.; Brewer, M.A.; Patankar, M.; Singh, V.; Eliceiri, K.W.; Campagnola, P.J. 3D texture analysis for classification of second harmonic generation images of human ovarian cancer. Sci. Rep.-UK 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Cicchi, R.; Kapsokalyvas, D.; De Giorgi, V.; Maio, V.; Van Wiechen, A.; Massi, D.; Lotti, T.; Pavone, F.S. Scoring of collagen organization in healthy and diseased human dermis by multiphoton microscopy. J. Biophotonics 2010, 3, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Drifka, C.R.; Eliceiri, K.W.; Weber, S.M.; Kao, W.J. A bioengineered heterotypic stroma-cancer microenvironment model to study pancreatic ductal adenocarcinoma. Lab Chip 2013, 13, 3965–3975. [Google Scholar] [CrossRef]
- Drifka, C.R.; Loeffler, A.G.; Mathewson, K.; Keikhosravi, A.; Eickhoff, J.C.; Liu, Y.; Weber, S.M.; Kao, W.J.; Eliceiri, K.W. Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection. Oncotarget 2016, 7, 76197. [Google Scholar] [CrossRef]
- Conklin, M.W.; Eickhoff, J.C.; Riching, K.M.; Pehlke, C.A.; Eliceiri, K.W.; Provenzano, P.P.; Friedl, A.; Keely, P.J. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am J. Pathol. 2011, 178, 1221–1232. [Google Scholar] [CrossRef]
- Rueden, C.T.; Conklin, M.W.; Provenzano, P.P.; Keely, P.J.; Eliceiri, K.W. Nonlinear optical microscopy and computational analysis of intrinsic signatures in breast cancer. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2009, 2009, 4077–4080. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Keely, P.J. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 2009, 28, 4326–4343. [Google Scholar] [CrossRef]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef]
- Brown, E.; McKee, T.; diTomaso, E.; Pluen, A.; Seed, B.; Boucher, Y.; Jain, R.K. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat. Med. 2003, 9, 796–800. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Nadiarnykh, O.; Lacomb, R.B.; Brewer, M.A.; Campagnola, P.J. Alterations of the extracellular matrix in ovarian cancer studied by Second Harmonic Generation imaging microscopy. BMC Cancer 2010, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Nadiarynkh, O.; Plotnikov, S.; Campagnola, P.J. Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure. Nat. Protoc. 2012, 7, 654–669. [Google Scholar] [CrossRef]
- Campagnola, P.J.; Dong, C.Y. Second harmonic generation microscopy: Principles and applications to disease diagnosis. Lasers Photonics Rev. 2011, 5, 13–26. [Google Scholar] [CrossRef]
- Pena, A.M.; Fabre, A.; Debarre, D.; Marchal-Somme, J.; Crestani, B.; Martin, J.L.; Beaurepaire, E.; Schanne-Klein, M.C. Three-dimensional investigation and scoring of extracellular matrix remodeling during lung fibrosis using multiphoton microscopy. Microsc. Res. Tech. 2007, 70, 162–170. [Google Scholar] [CrossRef]
- Sun, W.; Chang, S.; Tai, D.C.; Tan, N.; Xiao, G.; Tang, H.; Yu, H. Nonlinear optical microscopy: Use of second harmonic generation and two-photon microscopy for automated quantitative liver fibrosis studies. J. Biomed. Opt. 2008, 13, 064010. [Google Scholar] [CrossRef]
- Campagnola, P.J.; Loew, L.M. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat. Biotechnol. 2003, 21, 1356–1360. [Google Scholar] [CrossRef]
- Campbell, K.R.; Chaudhary, R.; Handel, J.M.; Patankar, M.S.; Campagnola, P.J. Polarization-resolved second harmonic generation imaging of human ovarian cancer. J. Biomed. Opt. 2018, 23, 1–8. [Google Scholar] [CrossRef]
- Tilbury, K.B.; Campbell, K.R.; Eliceiri, K.W.; Salih, S.M.; Patankar, M.; Campagnola, P.J. Stromal alterations in ovarian cancers via wavelength dependent Second Harmonic Generation microscopy and optical scattering. BMC Cancer 2017, 17, 102. [Google Scholar] [CrossRef]
- Lacomb, R.; Nadiarnykh, O.; Townsend, S.S.; Campagnola, P.J. Phase Matching considerations in Second Harmonic Generation from tissues: Effects on emission directionality, conversion efficiency and observed morphology. Opt. Commun. 2008, 281, 1823–1832. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE T Image Process 2004, 13, 600–612. [Google Scholar] [CrossRef] [Green Version]
- Nadiarnykh, O.; LaComb, R.B.; Campagnola, P.J.; Mohler, W.A. Coherent and incoherent SHG in fibrillar cellulose matrices. Opt. Express 2007, 15, 3348–3360. [Google Scholar] [CrossRef] [PubMed]
- Nadiarnykh, O.; Plotnikov, S.; Mohler, W.A.; Kalajzic, I.; Redford-Badwal, D.; Campagnola, P.J. Second Harmonic Generation imaging microscopy studies of Osteogenesis Imperfecta. J. Biomed. Opt. 2007, 12, 051805. [Google Scholar] [CrossRef]
- Campbell, K.R.; Chaudhary, R.; Montano, M.; Iozzo, R.V.; Bushman, W.A.; Campagnola, P.J. Second-harmonic generation microscopy analysis reveals proteoglycan decorin is necessary for proper collagen organization in prostate. J. Biomed. Opt. 2019, 24, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, B.L.; Brewer, M.A.; Nadiarnykh, O.; Hocker, J.; Singh, V.; Mackie, T.R.; Campagnola, P.J. Texture analysis applied to second harmonic generation image data for ovarian cancer classification. J. Biomed. Opt. 2014, 19, 096007. [Google Scholar] [CrossRef] [Green Version]
- Bredfeldt, J.S.; Liu, Y.; Pehlke, C.A.; Conklin, M.W.; Szulczewski, J.M.; Inman, D.R.; Keely, P.J.; Nowak, R.D.; Mackie, T.R.; Eliceiri, K.W. Computational segmentation of collagen fibers from second-harmonic generation images of breast cancer. J. Biomed. Opt. 2014, 19, 16007. [Google Scholar] [CrossRef]
- Stein, A.M.; Vader, D.A.; Jawerth, L.M.; Weitz, D.A.; Sander, L.M. An algorithm for extracting the network geometry of three-dimensional collagen gels. J. Microsc. 2008, 232, 463–475. [Google Scholar] [CrossRef]
- Ajeti, V.; Lara-Santiago, J.; Alkmin, S.; Campagnola, P.J. Ovarian and Breast Cancer Migration Dynamics on Laminin and Fibronectin Bidirectional Gradient Fibers Fabricated via Multiphoton Excited Photochemistry. Cell Mol. Bioeng. 2017, 10, 295–311. [Google Scholar] [CrossRef]
- Kuhn, E.; Kurman, R.J.; Vang, R.; Sehdev, A.S.; Han, G.; Soslow, R.; Wang, T.L.; Shih Ie, M. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma--evidence supporting the clonal relationship of the two lesions. J. Pathol. 2012, 226, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Marquez, R.T.; Baggerly, K.A.; Patterson, A.P.; Liu, J.; Broaddus, R.; Frumovitz, M.; Atkinson, E.N.; Smith, D.I.; Hartmann, L.; Fishman, D.; et al. Patterns of gene expression in different histotypes of epithelial ovarian cancer correlate with those in normal fallopian tube, endometrium, and colon. Clin. Cancer Res. 2005, 11, 6116–6126. [Google Scholar] [CrossRef] [Green Version]
- Vang, R.; Shih Ie, M.; Kurman, R.J. Ovarian low-grade and high-grade serous carcinoma: Pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv. Anta Pathol. 2009, 16, 267–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piek, J.M.; van Diest, P.J.; Verheijen, R.H. Ovarian carcinogenesis: An alternative hypothesis. Adv. Exp. Med. Biol. 2008, 622, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Vang, R.; Shih Ie, M.; Kurman, R.J. Fallopian tube precursors of ovarian low- and high-grade serous neoplasms. Histopathology 2013, 62, 44–58. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.C.; Leng, Y.X.; Xi, J.F.; Li, X.D. Scanning all-fiber-optic endomicroscopy system for 3D nonlinear optical imaging of biological tissues. Opt. Express 2009, 17, 7907–7915. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, G.N.; Sinha, S.; Liske, H.; Chen, X.; Nguyen, V.; Delp, S.L.; Schnitzer, M.J. In Vivo Imaging of Human Sarcomere Twitch Dynamics in Individual Motor Units. Neuron 2015, 88, 1109–1120. [Google Scholar] [CrossRef] [Green Version]
- Lien, C.H.; Tilbury, K.; Chen, S.J.; Campagnola, P.J. Precise, motion-free polarization control in Second Harmonic Generation microscopy using a liquid crystal modulator in the infinity space. Biomed. Opt. Express 2013, 4, 1991–2002. [Google Scholar] [CrossRef] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rentchler, E.C.; Gant, K.L.; Drapkin, R.; Patankar, M.; J. Campagnola, P. Imaging Collagen Alterations in STICs and High Grade Ovarian Cancers in the Fallopian Tubes by Second Harmonic Generation Microscopy. Cancers 2019, 11, 1805. https://doi.org/10.3390/cancers11111805
Rentchler EC, Gant KL, Drapkin R, Patankar M, J. Campagnola P. Imaging Collagen Alterations in STICs and High Grade Ovarian Cancers in the Fallopian Tubes by Second Harmonic Generation Microscopy. Cancers. 2019; 11(11):1805. https://doi.org/10.3390/cancers11111805
Chicago/Turabian StyleRentchler, Eric C., Kristal L. Gant, Ronny Drapkin, Manish Patankar, and Paul J. Campagnola. 2019. "Imaging Collagen Alterations in STICs and High Grade Ovarian Cancers in the Fallopian Tubes by Second Harmonic Generation Microscopy" Cancers 11, no. 11: 1805. https://doi.org/10.3390/cancers11111805




