Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells
Abstract
1. Introduction
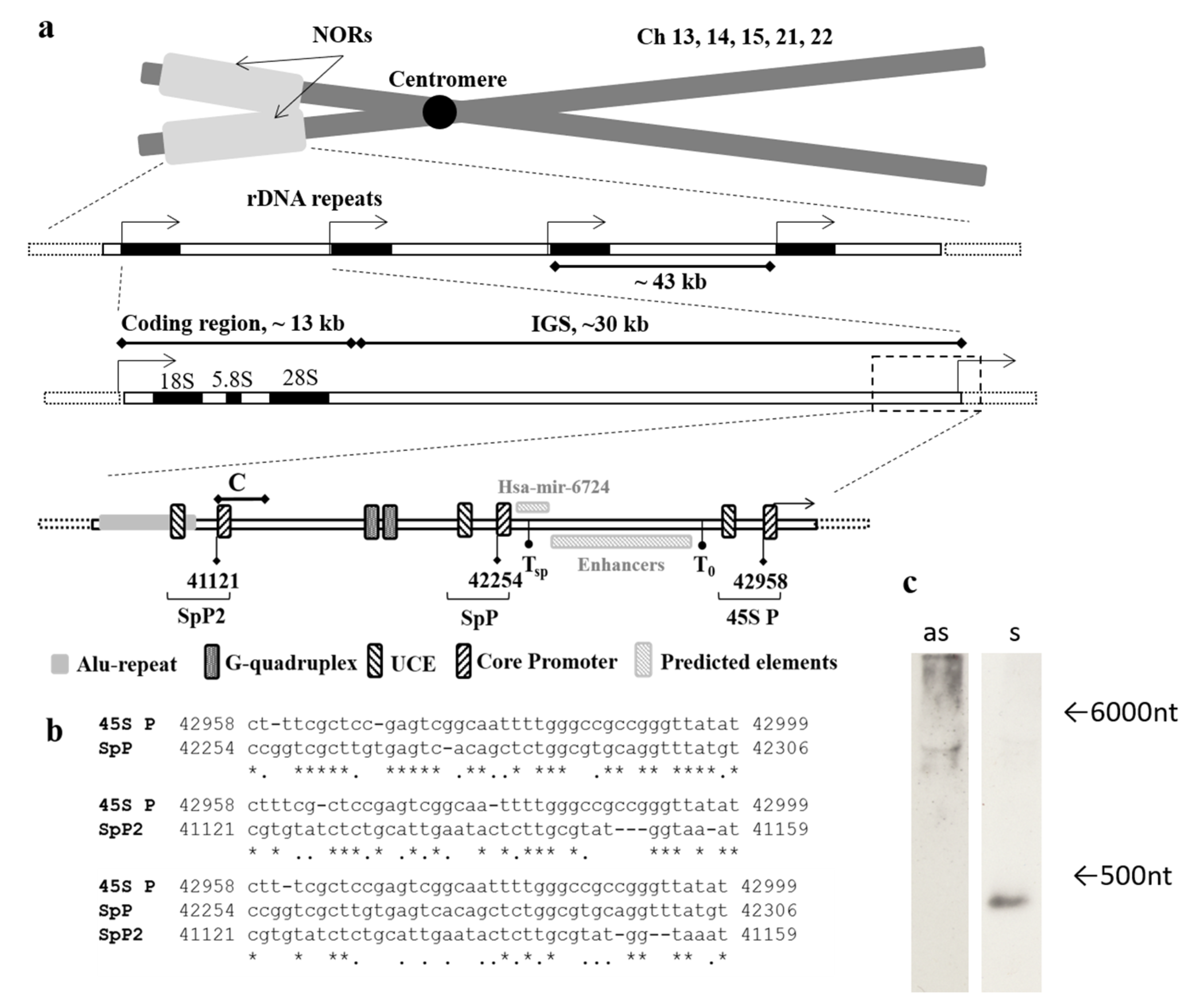
1.1. The Intergenic Spacer of rDNA Harbors Many Intriguing Features
1.2. Glioma and Glioblastoma Need Deep Molecular Phenotype Characterization
2. Results
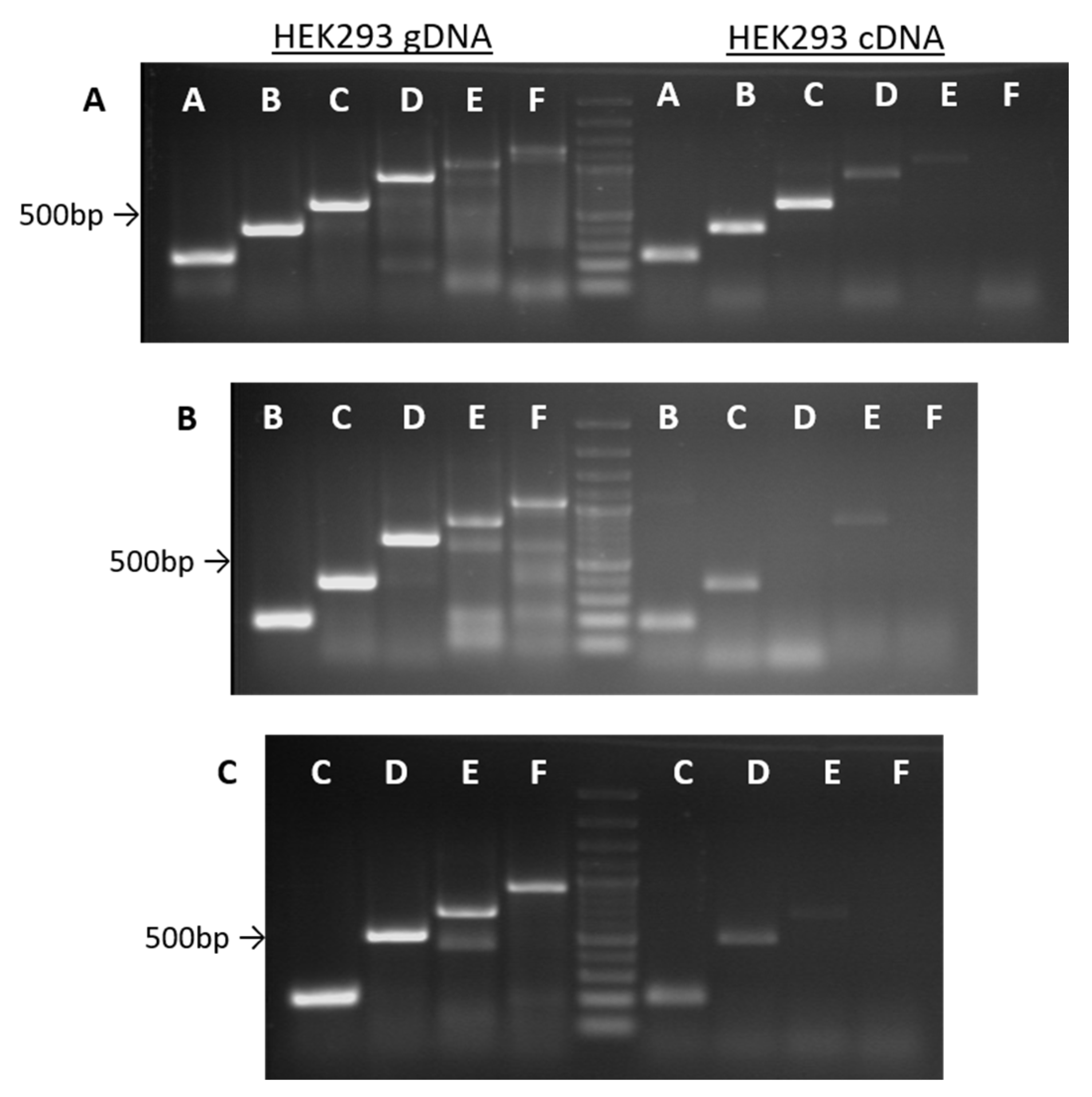
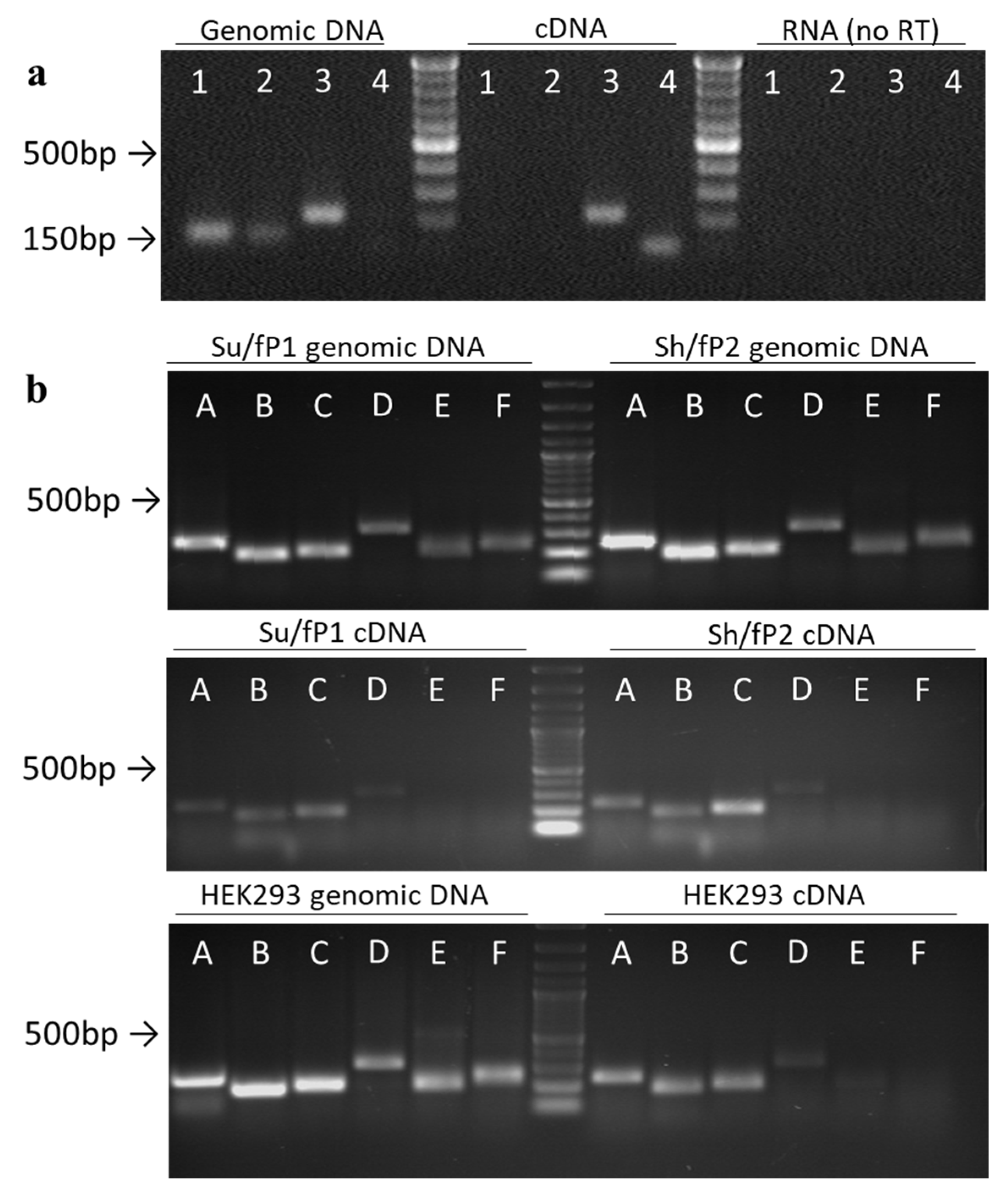
2.1. An Alu-Repeat Neighboring Region May Act as a Promoter
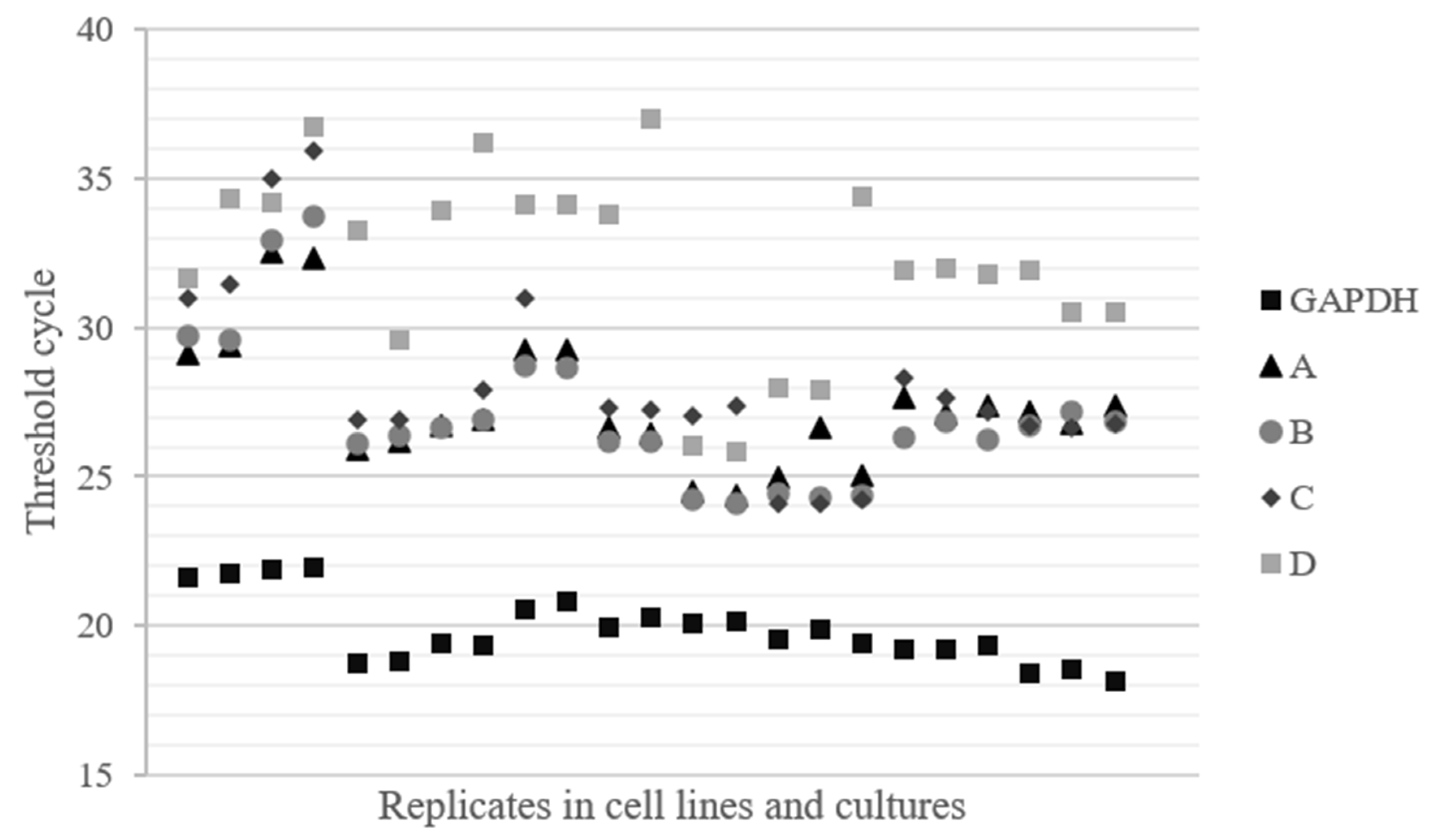
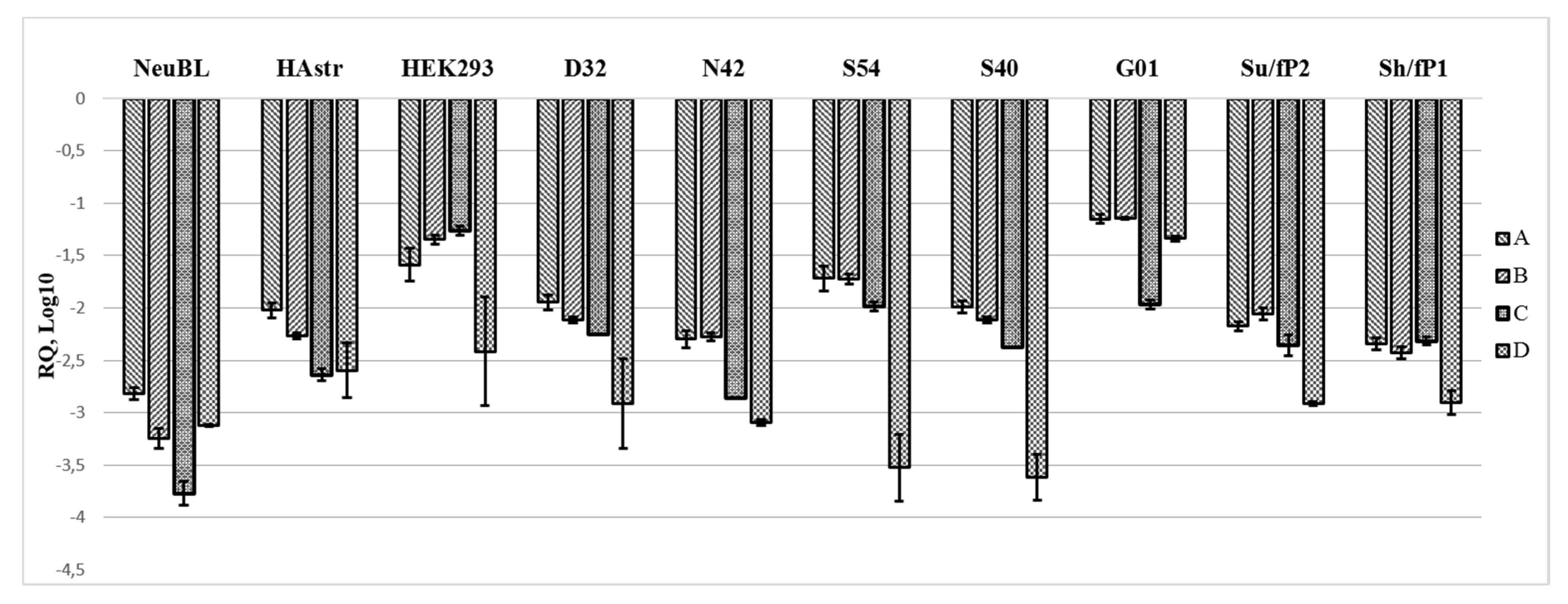
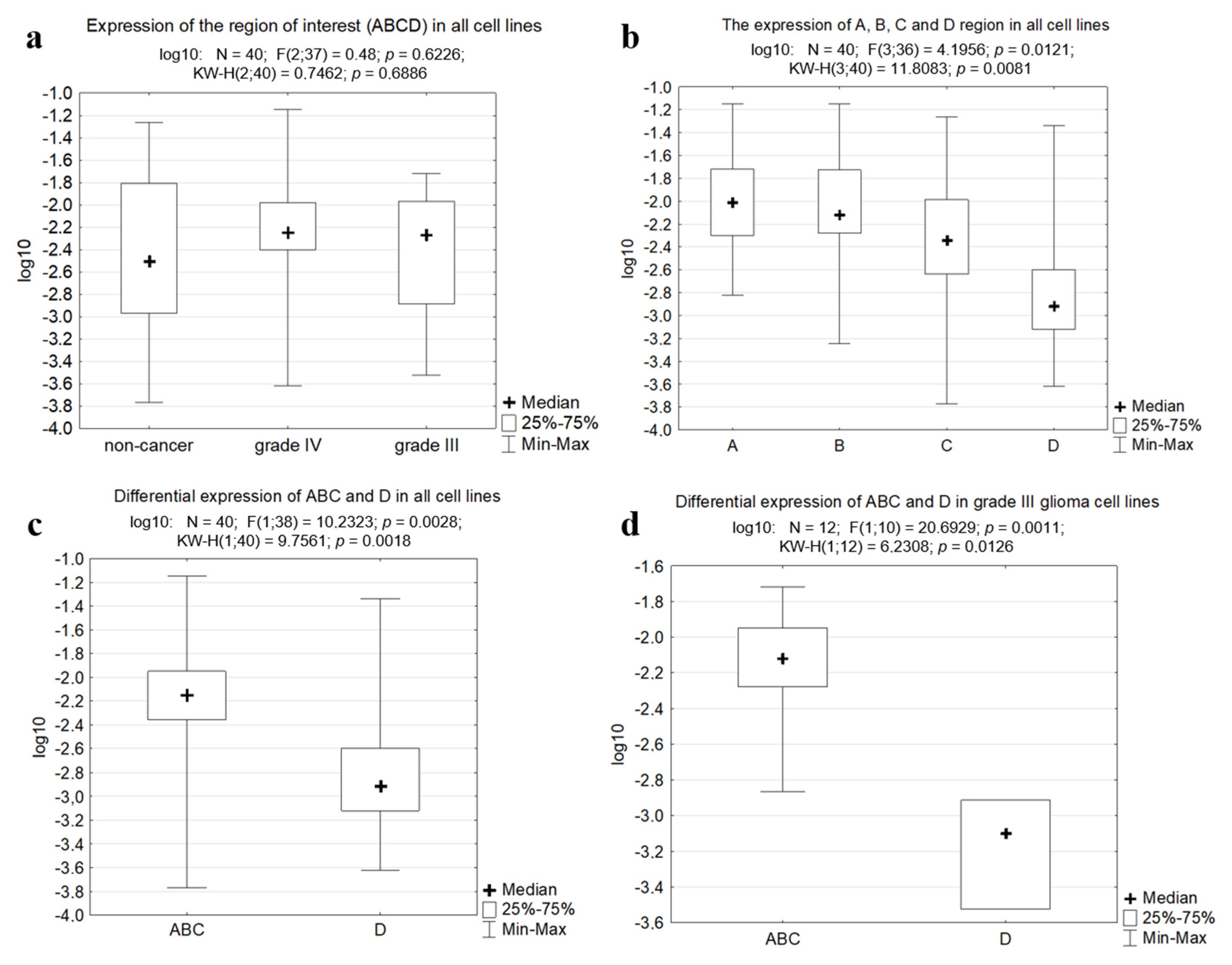
2.2. The Region Upstream of the rDNA Promoter is Differentially Expressed in Cells
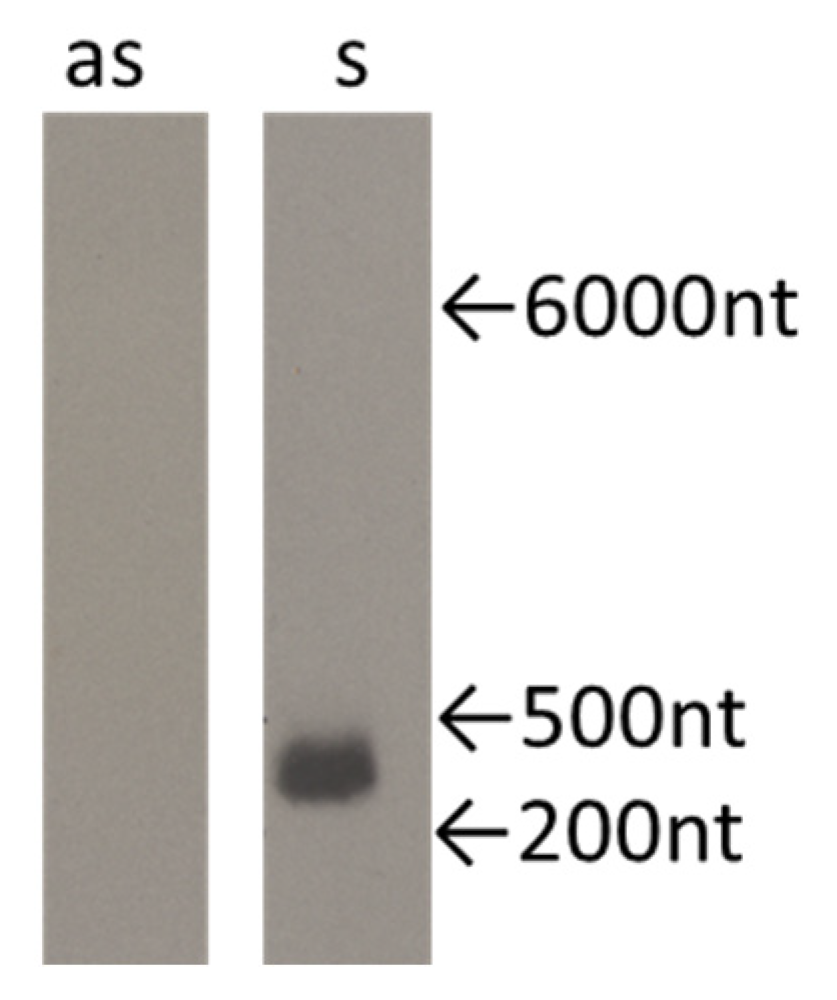
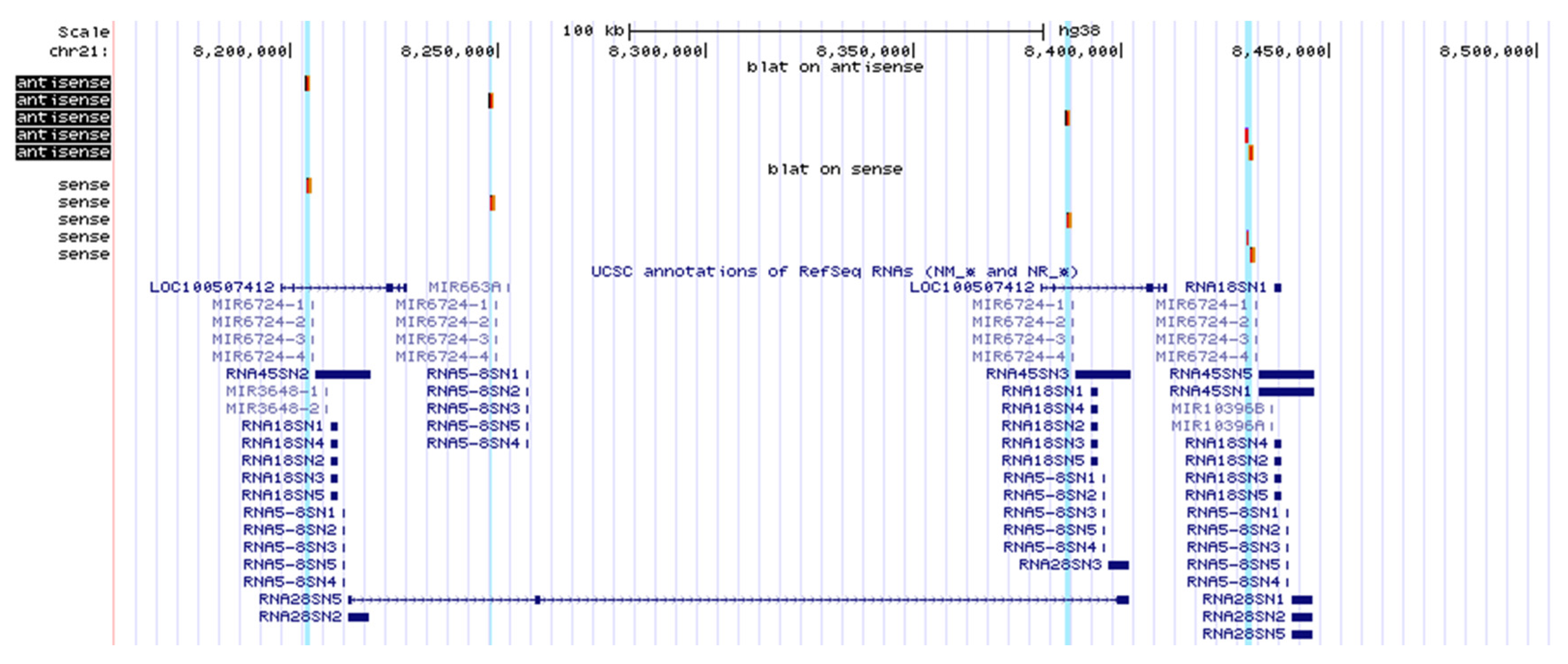
2.3. Both Sense and Antisense Transcripts are Synthesized
2.4. Database Analysis
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cultures Characteristics
4.2. RNA and DNA Extraction
4.3. Northern Blot Analysis
4.4. Primer Design
4.5. Primer Efficiency Determination
4.6. Reverse Transcription and qPCR
4.7. 3′ RACE
4.8. 5′ RACE
4.9. Southern-Blot Analysis
4.10. Bioinformatic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| bp | base pairs |
| cDNA | complementary DNA |
| ChIP-Seq | chromatin immunoprecipitation and sequencing |
| gDNA | genomic DNA |
| IGS | intergenic spacer |
| kb | kilo base pairs |
| NOR | nucleolar organizer region |
| pRNA | promoter RNA |
| PCR | polymerase chain reaction |
| qPCR | quantitative PCR |
| RACE | rapid amplification of cDNA ends |
| sncRNA | small non-coding RNA |
| WHO | World Health Organization |
Appendix A
| Name | Sequence (5′–3′), Orientation (f/r) | Melting Temperature, °C | Amplicon Size, bp |
|---|---|---|---|
| A | GTTTCGTCCTTTTGAGACAGAGT, f GTGGGCGCATCACAGGAGGTC, r | 59 66 | 254 |
| B | TCCAACTCCCGACCTCCTGT, f TGCAGAGATACACGTTGTCGTTG, r | 63 62 | 194 |
| C | CAACGACAACGTGTATCTCTGCA, f TACGCTCGGTTCATTTACACACA, r | 62 61 | 208 |
| D | ATGTGTGTAAATGAACCGAGCGTA, f TACACAAAGTAAACTTCTGAAACAC, r | 61 57 | 328 |
| E | GGAGTGTTTCAGAAGTTTACTTTGTG, f AGACACACGGAGAGGCAGAAT, r | 60 62 | 208 |
| F | CGTGTGTCTGCAGCGACC, f GAAAGCGAAACCGTGAGTCGA, r | 62 62 | 243 |
| 18S | TTTCTCGATTCCGTGGGTGG, f CCCGGACATCTAAGGGCATC, r | 60 60 | 211 |
| GAPDH | CCATGGGGAAGGTGAAGGTC, f AGTGATGGCATGGACTGTGG, r | 60 60 | 548 |
| GAPDH2 | ACCGTCAAGGCTGAGAACG, f GCATCGCCCCACTTGATTTT, r | 60 59 | 95 |
| IGS36 | GCACCTCTCCGGAAACATTG, f CTTAACCACGCACCCACGAA, r | 58 59 | 150 |
| IGS42 | GCGATCCTTTCTGGCGAGTC, f CGGGAGCCGGAAGCATTTTC, r | 60 60 | 143 |
| Name | 5′–3′ Sequence | |
|---|---|---|
| 3′ RACE | Adapter | GCGAGCACAGAATTAATACGACTCACTATAGGT12VN |
| Primer | GCGAGCACAGAATTAATACGACT | |
| 5′ RACE | Adapter | GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA |
| Outer Primer | GCTGATGGCGATGAATGAACACTG | |
| Inner Primer | CGCGGATCCGAACACTGCGTTTGCTGGCTTTGATG | |
References
- Gonzalez, I.L.; Sylvester, J.E. Complete sequence of the 43-kb human ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics 1995, 27, 320–328. [Google Scholar] [CrossRef] [PubMed]
- Jacob, M.D.; Audas, T.E.; Mullineux, S.; Lee, S. Where no RNA polymerase has gone before. Nucleus 2012, 3, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Netchvolodov, K.K.; Boiko, A.V.; Ryskov, A.P.; Kupriyanova, N.S. Evolutionary divergence of the pre-promotor region of ribosomal DNA in the great apes. DNA Seq. 2006, 17, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Agrawa, S.; Ganley, A.R.D. The conservation landscape of the human ribosomal RNA gene repeats. PLoS ONE 2018, 13, e0207531. [Google Scholar] [CrossRef]
- Morgan, G.T.; Reeder, R.H.; Bakken, A.H. Transcription in cloned spacers of Xenopus laevis ribosomal, DNA. Proc. Natl. Acad. Sci. USA 1983, 80, 6490–6694. [Google Scholar] [CrossRef]
- Kuhn, A.; Grummt, I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBOJ 1987, 6, 3487–3492. [Google Scholar] [CrossRef]
- Audas, T.E.; Jacob, M.D.; Lee, S. Immobilization of Proteins in the Nucleolus by Ribosomal Intergenic Spacer Noncoding, RNA. Mol. Cell 2012, 45, 147–157. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.Y.; Wan, F.C.; Liu, F.J.; Liu, J.; Zhang, N.; Jin, S.H.; Li, J.Y. Deep sequencing analysis of small non-coding RNAs reveals the diversity of microRNAs and piRNAs in the human epididymis. Gene 2012, 497, 330–335. [Google Scholar] [CrossRef]
- Mayer, C.; Schmitz, K.M.; Li, J.; Grummt, I.; Santoro, R. Intergenic Transcripts Regulate the Epigenetic State of rRNA Genes. Mol. Cell 2006, 22, 351–361. [Google Scholar] [CrossRef]
- McStay, B.; Grummt, I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 2008, 24, 131–157. [Google Scholar] [CrossRef]
- Mars, J.C.; Sabourin-Felix, M.; Tremblay, M.G.; Moss, T. A Deconvolution Protocol for ChIP-Seq Reveals Analogous Enhancer Structures on the Mouse and Human Ribosomal RNA Genes. G3 (Bethesda) 2018, 8, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Kupriyanova, N.S.; Ryskov, A.P. Discrepancy in the regulation of ribosomal RNA expression between primates and other vertebrates. Glob. J. Biochem. 2011, 2, 271–282. [Google Scholar]
- Shiao, Y.H.; Lupascu, S.T.; Gu, Y.D.; Kasprzak, W.; Hwang, C.J.; Fields, J.R.; Leighty, R.M.; Quiñones, O.; Shapiro, B.A.; Alvord, W.G.; et al. An intergenic non-coding rRNA correlated with expression of the rRNA and frequency of an rRNA single nucleotide polymorphism in lung cancer cells. PLoS ONE 2009, 4, e7505. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smoll, N.R.; Hamilton, B. Incidence and relative survival of anaplastic astrocytomas. Neuro. Oncol. 2014, 16, 1400–1407. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, J.; Quan, J.; Liu, W.; Tan, H.; Li, W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci. 2018, 109, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.M.; Kivinen, V.; Liu, Y.; Annala, M.; Cogdell, D.; Liu, X.; Liu, C.G.; Sawaya, R.; Yli-Harja, O.; Shmulevich, I.; et al. Transcriptome and Small RNA Deep Sequencing Reveals Deregulation of miRNA Biogenesis in Human Glioma. J. Pathol. 2013, 229, 449–459. [Google Scholar] [CrossRef]
- Goodenberger, M.L.; Jenkins, R.B. Genetics of adult glioma. Cancer Genet. 2012, 205, 613–621. [Google Scholar] [CrossRef]
- Rajesh, Y.; Pal, I.; Banik, P.; Chakraborty, S.; Borkar, S.A.; Dey, G.; Mukherjee, A.; Mandal, M. Insights into molecular therapy of glioma: Current challenges and next generation blueprint. Acta Pharm. Sin. 2017, 38, 591–613. [Google Scholar] [CrossRef]
- Bleeker, F.E.; Molenaar, R.J.; Leenstra, S. Recent advances in the molecular understanding of glioblastoma. J. Neurooncol. 2012, 108, 11–27. [Google Scholar] [CrossRef]
- Gallego, O. Nonsurgical treatment of recurrent glioblastoma. Curr. Oncol. 2015, 22, e273–e281. [Google Scholar] [CrossRef]
- Møller, H.G.; Rasmussen, A.P.; Andersen, H.H.; Johnsen, K.B.; Henriksen, M.; Duroux, M. A systematic review of microRNA in glioblastoma multiforme: Micro-modulators in the mesenchymal mode of migration and invasion. Mol. Neurobiol. 2013, 47, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Brower, J.V.; Clark, P.A.; Lyon, W.; Kuo, J.S. MicroRNAs in cancer: Glioblastoma and glioblastoma cancer stem cells. Neurochem. Int. 2014, 77, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Warmerdam, D.O.; Wolthuis, R.M.F. Keeping ribosomal DNA intact: A repeating challenge. Chromosome Res. 2019, 27, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Li, H.; Perry, J.M.; Singh, V.P.; Unruh, J.; Yu, Z.; Zakari, M.; McDowell, W.; Li, L.; Gerton, J.L. Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet. 2017, 13, e1006771. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lemos, B. Ribosomal DNA copy number amplification and loss in human cancers is linked to tumor genetic context, nucleolus activity, and proliferation. PLoS Genet. 2017, 13, e1006994. [Google Scholar] [CrossRef]
- Baskaran, S.; Mayrhofer, M.; Kultima, H.G.; Bergström, T.; Elfineh, L.; Cavelier, L.; Isaksson, A.; Nelander, S. Primary glioblastoma cells for precision medicine: A quantitative portrait of genomic (in)stability during the first 30 passages. Neuro. Oncol. 2018, 20, 1080–1091. [Google Scholar] [CrossRef]
- Belin, S.; Beghin, A.; Solano-Gonzàlez, E.; Bezin, L.; Brunet-Manquat, S.; Textoris, J.; Prats, A.C.; Mertani, H.C.; Dumontet, C.; Diaz, J.J. Dysregulation of ribosome biogenesis and translational capacity is associated with tumor progression of human breast cancer cells. PLoS ONE 2009, 4, e7147. [Google Scholar] [CrossRef]
- Olausson, K.H.; Nistér, M.; Lindström, M.S. Loss of nucleolar histone chaperone, NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase, DNMT3A to drive ribosomal DNA transcription. J. Biol. Chem. 2014, 289, 34601–34619. [Google Scholar] [CrossRef]
- Frescas, D.; Guardavaccaro, D.; Bassermann, F.; Koyama-Nasu, R.; Pagano, M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 2007, 450, 309–313. [Google Scholar] [CrossRef]
- Stępiński, D. The nucleolus, an ally, and an enemy of cancer cells. Histochem. Cell Biol. 2018, 150, 607–629. [Google Scholar] [CrossRef]
- Watanabe, T.; Nobusawa, S.; Kleihues, P.; Ohgaki, H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 2009, 174, 1149–1153. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and, IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef] [PubMed]
- Núñez, F.J.; Mendez, F.M.; Kadiyala, P.; Alghamri, M.S.; Savelieff, M.G.; Garcia-Fabiani, M.B.; Haase, S.; Koschmann, C.; Calinescu, A.A.; Kamran, N.; et al. IDH1-R132H acts as a tumor suppressor in glioma via epigenetic up-regulation of the DNA damage response. Sci. Transl. Med. 2019, 11, eaaq1427. [Google Scholar] [CrossRef] [PubMed]
- Vacík, T.; Kereïche, S.; Raška, I.; Cmarko, D.; Smirnov, E. Life time of some RNA products of rDNA intergenic spacer in HeLa cells. Histochem. Cell Biol. 2019, 152, 271–280. [Google Scholar] [CrossRef]
- Savić, N.; Bär, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in, ESCs. Cell Stem Cell 2014, 4, 15–720. [Google Scholar] [CrossRef]
- Yoshikawa, M.; Fujii, Y.R. Human Ribosomal, RNA-Derived Resident MicroRNAs as the Transmitter of Information upon the Cytoplasmic Cancer Stress. Biomed. Res. Int. 2016, 2016, 7562085. [Google Scholar] [CrossRef]
- Guetg, C.; Santoro, R. Formation of nuclear heterochromatin: The nucleolar point of view. Epigenetics 2012, 7, 811–814. [Google Scholar] [CrossRef]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef]
- Etchegaray, J.P.; Mostoslavsky, R. A sirtuin’s role in preventing senescence by protecting ribosomal, DNA. J. Biol. Chem. 2018, 293, 11251–11252. [Google Scholar] [CrossRef]
- van Sluis, M.; McStay, B. A localized nucleolar DNA damage response facilitates recruitment of the homology-directed repair machinery independent of cell cycle stage. Genes Dev. 2015, 29, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Akamatsu, Y.; Kobayashi, T. The Human RNA Polymerase I Transcription Terminator Complex Acts as a Replication Fork Barrier That Coordinates the Progress of Replication with rRNA Transcription Activity. Mol. Cell Biol. 2015, 35, 1871–1881. [Google Scholar] [CrossRef]
- Horigome, C.; Unozawa, E.; Ooki, T.; Kobayashi, T. Ribosomal RNA gene repeats associate with the nuclear pore complex for maintenance after DNA damage. PLoS Genet. 2019, 15, e1008103. [Google Scholar] [CrossRef] [PubMed]
- Malinovskaya, E.M.; Ershova, E.S.; Golimbet, V.E.; Porokhovnik, L.N.; Lyapunova, N.A.; Kutsev, S.I.; Veiko, N.N.; Kostyuk, S.V. Copy Number of Human Ribosomal Genes With Aging: Unchanged Mean, but Narrowed Range and Decreased Variance in Elderly Group. Front. Genet. 2018, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Salim, D.; Gerton, J.L. Ribosomal DNA instability and genome adaptability. Chromosome Res. 2019, 27, 73–87. [Google Scholar] [CrossRef]
- Porokhovnik, L.N.; Lyapunova, N.A. Dosage effects of human ribosomal genes (rDNA) in health and disease. Chromosome Res. 2019, 27, 5–17. [Google Scholar] [CrossRef]
- Gibbons, J.G.; Branco, A.T.; Yu, S.; Lemos, B. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat. Commun. 2014, 5, 4850. [Google Scholar] [CrossRef]
- Kim, J.H.; Dilthey, A.T.; Nagaraja, R.; Lee, H.S.; Koren, S.; Dudekula, D.; Wood, W.H., III; Piao, Y.; Ogurtsov, A.Y.; Utani, K.; et al. Variation in human chromosome 21 ribosomal RNA genes characterized by, TAR cloning and long-read sequencing. Nucleic Acids Res. 2018, 46, 6712–6725. [Google Scholar] [CrossRef]
- Vydzhak, O.; Luke, B.; Schindler, N. Non-coding RNAs at the Eukaryotic rDNA Locus: RNA-DNA Hybrids and Beyond. J. Mol. Biol. 2020, 432, 4287–4304. [Google Scholar] [CrossRef]
- Zentner, G.E.; Saiakhova, A.; Manaenkov, P.; Adams, M.D.; Scacheri, P.C. Integrative genomic analysis of human ribosomal, DNA. Nucleic Acids Res. 2011, 39, 4949–4960. [Google Scholar] [CrossRef]
- Yap, K.; Mukhina, S.; Zhang, G.; Tan, J.S.C.; Ong, H.S.; Makeyev, E.V. A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol. Cell 2018, 72, 525–540.e13. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Mueller, J.E.; Bryk, M. Sir2 represses endogenous polymerase, II. transcription units in the ribosomal DNA nontranscribed spacer. Mol. Biol. Cell 2006, 17, 3848–3859. [Google Scholar] [CrossRef][Green Version]
- Caudron-Herger, M.; Pankert, T.; Rippe, K. Regulation of nucleolus assembly by non-coding RNA polymerase, II. transcripts. Nucleus 2016, 7, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Khitrinskaia, I.Y.; Stepanov, V.A.; Puzyrev, V.P. Alu-povtory v genome cheloveka [Alu repeats in the human genome]. Mol. Biol. (Mosk.) 2003, 37, 382–391. (In Russian) [Google Scholar] [PubMed]
- Zhang, X.O.; Gingeras, T.R.; Weng, Z. Genome-wide analysis of polymerase, II. -transcribed Alu elements suggests cell-type-specific enhancer function. Genome Res. 2019, 29, 1402–1414. [Google Scholar] [CrossRef] [PubMed]
- Pustogarov, N.; Panteleev, D.; Goryaynov, S.A.; Ryabova, A.V.; Rybalkina, E.Y.; Revishchin, A.; Potapov, A.A.; Pavlova, G. Hiding in the Shadows: CPOX Expression and 5-ALA Induced Fluorescence in Human Glioma Cells. Mol. Neurobiol. 2017, 54, 5699–5708. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Knudsen, S. Promoter 2.0, for the recognition of PolII promoter sequences. Bioinformatics 1999, 15, 356–3361. [Google Scholar] [CrossRef]
- Solovyev, V.V.; Shahmuradov, I.A.; Salamov, A.A. Identification of promoter regions and regulatory sites. Methods Mol. Biol. 2010, 674, 57–83. [Google Scholar]
- Umarov, R.K.; Solovyev, V.V. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE 2017, 12, e0171410. [Google Scholar] [CrossRef]
- Lee, T.Y.; Chang, W.C.; Hsu, J.B.; Chang, T.H.; Shien, D.M. GPMiner: An integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genom. 2012, 13, S3. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at, UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J. BLAT—The, BLAST-like alignment tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]


| Healthy or Relatively Healthy Cells | Malignant Cells | ||
|---|---|---|---|
| HEK293 | Human embryo kidney | D32 | Glioma III |
| Hastr | Human astrocytes | N42 | Glioma III |
| Neubl | Neuroblasts | S54 | Glioma III |
| – | – | S40 | Glioblastoma IV |
| – | – | G01 | Glioblastoma IV |
| – | – | Su/fP1 | Glioblastoma IV |
| – | – | Sh/fP2 | Glioblastoma IV |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sadova, A.A.; Kupriyanova, N.S.; Pavlova, G.V. Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells. Cancers 2020, 12, 2090. https://doi.org/10.3390/cancers12082090
Sadova AA, Kupriyanova NS, Pavlova GV. Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells. Cancers. 2020; 12(8):2090. https://doi.org/10.3390/cancers12082090
Chicago/Turabian StyleSadova, Anastasia A., Natalia S. Kupriyanova, and Galina V. Pavlova. 2020. "Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells" Cancers 12, no. 8: 2090. https://doi.org/10.3390/cancers12082090
APA StyleSadova, A. A., Kupriyanova, N. S., & Pavlova, G. V. (2020). Mapping and Quantification of Non-Coding RNA Originating from the rDNA in Human Glioma Cells. Cancers, 12(8), 2090. https://doi.org/10.3390/cancers12082090





