Connectivity Map Analysis Indicates PI3K/Akt/mTOR Inhibitors as Potential Anti-Hypoxia Drugs in Neuroblastoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. CMap Analysis
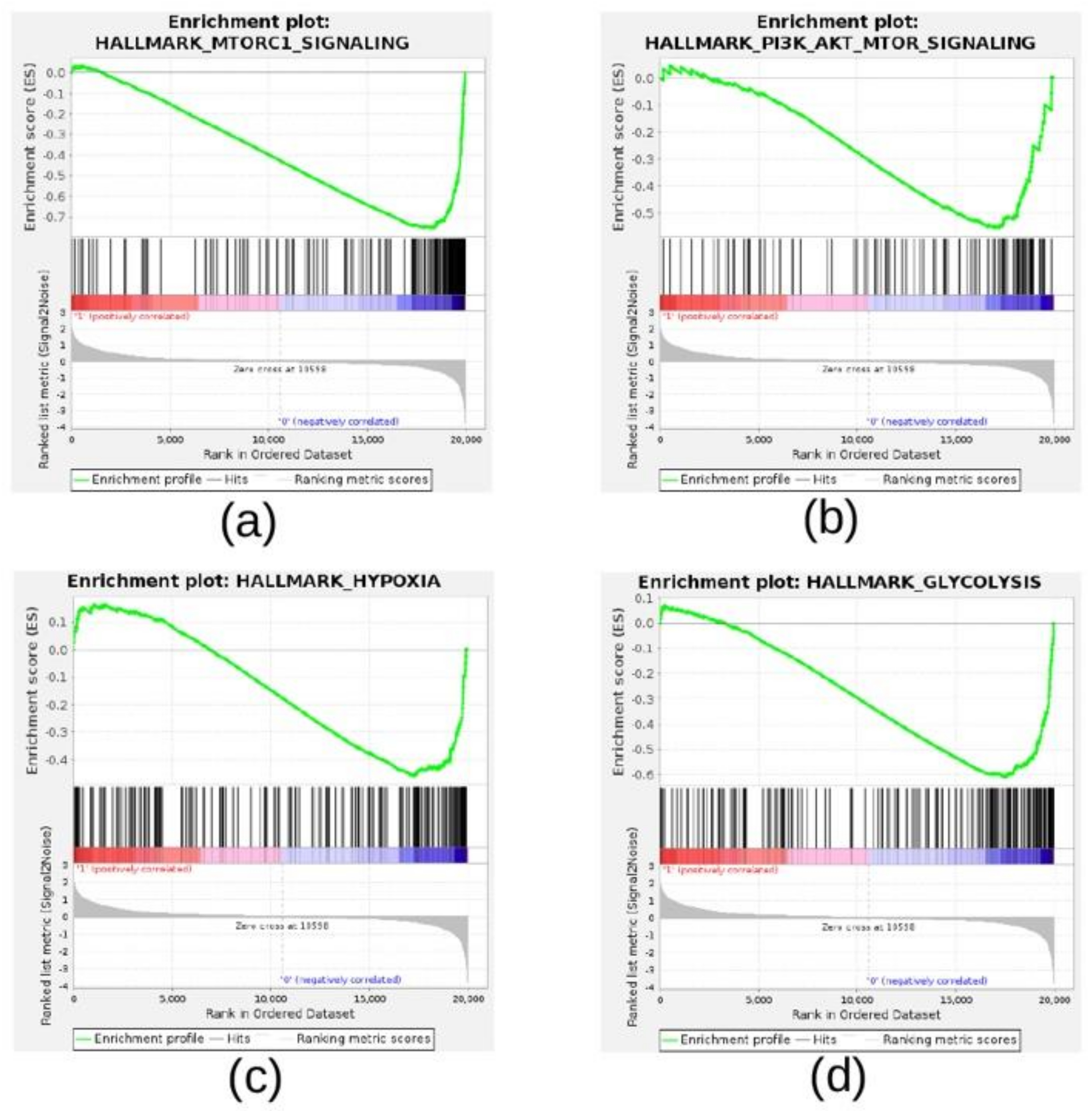
2.2. Gene Set Enrichment Analysis
3. Results
3.1. The CMap Analysis Identified Significant Connections Between Hypoxia-Modulated Genes and Compounds Belonging to the Class of PI3K/Akt/mTOR Onhibitors
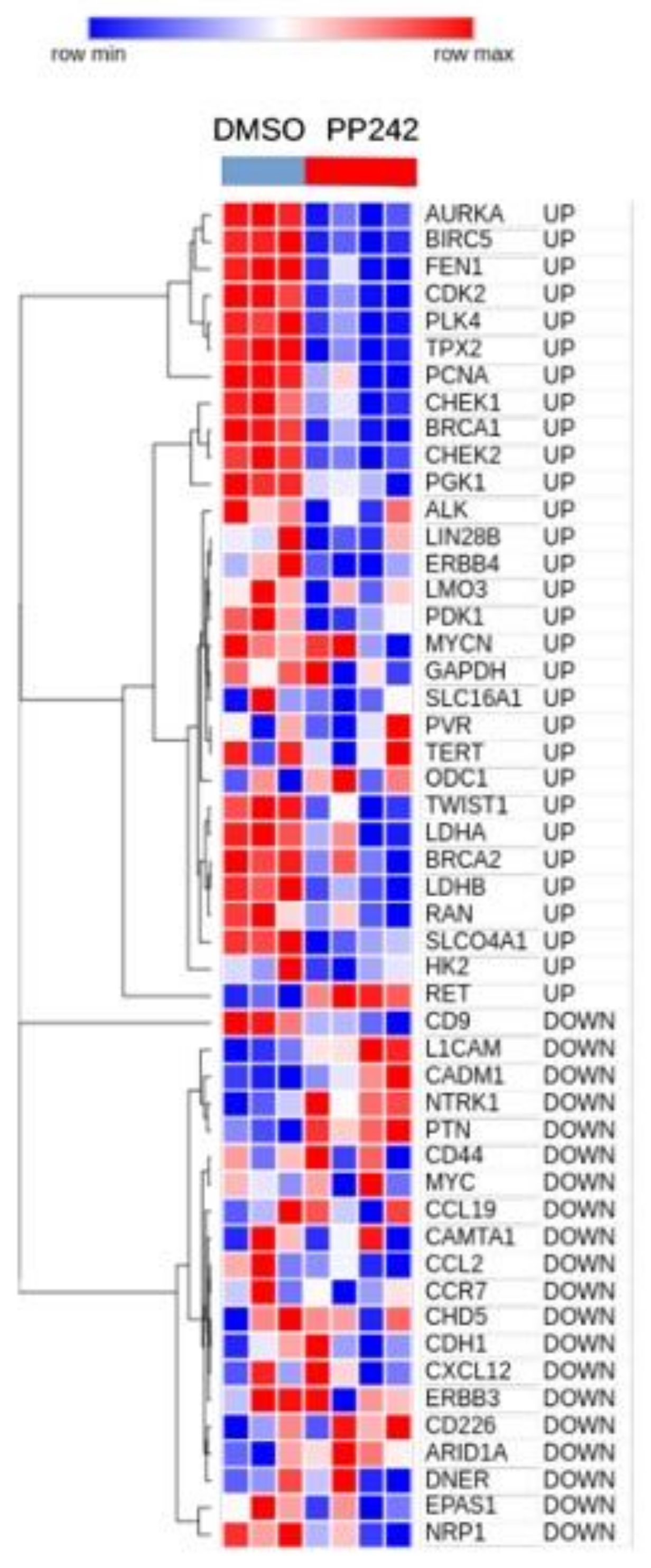
3.2. Validation of CMap Findings in Published Gene Expression Profiles of NB Cells Cultured under Hypoxia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cohn, S.L.; Pearson, A.D.J.; London, W.B.; Monclair, T.; Ambros, P.F.; Brodeur, G.M.; Faldum, A.; Hero, B.; Iehara, T.; Machin, D.; et al. The international neuroblastoma risk group (INRG) classification system: An INRG task force report. J. Clin. Oncol. 2009, 27, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.J.; Doral, M.Y.; DuBois, S.G.; Villablanca, J.G.; Yanik, G.A.; Matthay, K.K. Different outcomes for relapsed versus refractory neuroblastoma after therapy with 131I-metaiodobenzylguanidine (131I-MIBG). Eur. J. Cancer 2015, 51, 2465–2472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zage, P.E. Novel Therapies for Relapsed and Refractory Neuroblastoma. Children 2018, 5, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernauer, C.; Man, Y.K.S.; Chisholm, J.C.; Lepicard, E.Y.; Robinson, S.P.; Shipley, J.M. Hypoxia and its therapeutic possibilities in paediatric cancers. Br. J. Cancer 2021, 124, 539–551. [Google Scholar] [CrossRef]
- Huertas-Castaño, C.; Gómez-Muñoz, M.A.; Pardal, R.; Vega, F.M. Hypoxia in the Initiation and Progression of Neuroblastoma Tumours. Int. J. Mol. Sci. 2019, 21, 39. [Google Scholar] [CrossRef] [Green Version]
- Bosco, M.C.; D’Orazi, G.; Del Bufalo, D. Targeting hypoxia in tumor: A new promising therapeutic strategy. J. Exp. Clin. Cancer Res. 2020, 39, 8. [Google Scholar] [CrossRef]
- Corrado, C.; Fontana, S. Hypoxia and HIF Signaling: One Axis with Divergent Effects. Int. J. Mol. Sci. 2020, 21, 5611. [Google Scholar] [CrossRef]
- Cangelosi, D.; Morini, M.; Zanardi, N.; Sementa, A.R.; Muselli, M.; Conte, M.; Garaventa, A.; Pfeffer, U.; Bosco, M.C.; Varesio, L.; et al. Hypoxia Predicts Poor Prognosis in Neuroblastoma Patients and Associates with Biological Mechanisms Involved in Telomerase Activation and Tumor Microenvironment Reprogramming. Cancers 2020, 12, 2343. [Google Scholar] [CrossRef]
- Ognibene, M.; Cangelosi, D.; Morini, M.; Segalerba, D.; Bosco, M.C.; Sementa, A.R.; Eva, A.; Varesio, L. Immunohistochemical analysis of PDK1, PHD3 and HIF-1α expression defines the hypoxic status of neuroblastoma tumors. PLoS ONE 2017, 12, e0187206. [Google Scholar] [CrossRef] [Green Version]
- Fardin, P.; Barla, A.; Mosci, S.; Rosasco, L.; Verri, A.; Versteeg, R.; Caron, H.N.; Molenaar, J.J.; Øra, I.; Eva, A.; et al. A biology-driven approach identifies the hypoxia gene signature as a predictor of the outcome of neuroblastoma patients. Mol. Cancer 2010, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Applebaum, M.A.; Jha, A.; Kao, C.; Hernandez, K.M.; Dewane, G.; Salwen, H.R.; Chlenski, A.; Dobratic, M.; Mariani, C.J.; Godley, L.A.; et al. Integrative genomics reveals hypoxia inducible genes that are associated with a poor prognosis in neuroblastoma patients. Oncotarget 2016, 7, 76816–76826. [Google Scholar] [CrossRef] [Green Version]
- Cangelosi, D.; Pelassa, S.; Morini, M.; Conte, M.; Bosco, M.C.; Eva, A.; Sementa, A.R.; Varesio, L. Artificial neural network classifier predicts neuroblastoma patients’ outcome. BMC Bioinform. 2016, 17 (Suppl. 12), 347. [Google Scholar] [CrossRef] [Green Version]
- Cangelosi, D.; Muselli, M.; Parodi, S.; Blengio, F.; Becherini, P.; Versteeg, R.; Conte, M.; Varesio, L. Use of Attribute Driven Incremental Discretization and Logic Learning Machine to build a prognostic classifier for neuroblastoma patients. BMC Bioinform. 2014, 15 (Suppl. 5), S4. [Google Scholar] [CrossRef] [Green Version]
- Mousavi, S.Z.; Rahmanian, M.; Sami, A. A connectivity map-based drug repurposing study and integrative analysis of transcriptomic profiling of SARS-CoV-2 infection. Infect. Genet. Evol. 2020, 86, 104610. [Google Scholar] [CrossRef]
- Musa, A.; Ghoraie, L.S.; Zhang, S.-D.; Galzko, G.; Yli-Harja, O.; Dehmer, M.; Haibe-Kains, B.; Emmert-Streib, F. A review of connectivity map and computational approaches in pharmacogenomics. Brief. Bioinform. 2017, 19, 506–523. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452. [Google Scholar] [CrossRef]
- Xu, F.; Na, L.; Li, Y.; Chen, L. Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ortega, M.A.; Fraile-Martínez, O.; Asúnsolo, Á.; Buján, J.; García-Honduvilla, N.; Coca, S. Signal Transduction Pathways in Breast Cancer: The Important Role of PI3K/Akt/mTOR. J. Oncol. 2020, 2020, 9258396. [Google Scholar] [CrossRef] [Green Version]
- CMap Website. Available online: https://clue.io/ (accessed on 15 February 2021).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Mohlin, S.; Hamidian, A.; Von Stedingk, K.; Bridges, E.; Wigerup, C.; Bexell, D.; Påhlman, S. PI3K–mTORC2 but not PI3K–mTORC1 Regulates Transcription of HIF2A/EPAS1 and Vascularization in Neuroblastoma. Cancer Res. 2015, 75, 4617–4628. [Google Scholar] [CrossRef] [Green Version]
- R2: Genomics Analysis and Visualization Platform. Available online: http://r2.amc.nl (accessed on 18 November 2020).
- The US National Library of Medicine. ClinicalTrials.gov. Available online: https://www.clinicaltrials.gov/ (accessed on 28 November 2020).
- Johnsen, J.I.; Segerström, L.; Orrego, A.; Elfman, L.; Henriksson, M.; Kågedal, B.; Eksborg, S.; Sveinbjörnsson, B.; Kogner, P. Inhibitors of mammalian target of rapamycin downregulate MYCN protein expression and inhibit neuroblastoma growth in vitro and in vivo. Oncogene 2007, 27, 2910–2922. [Google Scholar] [CrossRef] [Green Version]
- Wilson, W.R.; Hay, M. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Vanichapol, T.; Chutipongtanate, S.; Anurathapan, U.; Hongeng, S. Immune Escape Mechanisms and Future Prospects for Immunotherapy in Neuroblastoma. BioMed Res. Int. 2018, 2018, 1812535. [Google Scholar] [CrossRef]
- Puppo, M.; Battaglia, F.; Ottaviano, C.; Delfino, S.; Ribatti, D.; Varesio, L.; Bosco, M.C. Topotecan inhibits vascular endothelial growth factor production and angiogenic activity induced by hypoxia in human neuroblastoma by targeting hypoxia-inducible factor-1α and -2α. Mol. Cancer Ther. 2008, 7, 1974–1984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S. Targeting the Tumor Microenvironment in Neuroblastoma: Recent Advances and Future Directions. Cancers 2020, 12, 2057. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.S.L.; Cui, W. Proliferation, survival and metabolism: The role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 2016, 143, 3050–3060. [Google Scholar] [CrossRef] [Green Version]
- Khan, K.H.; Yap, T.A.; Yan, L.; Cunningham, D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin. J. Cancer 2013, 32, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Alzahrani, A.S. PI3K/Akt/mTOR inhibitors in cancer: At the bench and bedside. Semin. Cancer Biol. 2019, 59, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wouters, B.G.; Koritzinsky, M. Hypoxia signalling through mTOR and the unfolded protein response in cancer. Nat. Rev. Cancer 2008, 8, 851–864. [Google Scholar] [CrossRef] [PubMed]
- Kilic-Eren, M.; Boylu, T.; Tabor, V. Targeting PI3K/Akt represses Hypoxia inducible factor-1α activation and sensitizes Rhabdomyosarcoma and Ewing’s sarcoma cells for apoptosis. Cancer Cell Int. 2013, 13, 36. [Google Scholar] [CrossRef] [Green Version]
- Kelly, C.J.; Hussien, K.; Fokas, E.; Kannan, P.; Shipley, R.; Ashton, T.M.; Stratford, M.; Pearson, N.; Muschel, R.J. Regulation of O2 consumption by the PI3K and mTOR pathways contributes to tumor hypoxia. Radiother. Oncol. 2014, 111, 72–80. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Ding, X.; Ye, H.; Wang, J.; Shao, J.; Huang, T. Hypoxia enhances the migration and invasion of human glioblastoma U87 cells through PI3K/Akt/mTOR/HIF-1α pathway. NeuroReport 2018, 29, 1578–1585. [Google Scholar] [CrossRef]
- Asnaghi, L.; Lin, M.H.; Lim, K.S.; Lim, K.J.; Tripathy, A.; Wendeborn, M.; Merbs, S.L.; Handa, J.T.; Sodhi, A.; Bar, E.E.; et al. Hypoxia Promotes Uveal Melanoma Invasion through Enhanced Notch and MAPK Activation. PLoS ONE 2014, 9, e105372. [Google Scholar] [CrossRef] [Green Version]
- Hudson, C.C.; Liu, M.; Chiang, G.; Otterness, D.M.; Loomis, D.C.; Kaper, F.; Giaccia, A.J.; Abraham, R.T. Regulation of Hypoxia-Inducible Factor 1α Expression and Function by the Mammalian Target of Rapamycin. Mol. Cell. Biol. 2002, 22, 7004–7014. [Google Scholar] [CrossRef] [Green Version]
- Fulda, S. The PI3K/Akt/mTOR Pathway as Therapeutic Target in Neuroblastoma. Curr. Cancer Drug Targets 2009, 9, 729–737. [Google Scholar] [CrossRef]
- Zafar, A.; Wang, W.; Liu, G.; Wang, X.; Xian, W.; McKeon, F.; Foster, J.; Zhou, J.; Zhang, R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med. Res. Rev. 2021, 41, 961–1021. [Google Scholar] [CrossRef]
- Segerström, L.; Baryawno, N.; Sveinbjornsson, B.; Wickström, M.; Elfman, L.; Kogner, P.; Johnsen, J.I. Effects of small molecule inhibitors of PI3K/Akt/mTOR signaling on neuroblastoma growth in vitro and in vivo. Int. J. Cancer 2011, 129, 2958–2965. [Google Scholar] [CrossRef] [PubMed]
- King, D.; Yeomanson, D.; Bryant, H.E. PI3King the Lock. J. Pediatr. Hematol. Oncol. 2015, 37, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Amoroso, L.; Haupt, R.; Garaventa, A.; Ponzoni, M. Investigational drugs in phase II clinical trials for the treatment of neuroblastoma. Expert Opin. Investig. Drugs 2017, 26, 1281–1293. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.R.; Aveic, S.; Seydel, A.; Tonini, G.P. Neuroblastoma treatment in the post-genomic era. J. Biomed. Sci. 2017, 24, 14. [Google Scholar] [CrossRef] [Green Version]
- Beppu, K.; Nakamura, K.; Linehan, W.M.; Rapisarda, A.; Thiele, C.J.; van Gisbergen, K.P.; Aarnoudse, C.A.; Meijer, G.A.; Geijtenbeek, T.B.; van Kooyk, Y. Topotecan Blocks Hypoxia-Inducible Factor-1α and Vascular Endothelial Growth Factor Expression Induced by Insulin-Like Growth Factor-I in Neuroblastoma Cells. Cancer Res. 2005, 65, 4775–4781. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.; Rychahou, P.G.; Ishola, T.A.; Mourot, J.M.; Evers, B.M.; Chung, D.H. N-myc is a novel regulator of PI3K-mediated VEGF expression in neuroblastoma. Oncogene 2008, 27, 3999–4007. [Google Scholar] [CrossRef] [Green Version]
- Mei, H.; Wang, Y.; Lin, Z.; Tong, Q. The mTOR Signaling Pathway in Pediatric Neuroblastoma. Pediatr. Hematol. Oncol. 2013, 30, 605–615. [Google Scholar] [CrossRef]
- Valter, K.; Zhivotovsky, B.; Gogvadze, V. Cell death-based treatment of neuroblastoma. Cell Death Dis. 2018, 9, 113. [Google Scholar] [CrossRef] [Green Version]
- Smith, V.; Foster, J. High-Risk Neuroblastoma Treatment Review. Children 2018, 5, 114. [Google Scholar] [CrossRef] [Green Version]
- Amoroso, L.; Erminio, G.; Makin, G.; Pearson, A.D.J.; Brock, P.; Valteau-Couanet, D.; Castel, V.; Pasquet, M.; Laureys, G.; Thomas, C.; et al. Topotecan-Vincristine-Doxorubicin in Stage 4 High-Risk Neuroblastoma Patients Failing to Achieve a Complete Metastatic Response to Rapid COJEC: A SIOPEN Study. Cancer Res. Treat. 2018, 50, 148–155. [Google Scholar] [CrossRef]
- Tian, T.; Li, X.; Zhang, J. mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int. J. Mol. Sci. 2019, 20, 755. [Google Scholar] [CrossRef] [Green Version]
- Toschi, A.; Lee, E.; Gadir, N.; Ohh, M.; Foster, D.A. Differential Dependence of Hypoxia-inducible Factors 1α and 2α on mTORC1 and mTORC2. J. Biol. Chem. 2008, 283, 34495–34499. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Berel, O.; Wang, Y.; Li, P.; Bhowmick, N.A.; Figlin, R.A.; Kim, H.L. A Comparison of Ku0063794, a Dual mTORC1 and mTORC2 Inhibitor, and Temsirolimus in Preclinical Renal Cell Carcinoma Models. PLoS ONE 2013, 8, e54918. [Google Scholar] [CrossRef] [Green Version]
- Mateo, J.; Olmos, D.; Dumez, H.; Poondru, S.; Samberg, N.L.; Barr, S.; van Tornout, J.M.; Jie, F.; Sandhu, S.; Tan, D.S.; et al. A first in man, dose-finding study of the mTORC1/mTORC2 inhibitor OSI-027 in patients with advanced solid malignancies. Br. J. Cancer 2016, 114, 889–896. [Google Scholar] [CrossRef]
- Willems, L.; Chapuis, N.; Puissant, A.; Maciel, T.T.; Green, A.S.; Jacque, N.; Vignon, C.; Park, S.; Guichard, S.; Herault, O.; et al. The dual mTORC1 and mTORC2 inhibitor AZD8055 has anti-tumor activity in acute myeloid leukemia. Leukemia 2012, 26, 1195–1202. [Google Scholar] [CrossRef] [Green Version]
- Yu, K.; Shi, C.; Toral-Barza, L.; Lucas, J.; Shor, B.; Kim, J.E.; Zhang, W.-G.; Mahoney, R.; Gaydos, C.; Tardio, L.; et al. Beyond Rapalog Therapy: Preclinical Pharmacology and Antitumor Activity of WYE-125132, an ATP-Competitive and Specific Inhibitor of mTORC1 and mTORC2. Cancer Res. 2010, 70, 621–631. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Jin, Z.; Yu, K.; Liu, Q. NVP-BEZ235-induced autophagy as a potential therapeutic approach for multiple myeloma. Am. J. Transl. Res. 2019, 11, 87–105. [Google Scholar]
- Ballou, L.M.; Lin, R.Z. Rapamycin and mTOR kinase inhibitors. J. Chem. Biol. 2008, 1, 27–36. [Google Scholar] [CrossRef] [Green Version]
- Anderson, L.L.; Mao, X.; Scott, B.A.; Crowder, C.M. Survival from Hypoxia in C. elegans by Inactivation of Aminoacyl-tRNA Synthetases. Science 2009, 323, 630–633. [Google Scholar] [CrossRef] [Green Version]






| Id a | Name b | Description c | Target d | median_tau_score e | FDA- Approved f | Clinical Phase g | Clinical Trial on NB h |
|---|---|---|---|---|---|---|---|
| BRD-K67566344 | KU-0063794 | mTOR inhibitor | MTOR | −97.04 | No | Preclinical | No |
| BRD-A75409952 | wortmannin | PI3K inhibitor, ATM kinase inhibitor, ATR kinase inhibitor, DNA dependent protein kinase inhibitor, mTOR inhibitor, phosphatidylinositol 3-kinase (PI3K) inhibitor, and PLK inhibitor | PIK3CA, PIK3CG, PLK1, ATM, ATR, MTOR, PI4KA, PI4KB, PIK3CD, PIK3R1, PLK3, and PRKDC | −95.66 | No | Preclinical | No |
| BRD-K94294671 | OSI-027 | mTOR inhibitor | MTOR | −94.93 | No | 1 | No |
| BRD-K69932463 | AZD-8055 | mTOR inhibitor | MTOR | −94.49 | No | 1 | No |
| BRD-A45498368 | WYE-125132 | mTOR inhibitor and PI3K inhibitor | MTOR and PIK3CA | −94.32 | No | Preclinical | No |
| BRD-A62025033 | temsirolimus | mTOR inhibitor, cell cycle inhibitor, immunosuppressant, and protein kinase inhibitor | MTOR and PTEN | −93.9 | Yes | Launched | Yes |
| BRD-K78431006 | crizotinib | ALK tyrosine kinase receptor inhibitor, c-Met inhibitor, hepatocyte growth factor receptor inhibitor, and tyrosine kinase inhibitor | ALK, MET, CYP2B6, CYP3A5, MST1R, and ROS1 | −93.21 | Yes | Launched | Yes |
| BRD-A15079084 | phorbol-12-myristate-13-acetate | PKC activator and CD antagonist | CD4, KCNT2, PRKCA, and TRPV4 | −93.05 | No | 2 | No |
| BRD-A42649439 | triciribine | AKT inhibitor | AKT1, AKT2, and AKT3 | −93.04 | No | 1 and 2 | No |
| BRD-U82589721 | HG-5-113-01 | Protein kinase inhibitor | ABL1, LTK, and STK10 | −92.49 | No | N.D. | No |
| BRD-K13049116 | BMS-754807 | Insulin growth factor receptor inhibitor | IGF1R and AKT1 | −92.34 | No | 2 | No |
| BRD-K12184916 | NVP-BEZ235 | mTOR inhibitor, PI3K inhibitor, and protein kinase inhibitor | MTOR, PIK3CA, PIK3CG, PIK3CD, ATR, and PIK3CB | −91.92 | No | 3 | No |
| BRD-K91370081 | anisomycin | DNA synthesis inhibitor | NHP2L1, RPL10L, RPL11, RPL13A, RPL15, RPL19, RPL23, RPL23A, RPL26L1, RPL3, RPL37, RPL8, and RSL24D1 | −91.61 | No | Preclinical | No |
| BRD-A48570745 | ivermectin | GABA receptor agonist, acetylcholine receptor allosteric modulator, GABA release stimulant, and purinergic receptor allosteric modulator | CHRNA7, GABRB3, GLRA3, and P2RX7 | −91.41 | Yes | Launched | No |
| BRD-A60414806 | vincristine | Tubulin inhibitor, microtubule inhibitor, microtubule polymerization inhibitor, tubulin polymerisation inhibitor, and vinca alkaloid | TUBB and TUBA4A | −90.83 | Yes | Launched | Yes |
| BRD-A11678676 | wortmannin | PI3K inhibitor, ATM kinase inhibitor, ATR kinase inhibitor, DNA dependent protein kinase inhibitor, mTOR inhibitor, phosphatidylinositol 3-kinase (PI3K) inhibitor, and PLK inhibitor | PIK3CA, PIK3CG, PLK1, ATM, ATR, MTOR, PI4KA, PI4KB, PIK3CD, PIK3R1, PLK3, and PRKDC | −90.73 | No | Preclinical | No |
| BRD-A52650764 | ingenol | PKC activator | PRKCD and PRKCE | −90.37 | Yes | Launched | No |
| BRD-M86331534 | pyrvinium-pamoate | AKT inhibitor, androgen receptor (AR) inhibitor, and androgen receptor antagonist | AR | −90.28 | Yes | Launched | No |
| BRD-K87343924 | wortmannin | PI3K inhibitor, ATM kinase inhibitor, ATR kinase inhibitor, DNA dependent protein kinase inhibitor, mTOR inhibitor, phosphatidylinositol 3-kinase (PI3K) inhibitor, and PLK inhibitor | PIK3CA, PIK3CG, PLK1, ATM, ATR, MTOR, PI4KA, PI4KB, PIK3D, PIK3R, PLK3, and PRKDC | −90.06 | No | Preclinical | No |
| Gene Set a | ES b | NES c | NOM p-Value d | FDR q-Value e |
|---|---|---|---|---|
| HALLMARK_HYPOXIA | 0.74 | 2.96 | 0.00 | 0.00 |
| HALLMARK_GLYCOLYSIS | 0.65 | 2.64 | 0.00 | 0.00 |
| HALLMARK_MTORC1_SIGNALING | 0.65 | 2.60 | 0.00 | 0.00 |
| HALLMARK_ANGIOGENESIS | 0.65 | 1.92 | 0.00 | 0.00 |
| HALLMARK_PROTEIN_SECRETION | 0.47 | 1.73 | 0.00 | 0.01 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | 0.51 | 1.71 | 0.00 | 0.01 |
| HALLMARK_FATTY_ACID_METABOLISM | 0.44 | 1.70 | 0.00 | 0.01 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 0.45 | 1.64 | 0.00 | 0.01 |
| HALLMARK_ADIPOGENESIS | 0.40 | 1.56 | 0.00 | 0.02 |
| HALLMARK_G2M_CHECKPOINT | −0.43 | −1.62 | 0.00 | 0.02 |
| HALLMARK_MITOTIC_SPINDLE | −0.58 | −2.19 | 0.00 | 0.00 |
| Gene Set a | ES b | NES c | NOM p-Value d | FDR q-Value e |
|---|---|---|---|---|
| HALLMARK_E2F_TARGETS | −0.87 | −2.88 | 0.00 | 0.00 |
| HALLMARK_G2M_CHECKPOINT | −0.80 | −2.62 | 0.00 | 0.00 |
| HALLMARK_MTORC1_SIGNALING | −0.75 | −2.50 | 0.00 | 0.00 |
| HALLMARK_GLYCOLYSIS | −0.61 | −2.07 | 0.00 | 0.00 |
| HALLMARK_MYC_TARGETS_V1 | −0.61 | −2.05 | 0.00 | 0.00 |
| HALLMARK_ESTROGEN_RESPONSE_LATE | −0.59 | −1.98 | 0.00 | 0.00 |
| HALLMARK_CHOLESTEROL_HOMEOSTASIS | −0.66 | −1.95 | 0.00 | 0.00 |
| HALLMARK_UV_RESPONSE_UP | −0.58 | −1.91 | 0.00 | 0.00 |
| HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY | −0.69 | −1.87 | 0.00 | 0.00 |
| HALLMARK_MITOTIC_SPINDLE | −0.54 | −1.82 | 0.00 | 0.00 |
| HALLMARK_PI3K_AKT_MTOR_SIGNALING | −0.55 | −1.76 | 0.00 | 0.00 |
| HALLMARK_SPERMATOGENESIS | −0.55 | −1.74 | 0.00 | 0.00 |
| HALLMARK_UNFOLDED_PROTEIN_RESPONSE | −0.54 | −1.71 | 0.00 | 0.00 |
| HALLMARK_OXIDATIVE_PHOSPHORYLATION | −0.51 | −1.68 | 0.00 | 0.00 |
| HALLMARK_DNA_REPAIR | −0.52 | −1.66 | 0.00 | 0.00 |
| HALLMARK_FATTY_ACID_METABOLISM | −0.49 | −1.61 | 0.00 | 0.00 |
| HALLMARK_ANDROGEN_RESPONSE | −0.51 | −1.57 | 0.00 | 0.01 |
| HALLMARK_MYC_TARGETS_V2 | −0.54 | −1.54 | 0.01 | 0.01 |
| HALLMARK_HYPOXIA | −0.46 | −1.53 | 0.00 | 0.01 |
| HALLMARK_XENOBIOTIC_METABOLISM | −0.45 | −1.49 | 0.00 | 0.01 |
| HALLMARK_APOPTOSIS | −0.44 | −1.46 | 0.01 | 0.02 |
| HALLMARK_ADIPOGENESIS | −0.43 | −1.44 | 0.00 | 0.02 |
| HALLMARK_PEROXISOME | −0.46 | −1.41 | 0.02 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uva, P.; Bosco, M.C.; Eva, A.; Conte, M.; Garaventa, A.; Amoroso, L.; Cangelosi, D. Connectivity Map Analysis Indicates PI3K/Akt/mTOR Inhibitors as Potential Anti-Hypoxia Drugs in Neuroblastoma. Cancers 2021, 13, 2809. https://doi.org/10.3390/cancers13112809
Uva P, Bosco MC, Eva A, Conte M, Garaventa A, Amoroso L, Cangelosi D. Connectivity Map Analysis Indicates PI3K/Akt/mTOR Inhibitors as Potential Anti-Hypoxia Drugs in Neuroblastoma. Cancers. 2021; 13(11):2809. https://doi.org/10.3390/cancers13112809
Chicago/Turabian StyleUva, Paolo, Maria Carla Bosco, Alessandra Eva, Massimo Conte, Alberto Garaventa, Loredana Amoroso, and Davide Cangelosi. 2021. "Connectivity Map Analysis Indicates PI3K/Akt/mTOR Inhibitors as Potential Anti-Hypoxia Drugs in Neuroblastoma" Cancers 13, no. 11: 2809. https://doi.org/10.3390/cancers13112809
APA StyleUva, P., Bosco, M. C., Eva, A., Conte, M., Garaventa, A., Amoroso, L., & Cangelosi, D. (2021). Connectivity Map Analysis Indicates PI3K/Akt/mTOR Inhibitors as Potential Anti-Hypoxia Drugs in Neuroblastoma. Cancers, 13(11), 2809. https://doi.org/10.3390/cancers13112809







