A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohorts and Biospecimen Collection
2.2. RNA/DNA Extraction and p16 Immunohistochemistry
2.3. Functional RNA Quantification and RNA Reverse Transcription
2.4. HPV Testing
2.5. Next Generation Sequencing (NGS)
2.6. Gene Expression Analyses
2.7. Creating a Six-Gene Signature
2.8. Cancer Cohort
2.8.1. Patient Characteristics and Biospecimen Collection
2.8.2. Gene Expression Profiling
2.9. Statistical Analyses
| Cone Excision Diagnosis | |||
|---|---|---|---|
| CIN3 Regression | Persistent CIN3 | p-Value | |
| n = 21 | n = 28 | ||
| Last cytology before biopsy | 0.71 a | ||
| AGUS | 0 (0) | 1 (100) | |
| ASC-H | 4 (36) | 7 (64) | |
| ASCUS | 0 (0) | 1 (100) | |
| HSIL | 10 (42) | 14 (58) | |
| LSIL | 6 (60) | 4 (40) | |
| Normal | 1 (50) | 1 (50) | |
| HPV Type in Biopsy | 0.79 a | ||
| HPV 16 | 9 (39) | 14 (61) | |
| HPV18 | 2 (40) | 3 (60) | |
| HPV 31 | 1 (50) | 1 (50) | |
| HPV 33 | 4 (36) | 7 (64) | |
| HPV 35 | 2 (100) | 0 (0) | |
| HPV 39 | 1 (50) | 1 (50) | |
| HPV 52 | 2 (50) | 2 (50) | |
| Age at diagnosis | 0.19 b | ||
| ≤29 | 8 (33) | 16 (67) | |
| >29 | 13 (52) | 12 (48) | |
| Interval between cytology and biopsy | 0.32 b | ||
| ≤41 | 12 (50) | 12 (50) | |
| >41 | 9 (36) | 16 (64) | |
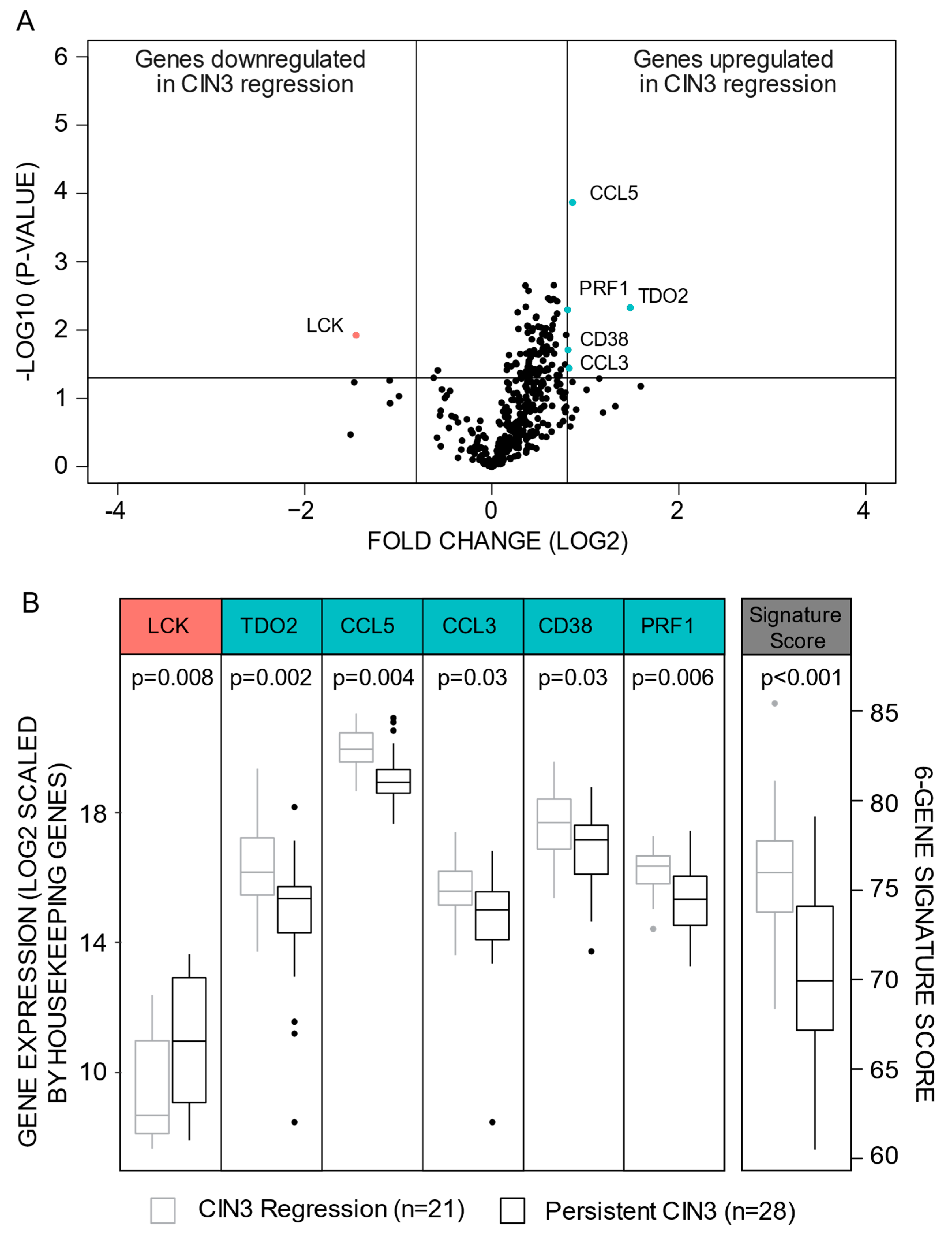
3. Results
3.1. A Six-Gene Signature Predicting CIN3 Regression
3.2. Persistent CIN3 Associates to Proliferation

3.3. High Regression Signature Score Associates to Favorable Survival and Less Aggressive Features within Cervical Cancer Patients
| Variables (n) a | Regression Signature Score | p-Value b | |
|---|---|---|---|
| Low Score (n = 91) | High Score (n = 148) | ||
| Median age (n = 239) | 0.98 | ||
| <44 years | 42 (38) | 68 (62) | |
| ≥44 years | 49 (38) | 80 (62) | |
| BMI (n = 236) | 0.17 | ||
| <25 | 42 (34) | 83 (66) | |
| ≥25 | 47 (42) | 64 (58) | |
| FIGO-09 stage (n = 239) | 0.27 | ||
| I-IB1 | 40 (34) | 76 (66) | |
| IB2-IV | 51 (42) | 72 (58) | |
| Histologic type (n = 239) | 0.15 | ||
| Squamous cell carcinoma | 59 (35) | 112 (65) | |
| Adenocarcinoma | 24 (50) | 24 (50) | |
| Other histologic type | 8 (40) | 12 (60) | |
| Histologic grade (n = 225) | 0.06 | ||
| Grade 1/2 | 58 (34) | 111 (66) | |
| Grade 3 | 27 (48) | 29 (52) | |
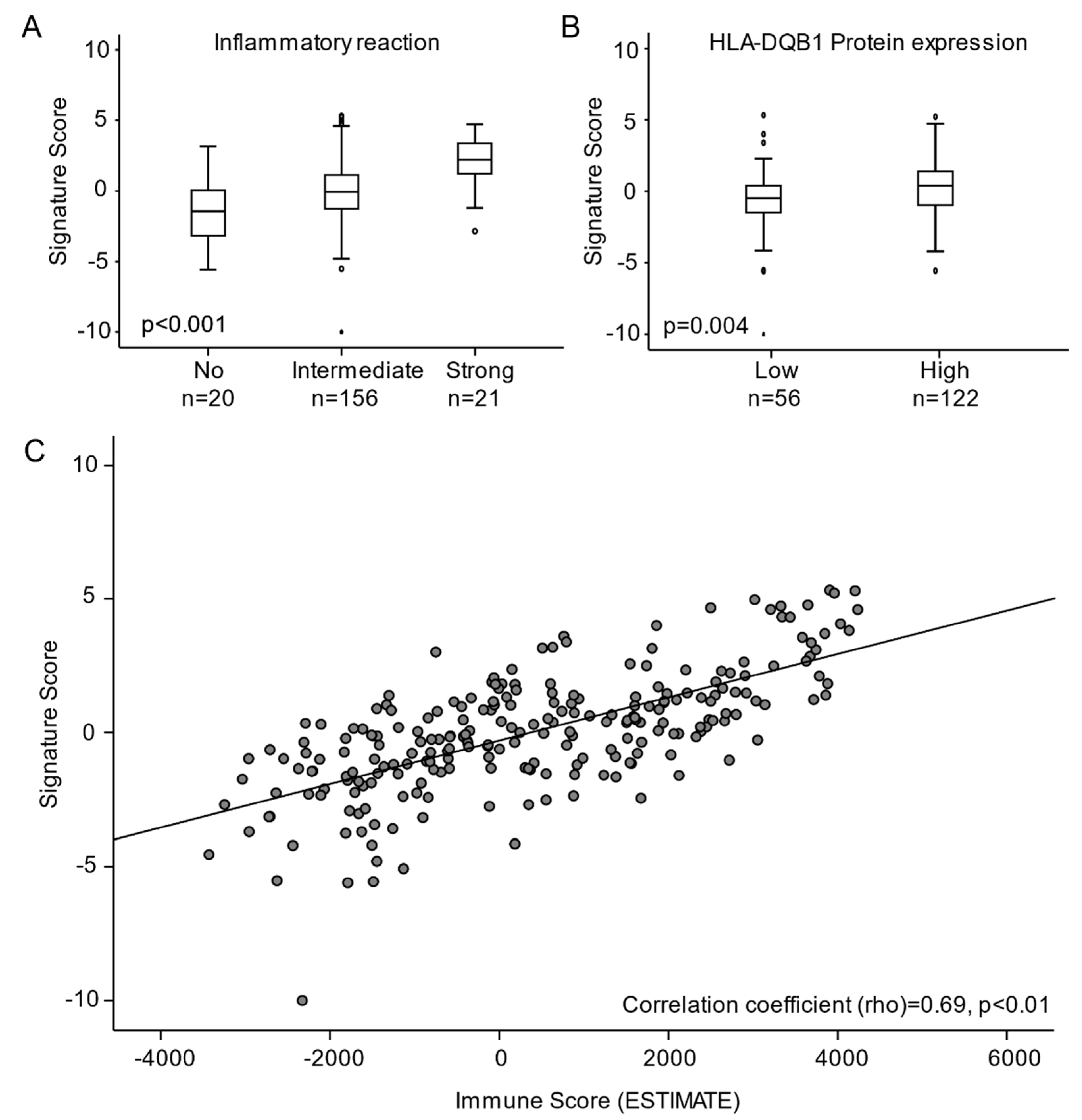
3.4. High Regression Signature Score Is Significantly Associated with Immune Activation in Cervical Carcinomas

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schiffman, M.; Castle, P.E.; Jeronimo, J.; Rodriguez, A.C.; Wacholder, S. Human papillomavirus and cervical cancer. Lancet 2007, 370, 890–907. [Google Scholar] [CrossRef]
- Rodríguez, A.C.; Schiffman, M.; Herrero, R.; Wacholder, S.; Hildesheim, A.; Castle, P.E.; Solomon, D.; Burk, R. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J. Natl. Cancer Inst. 2008, 100, 513–517. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.J.; Castle, P.E.; Lorincz, A.T.; Wacholder, S.; Sherman, M.; Scott, D.R.; Rush, B.B.; Glass, A.G.; Schiffman, M. The elevated 10-year risk of cervical precancer and cancer in women with human papillomavirus (HPV) type 16 or 18 and the possible utility of type-specific HPV testing in clinical practice. J. Natl. Cancer Inst. 2005, 97, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- McCredie, M.R.; Sharples, K.J.; Paul, C.; Baranyai, J.; Medley, G.; Jones, R.W.; Skegg, D.C. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: A retrospective cohort study. Lancet Oncol. 2008, 9, 425–434. [Google Scholar] [CrossRef]
- Tjalma, W.A.; Fiander, A.; Reich, O.; Powell, N.; Nowakowski, A.M.; Kirschner, B.; Koiss, R.; O’Leary, J.; Joura, E.A.; Rosenlund, M.; et al. Differences in human papillomavirus type distribution in high-grade cervical intraepithelial neoplasia and invasive cervical cancer in Europe. Int. J. Cancer 2013, 132, 854–867. [Google Scholar] [CrossRef] [PubMed]
- Ostör, A.G. Natural history of cervical intraepithelial neoplasia: A critical review. Int. J. Gynecol. Pathol. 1993, 12, 186–192. [Google Scholar] [CrossRef]
- Munk, A.C.; Gudlaugsson, E.; Ovestad, I.T.; Lovslett, K.; Fiane, B.; Hidle, B.; Kruse, A.J.; Skaland, I.; Janssen, E.A.; Baak, J.P. Interaction of epithelial biomarkers, local immune response and condom use in cervical intraepithelial neoplasia 2-3 regression. Gynecol Oncol. 2012, 127, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Ovestad, I.T.; Gudlaugsson, E.; Skaland, I.; Malpica, A.; Munk, A.C.; Janssen, E.A.; Baak, J.P. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. J. Clin. Pathol. 2011, 64, 303–307. [Google Scholar] [CrossRef]
- Trimble, C.L.; Piantadosi, S.; Gravitt, P.; Ronnett, B.; Pizer, E.; Elko, A.; Wilgus, B.; Yutzy, W.; Daniel, R.; Shah, K.; et al. Spontaneous regression of high-grade cervical dysplasia: Effects of human papillomavirus type and HLA phenotype. Clin. Cancer Res. 2005, 11, 4717–4723. [Google Scholar] [CrossRef] [Green Version]
- Hagen, B.; Skjeldestad, F.E.; Bratt, H.; Tingulstad, S.; Lie, A.K. CO2 laser conization for cervical intraepithelial neoplasia grade II-III: Complications and efficacy. Acta Obstet. Gynecol. Scand. 1998, 77, 558–563. [Google Scholar] [CrossRef] [PubMed]
- Santesso, N.; Mustafa, R.A.; Wiercioch, W.; Kehar, R.; Gandhi, S.; Chen, Y.; Cheung, A.; Hopkins, J.; Khatib, R.; Ma, B.; et al. Systematic reviews and meta-analyses of benefits and harms of cryotherapy, LEEP, and cold knife conization to treat cervical intraepithelial neoplasia. Int. J. Gynaecol. Obstet. 2016, 132, 266–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albrechtsen, S.; Rasmussen, S.; Thoresen, S.; Irgens, L.M.; Iversen, O.E. Pregnancy outcome in women before and after cervical conisation: Population based cohort study. BMJ 2008, 337, a1343. [Google Scholar] [CrossRef] [Green Version]
- Arbyn, M.; Kyrgiou, M.; Simoens, C.; Raifu, A.O.; Koliopoulos, G.; Martin-Hirsch, P.; Prendiville, W.; Paraskevaidis, E. Perinatal mortality and other severe adverse pregnancy outcomes associated with treatment of cervical intraepithelial neoplasia: Meta-analysis. BMJ 2008, 337, a1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kyrgiou, M.; Athanasiou, A.; Paraskevaidi, M.; Mitra, A.; Kalliala, I.; Martin-Hirsch, P.; Arbyn, M.; Bennett, P.; Paraskevaidis, E. Adverse obstetric outcomes after local treatment for cervical preinvasive and early invasive disease according to cone depth: Systematic review and meta-analysis. BMJ 2016, 354, i3633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maver, P.J.; Poljak, M. Primary HPV-based cervical cancer screening in Europe: Implementation status, challenges, and future plans. Clin. Microbiol. Infect. 2020, 26, 579–583. [Google Scholar] [CrossRef]
- Arbyn, M.; Ronco, G.; Anttila, A.; Meijer, C.J.; Poljak, M.; Ogilvie, G.; Koliopoulos, G.; Naucler, P.; Sankaranarayanan, R.; Peto, J. Evidence regarding human papillomavirus testing in secondary prevention of cervical cancer. Vaccine 2012, 30, F88–F99. [Google Scholar] [CrossRef]
- Ronco, G.; Dillner, J.; Elfström, K.M.; Tunesi, S.; Snijders, P.J.; Arbyn, M.; Kitchener, H.; Segnan, N.; Gilham, C.; Giorgi-Rossi, P.; et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: Follow-up of four European randomised controlled trials. Lancet 2014, 383, 524–532. [Google Scholar] [CrossRef]
- Orumaa, M.; Leinonen, M.K.; Campbell, S.; Møller, B.; Myklebust, T.; Nygård, M. Recent increase in incidence of cervical precancerous lesions in Norway: Nationwide study from 1992 to 2016. Int. J. Cancer 2019, 145, 2629–2638. [Google Scholar] [CrossRef] [Green Version]
- Guleria, S.; Faber, M.T.; Hansen, B.T.; Arnheim-Dahlström, L.; Liaw, K.L.; Munk, C.; Nygård, M.; Kjær, S.K. Self-perceived risk of STIs in a population-based study of Scandinavian women. Sex. Transm Infect. 2018, 94, 522–527. [Google Scholar] [CrossRef]
- Liu, G.; Hariri, S.; Bradley, H.; Gottlieb, S.L.; Leichliter, J.S.; Markowitz, L.E. Trends and patterns of sexual behaviors among adolescents and adults aged 14 to 59 years, United States. Sex. Transm Dis. 2015, 42, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Stigum, H.; Samuelsen, S.O.; Traeen, B. Analysis of first coitus. Arch. Sex. Behav. 2010, 39, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Waxman, A.G.; Chelmow, D.; Darragh, T.M.; Lawson, H.; Moscicki, A.B. Revised terminology for cervical histopathology and its implications for management of high-grade squamous intraepithelial lesions of the cervix. Obstet. Gynecol. 2012, 120, 1465–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castle, P.E.; Adcock, R.; Cuzick, J.; Wentzensen, N.; Torrez-Martinez, N.E.; Torres, S.M.; Stoler, M.H.; Ronnett, B.M.; Joste, N.E.; Darragh, T.M.; et al. Relationships of p16 Immunohistochemistry and Other Biomarkers with Diagnoses of Cervical Abnormalities: Implications for LAST Terminology. Arch. Pathol. Lab. Med. 2020, 144, 725–734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 169–182. [Google Scholar] [CrossRef]
- Shamseddine, A.A.; Burman, B.; Lee, N.Y.; Zamarin, D.; Riaz, N. Tumor Immunity and Immunotherapy for HPV-Related Cancers. Cancer Discov. 2021, 11, 1896–1912. [Google Scholar] [CrossRef]
- Munk, A.C.; Gudlaugsson, E.; Malpica, A.; Fiane, B.; Løvslett, K.I.; Kruse, A.J.; Øvestad, I.T.; Voorhorst, F.; Janssen, E.A.; Baak, J.P. Consistent condom use increases the regression rate of cervical intraepithelial neoplasia 2-3. PLoS ONE 2012, 7, e45114. [Google Scholar] [CrossRef] [Green Version]
- Munk, A.C.; Ovestad, I.T.; Gudlaugsson, E.; Løvslett, K.; Fiane, B.; van Diermen-Hidle, B.; Kruse, A.J.; Skaland, I.; Janssen, E.A.; Baak, J.P. Consistent condom use increases spontaneous regression in high-risk non-HPV16 but not in HPV16 CIN2-3 lesions, a prospective population-based cohort study. Infect. Agent Cancer 2012, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Dysvik, B.; Jonassen, I. J-Express: Exploring gene expression data using Java. Bioinformatics 2001, 17, 369–370. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Halle, M.K.; Sodal, M.; Forsse, D.; Engerud, H.; Woie, K.; Lura, N.G.; Wagner-Larsen, K.S.; Trovik, J.; Bertelsen, B.I.; Haldorsen, I.S.; et al. A 10-gene prognostic signature points to LIMCH1 and HLA-DQB1 as important players in aggressive cervical cancer disease. Br. J. Cancer 2021. [Google Scholar] [CrossRef]
- Halle, M.K.; Ojesina, A.I.; Engerud, H.; Woie, K.; Tangen, I.L.; Holst, F.; Hoivik, E.; Kusonmano, K.; Haldorsen, I.S.; Vintermyr, O.K.; et al. Clinicopathologic and molecular markers in cervical carcinoma: A prospective cohort study. Am. J. Obstet. Gynecol. 2017, 217, 432.e1–432.e17. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Youden, W.J. Index for rating diagnostic tests. Cancer 1950, 3, 32–35. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Tainio, K.; Athanasiou, A.; Tikkinen, K.A.O.; Aaltonen, R.; Cárdenas, J.; Hernándes; Glazer-Livson, S.; Jakobsson, M.; Joronen, K.; Kiviharju, M.; et al. Clinical course of untreated cervical intraepithelial neoplasia grade 2 under active surveillance: Systematic review and meta-analysis. BMJ 2018, 360, k499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bommhardt, U.; Schraven, B.; Simeoni, L. Beyond TCR Signaling: Emerging Functions of Lck in Cancer and Immunotherapy. Int. J. Mol. Sci. 2019, 20, 3500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, R.K.; Yoon, C.H.; Hyun, K.H.; Lee, H.; An, S.; Park, M.J.; Kim, M.J.; Lee, S.J. Role of lymphocyte-specific protein tyrosine kinase (LCK) in the expansion of glioma-initiating cells by fractionated radiation. Biochem. Biophys. Res. Commun. 2010, 402, 631–636. [Google Scholar] [CrossRef] [PubMed]
- Elsberger, B.; Fullerton, R.; Zino, S.; Jordan, F.; Mitchell, T.J.; Brunton, V.G.; Mallon, E.A.; Shiels, P.G.; Edwards, J. Breast cancer patients’ clinical outcome measures are associated with Src kinase family member expression. Br. J. Cancer 2010, 103, 899–909. [Google Scholar] [CrossRef]
- Robinson, D.; He, F.; Pretlow, T.; Kung, H.J. A tyrosine kinase profile of prostate carcinoma. Proc. Natl. Acad. Sci. USA 1996, 93, 5958–5962. [Google Scholar] [CrossRef] [Green Version]
- Veillette, A.; Foss, F.M.; Sausville, E.A.; Bolen, J.B.; Rosen, N. Expression of the lck tyrosine kinase gene in human colon carcinoma and other non-lymphoid human tumor cell lines. Oncogene Res. 1987, 1, 357–374. [Google Scholar]
- Chakraborty, G.; Rangaswami, H.; Jain, S.; Kundu, G.C. Hypoxia regulates cross-talk between Syk and Lck leading to breast cancer progression and angiogenesis. J. Biol. Chem. 2006, 281, 11322–11331. [Google Scholar] [CrossRef] [Green Version]
- Pilotte, L.; Larrieu, P.; Stroobant, V.; Colau, D.; Dolusic, E.; Frédérick, R.; De Plaen, E.; Uyttenhove, C.; Wouters, J.; Masereel, B.; et al. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 2012, 109, 2497–2502. [Google Scholar] [CrossRef] [Green Version]
- Ferns, D.M.; Kema, I.P.; Buist, M.R.; Nijman, H.W.; Kenter, G.G.; Jordanova, E.S. Indoleamine-2,3-dioxygenase (IDO) metabolic activity is detrimental for cervical cancer patient survival. Oncoimmunology 2015, 4, e981457. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Nollen, E.A.A.; Röhrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug. Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef] [PubMed]
- Venancio, P.A.; Consolaro, M.E.L.; Derchain, S.F.; Boccardo, E.; Villa, L.L.; Maria-Engler, S.S.; Campa, A.; Discacciati, M.G. Indoleamine 2,3-dioxygenase and tryptophan 2,3-dioxygenase expression in HPV infection, SILs, and cervical cancer. Cancer Cytopathol. 2019, 127, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Tuong, Z.K.; Frazer, I.H. Papillomavirus Immune Evasion Strategies Target the Infected Cell and the Local Immune System. Front. Oncol. 2019, 9, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konen, J.M.; Fradette, J.J.; Gibbons, D.L. The Good, the Bad and the Unknown of CD38 in the Metabolic Microenvironment and Immune Cell Functionality of Solid Tumors. Cells 2019, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Liao, S.; Xiao, S.; Chen, H.; Zhang, M.; Chen, Z.; Long, Y.; Gao, L.; Zhu, G.; He, J.; Peng, S.; et al. CD38 enhances the proliferation and inhibits the apoptosis of cervical cancer cells by affecting the mitochondria functions. Mol. Carcinog. 2017, 56, 2245–2257. [Google Scholar] [CrossRef]
- Liao, S.; Liang, L.; Yue, C.; He, J.; He, Z.; Jin, X.; Luo, G.; Zhou, Y. CD38 is involved in cell energy metabolism via activating the PI3K/AKT/mTOR signaling pathway in cervical cancer cells. Int. J. Oncol. 2020, 57, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.H.; Ng, H.H.M.; Lim, C.J.; Sim, X.N.; Malavasi, F.; Li, H.; Loh, J.J.H.; Sabai, K.; Kim, J.K.; Ong, C.C.H.; et al. Expression of CD38 on Macrophages Predicts Improved Prognosis in Hepatocellular Carcinoma. Front. Immunol. 2019, 10, 2093. [Google Scholar] [CrossRef] [Green Version]
- Ng, H.H.M.; Lee, R.Y.; Goh, S.; Tay, I.S.Y.; Lim, X.; Lee, B.; Chew, V.; Li, H.; Tan, B.; Lim, S.; et al. Immunohistochemical scoring of CD38 in the tumor microenvironment predicts responsiveness to anti-PD-1/PD-L1 immunotherapy in hepatocellular carcinoma. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Akamatsu, H.; Niwa, H.; Sumi, H.; Ozaki, Y.; Abe, A. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin. Cancer Res. 2001, 7, 285–289. [Google Scholar] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Sheu, B.C.; Chiou, S.H.; Lin, H.H.; Chow, S.N.; Huang, S.C.; Ho, H.N.; Hsu, S.M. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Res. 2005, 65, 2921–2929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, A.T.; da Rocha, N.P.; Avvad, E.; Grinsztejn, B.J.; Russomano, F.; Tristão, A.; Quintana Mde, S.; Perez, M.A.; Conceição-Silva, F.; Bonecini-Almeida Mda, G. Balance of apoptotic and anti-apoptotic marker and perforin granule release in squamous intraepithelial lesions. HIV infection leads to a decrease in perforin degranulation. Exp. Mol. Pathol. 2013, 95, 166–173. [Google Scholar] [CrossRef]
- zur Hausen, H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 2000, 92, 690–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahajan, A. Practical issues in the application of p16 immunohistochemistry in diagnostic pathology. Hum. Pathol. 2016, 51, 64–74. [Google Scholar] [CrossRef]
- Wang, S.S.; Trunk, M.; Schiffman, M.; Herrero, R.; Sherman, M.E.; Burk, R.D.; Hildesheim, A.; Bratti, M.C.; Wright, T.; Rodriguez, A.C.; et al. Validation of p16INK4a as a marker of oncogenic human papillomavirus infection in cervical biopsies from a population-based cohort in Costa Rica. Cancer Epidemiol. Biomarkers Prev. 2004, 13, 1355–1360. [Google Scholar]
- Ronsini, C.; Anchora, L.P.; Restaino, S.; Fedele, C.; Arciuolo, D.; Teodorico, E.; Bizzarri, N.; Zannoni, G.F.; Ferrandina, G.; Scambia, G.; et al. The role of semiquantitative evaluation of lympho-vascular space invasion in early stage cervical cancer patients. Gynecol. Oncol. 2021, 162, 299–307. [Google Scholar] [CrossRef]
- Holowaty, P.; Miller, A.B.; Rohan, T.; To, T. Natural history of dysplasia of the uterine cervix. J. Natl. Cancer Inst. 1999, 91, 252–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Oortmarssen, G.J.; Habbema, J.D. Epidemiological evidence for age-dependent regression of pre-invasive cervical cancer. Br. J. Cancer 1991, 64, 559–565. [Google Scholar] [CrossRef] [Green Version]
- Appleby, P.; Beral, V.; Berrington de González, A.; Colin, D.; Franceschi, S.; Goodill, A.; Green, J.; Peto, J.; Plummer, M.; Sweetland, S. Carcinoma of the cervix and tobacco smoking: Collaborative reanalysis of individual data on 13,541 women with carcinoma of the cervix and 23,017 women without carcinoma of the cervix from 23 epidemiological studies. Int. J. Cancer 2006, 118, 1481–1495. [Google Scholar] [CrossRef]
- Sand, F.L.; Munk, C.; Frederiksen, K.; Junge, J.; Iftner, T.; Dehlendorff, C.; Kjaer, S.K. Risk of CIN3 or worse with persistence of 13 individual oncogenic HPV types. Int. J. Cancer 2019, 144, 1975–1982. [Google Scholar] [CrossRef]
- International Collaboration of Epidemiological Studies of Cervical Cancer; Appleby, P.; Beral, V.; Berrington de Gonzalez, A.; Colin, D.; Franceschi, S.; Goodhill, A.; Green, J.; Peto, J.; Plummer, M.; et al. Cervical cancer and hormonal contraceptives: Collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet 2007, 370, 1609–1621. [Google Scholar] [CrossRef]
- International Collaboration of Epidemiological Studies of Cervical Cancer. Cervical carcinoma and reproductive factors: Collaborative reanalysis of individual data on 16,563 women with cervical carcinoma and 33,542 women without cervical carcinoma from 25 epidemiological studies. Int. J. Cancer 2006, 119, 1108–1124. [Google Scholar] [CrossRef]
- Salvadó, A.; Miralpeix, E.; Solé-Sedeno, J.M.; Kanjou, N.; Lloveras, B.; Duran, X.; Mancebo, G. Predictor factors for conservative management of cervical intraepithelial neoplasia grade 2: Cytology and HPV genotyping. Gynecol. Oncol. 2021, 162, 569–574. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Ntritsos, G.; Smith, A.; Tsilidis, K.K.; Marchesi, J.R.; Bennett, P.R.; Moscicki, A.B.; Kyrgiou, M. The vaginal microbiota associates with the regression of untreated cervical intraepithelial neoplasia 2 lesions. Nat. Commun. 2020, 11, 1999. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Lu, C.X. Spontaneous Regression of Cervical Intraepithelial Neoplasia 2: A Meta-analysis. Gynecol. Obstet. Invest. 2019, 84, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Koeneman, M.M.; Hendriks, N.; Kooreman, L.F.; Winkens, B.; Kruitwagen, R.F.; Kruse, A.J. Prognostic factors for spontaneous regression of high-risk human papillomavirus-positive cervical intra-epithelial neoplasia grade 2. Int. J. Gynecol. Cancer 2019, 29, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Skorstengaard, M.; Lynge, E.; Suhr, J.; Napolitano, G. Conservative management of women with cervical intraepithelial neoplasia grade 2 in Denmark: A cohort study. BJOG 2020, 127, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Andersen, E.S.; Nielsen, K.; Pedersen, B. The reliability of preconization diagnostic evaluation in patients with cervical intraepithelial neoplasia and microinvasive carcinoma. Gynecol. Oncol. 1995, 59, 143–147. [Google Scholar] [CrossRef]
- Mutombo, A.B.; Simoens, C.; Tozin, R.; Bogers, J.; Van Geertruyden, J.P.; Jacquemyn, Y. Efficacy of commercially available biological agents for the topical treatment of cervical intraepithelial neoplasia: A systematic review. Syst. Rev. 2019, 8, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonseca, B.O.; Possati-Resende, J.C.; Salcedo, M.P.; Schmeler, K.M.; Accorsi, G.S.; Fregnani, J.; Antoniazzi, M.; Pantano, N.P.; Santana, I.V.V.; Matsushita, G.M.; et al. Topical Imiquimod for the Treatment of High-Grade Squamous Intraepithelial Lesions of the Cervix: A Randomized Controlled Trial. Obstet. Gynecol. 2021, 137, 1043–1053. [Google Scholar] [CrossRef]
- Xiong, Y.; Cui, L.; Bian, C.; Zhao, X.; Wang, X. Clearance of human papillomavirus infection in patients with cervical intraepithelial neoplasia: A systemic review and meta-analysis. Medicine 2020, 99, e23155. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Halle, M.K.; Munk, A.C.; Engesæter, B.; Akbari, S.; Frafjord, A.; Hoivik, E.A.; Forsse, D.; Fasmer, K.E.; Woie, K.; Haldorsen, I.S.; et al. A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival. Cancers 2021, 13, 5737. https://doi.org/10.3390/cancers13225737
Halle MK, Munk AC, Engesæter B, Akbari S, Frafjord A, Hoivik EA, Forsse D, Fasmer KE, Woie K, Haldorsen IS, et al. A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival. Cancers. 2021; 13(22):5737. https://doi.org/10.3390/cancers13225737
Chicago/Turabian StyleHalle, Mari K., Ane Cecilie Munk, Birgit Engesæter, Saleha Akbari, Astri Frafjord, Erling A. Hoivik, David Forsse, Kristine E. Fasmer, Kathrine Woie, Ingfrid S. Haldorsen, and et al. 2021. "A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival" Cancers 13, no. 22: 5737. https://doi.org/10.3390/cancers13225737
APA StyleHalle, M. K., Munk, A. C., Engesæter, B., Akbari, S., Frafjord, A., Hoivik, E. A., Forsse, D., Fasmer, K. E., Woie, K., Haldorsen, I. S., Bertelsen, B. I., Janssen, E. A. M., Gudslaugsson, E., Krakstad, C., & Øvestad, I. T. (2021). A Gene Signature Identifying CIN3 Regression and Cervical Cancer Survival. Cancers, 13(22), 5737. https://doi.org/10.3390/cancers13225737






