Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
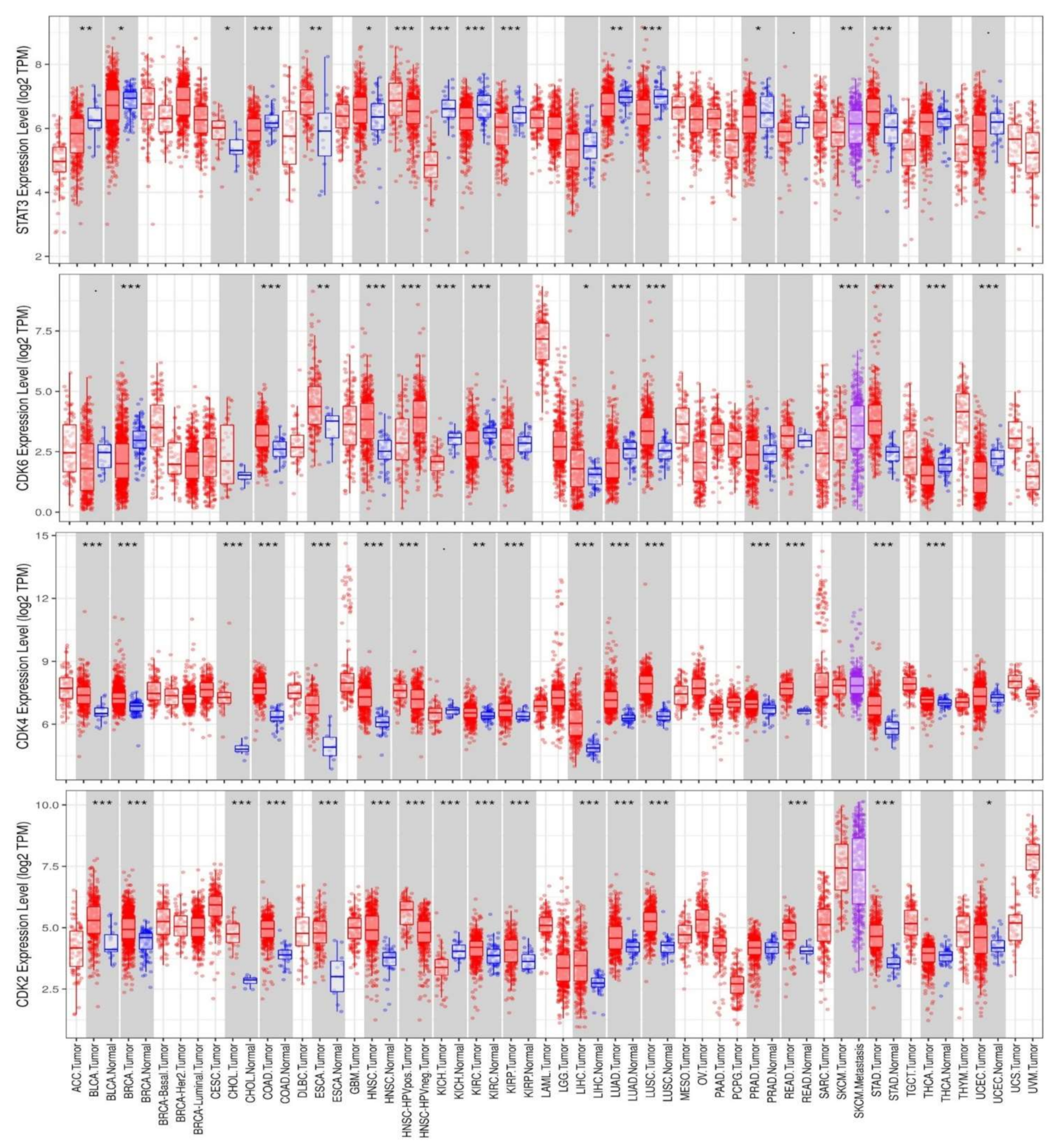
2.1. Differential Expression Analysis of STAT3/CDK2/D/6 Signatures in a Panel of Human Cancers
2.2. Survival Analysis of STAT3/CDK2/D/6 Signature in a Panel of Human Cancers
2.3. Protein-Protein Interaction and Functional Enrichment Analysis
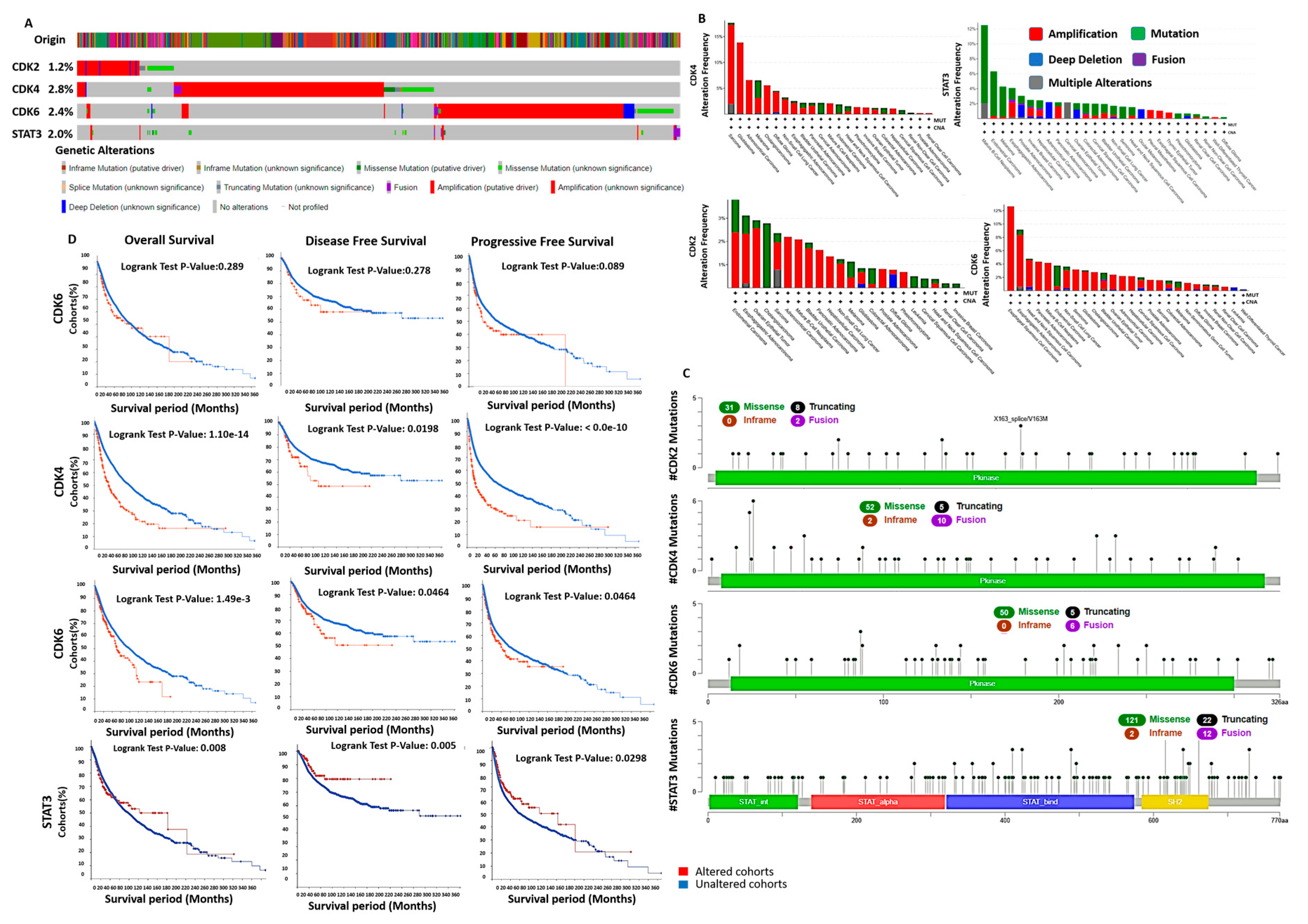
2.4. Analysis of STAT3/CDK2/4/6 Genetic Alterations and Its Prognostic Relevance in Multiple Cancers
2.5. Analysis of STAT3/CDK2/4/6 Association with Infiltrations of Cancer-Associated Fibroblast and Various Immune Cells
2.6. Analysis of STAT3/CDK2/4/6 Association with Dysfunctional T-Cells and Clinical Outcome of Immunotherapy
2.7. Statistical Analysis
3. Results
3.1. Overexpression of STAT3/CDK2/4/6 Signaling Networks Is Associated with Poor Prognoses of Multiple Cancers
3.2. STAT3/CDK2/4/6 Are Enriched in Cancer and Immune Associated Signaling Networks
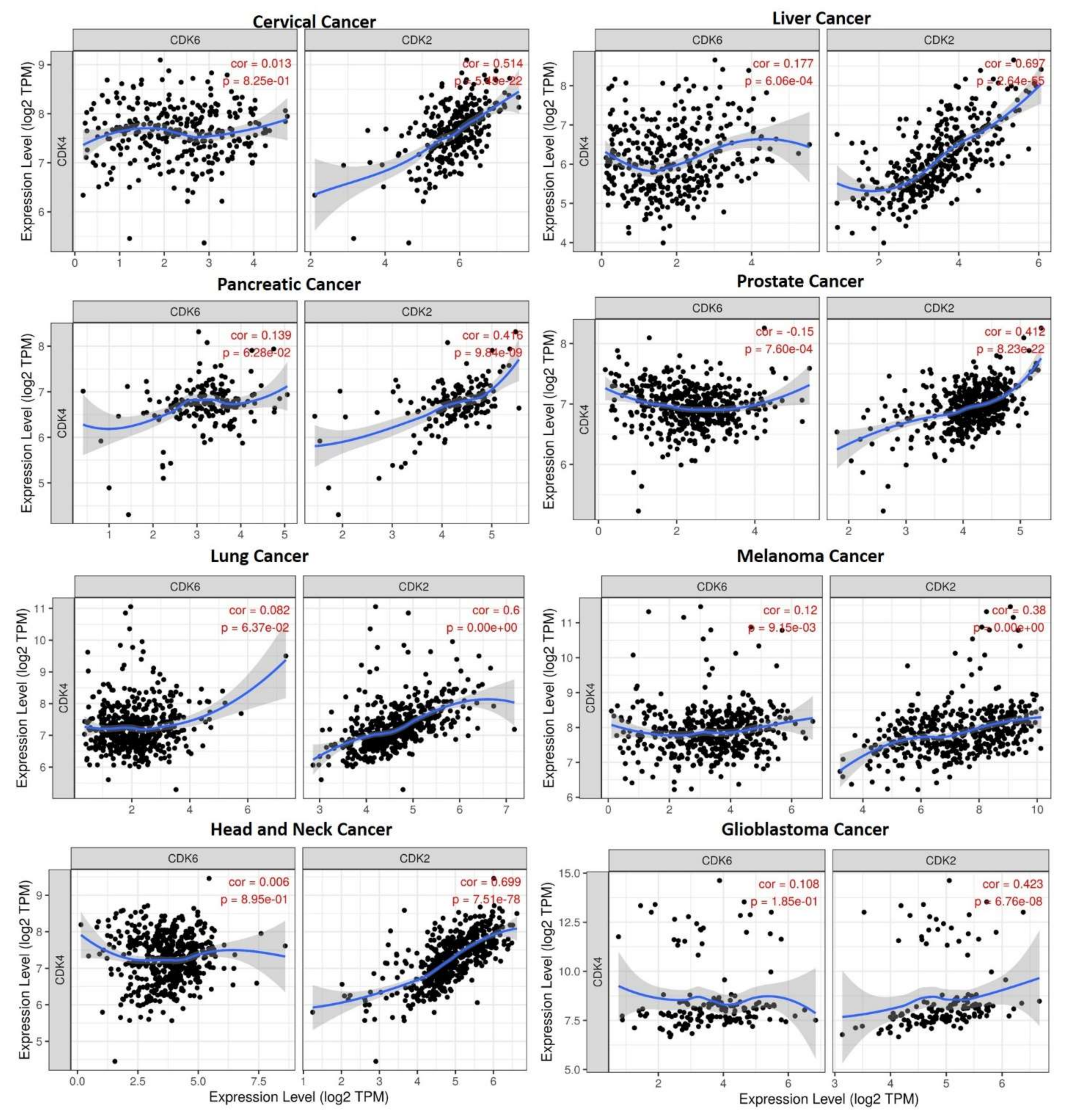
3.3. STAT3/CDK2/4/6 Expressions Are Associated with Tumor Immune Infiltrations
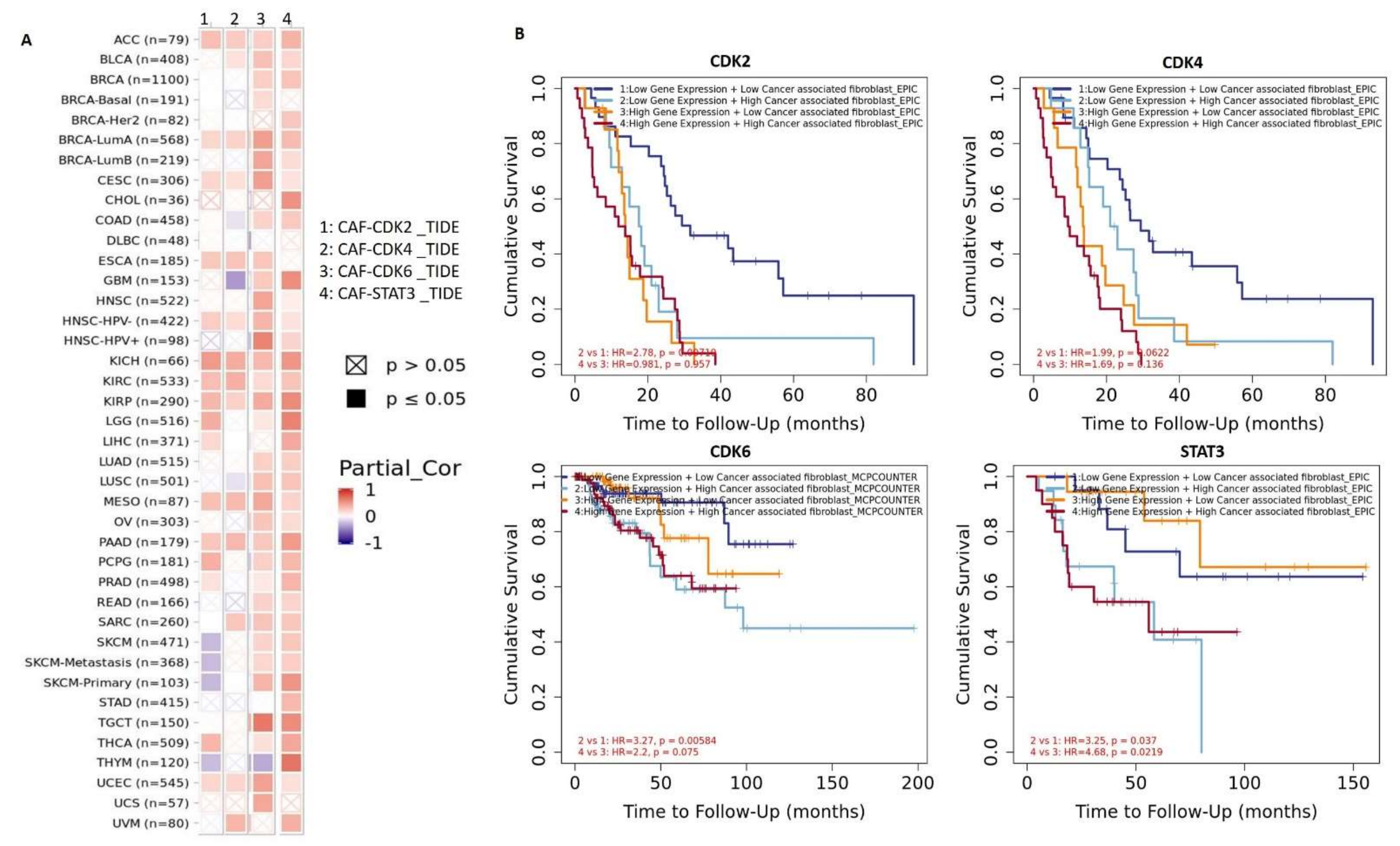
3.4. STAT3/CDK2/4/6 Are Associated with Cancer-Associated Fibroblast (CAF) Infiltration
3.5. Genetic Alterations of STAT3/CDK2/4/6 Are Associated with Poor Prognosis
3.6. Enrichment of Genes Alteration Co-Occurrence in Cancer Cohorts with STAT3/CDK2/4/6 Alterations
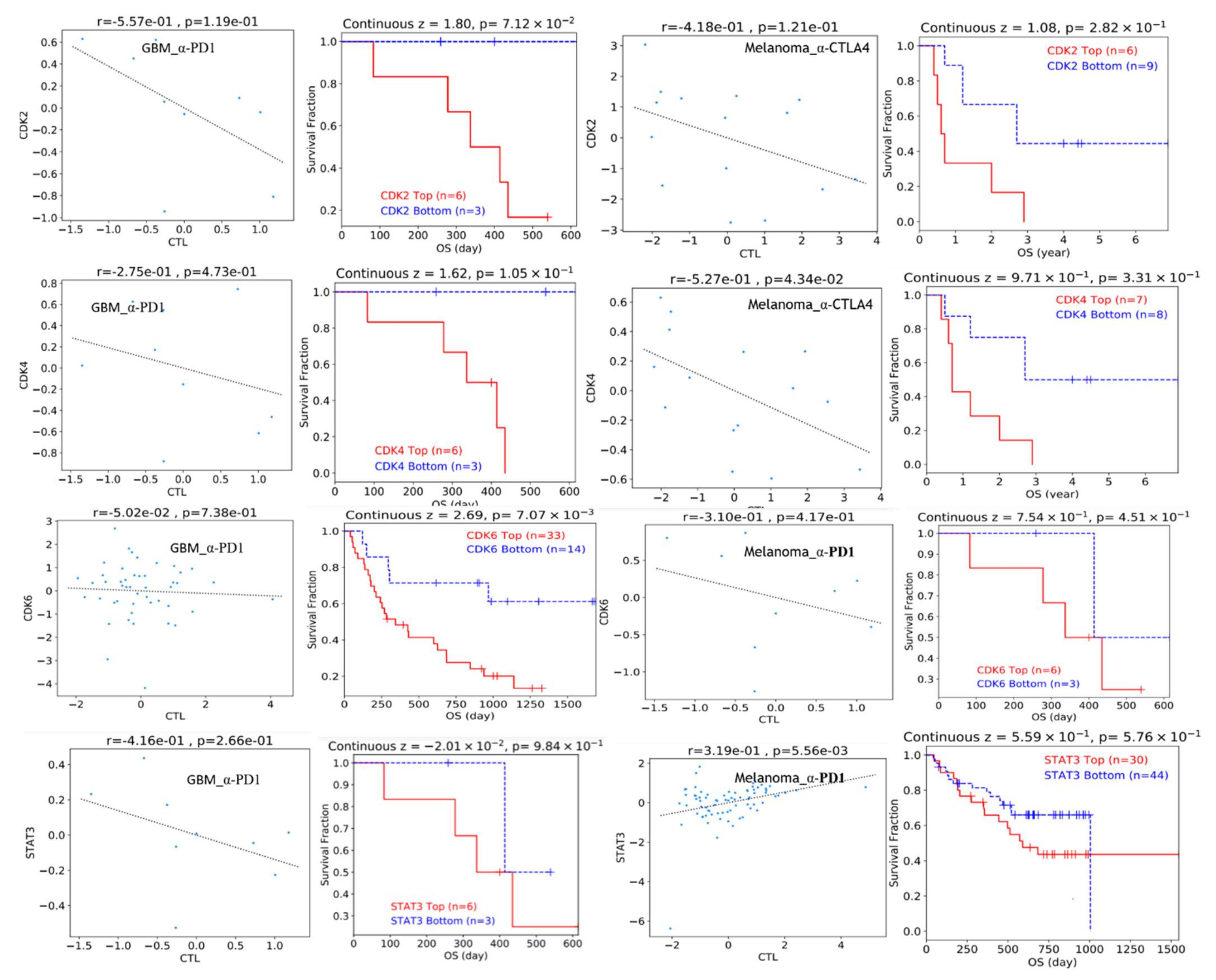
3.7. DNA Methylation and Copy Number Alterations of STAT3/CDK2/4/6 Are Associated with Dysfunctional T-Cell Phenotypes and Are of Prognostic Relevance in Multiple Cancers
3.8. STAT3/CDK2/4/6 Overexpression Predicts Poor Clinical Benefit to Immune Checkpoint Blockade Therapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mandal, M.; Sahoo, S.K.; Patra, P.; Mallik, S.; Zhao, Z. In silico ranking of phenolics for therapeutic effectiveness on cancer stem cells. BMC Bioinform. 2020, 21, 499. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; He, Y.-Y. Targeting the AMP-Activated Protein Kinase for Cancer Prevention and Therapy. Front. Oncol. 2013, 3. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Nakaoka, T.; Saito, Y.; Saito, H. Aberrant DNA methylation as a biomarker and a therapeutic target of cholangiocarcinoma. Int. J. Mol. Sci. 2017, 18, 1111. [Google Scholar] [CrossRef] [Green Version]
- Micevic, G.; Theodosakis, N.; Bosenberg, M. Aberrant DNA methylation in melanoma: Biomarker and therapeutic opportunities. Clin. Epigenet. 2017, 9, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Leygo, C.; Williams, M.; Jin, H.C.; Chan, M.W.; Chu, W.K.; Grusch, M.; Cheng, Y.Y. DNA methylation as a noninvasive epigenetic biomarker for the detection of cancer. Dis Markers 2017, 2017, 3726595. [Google Scholar] [CrossRef]
- Wu, T.; Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 2017, 387, 61–68. [Google Scholar] [CrossRef]
- Pitt, J.; Marabelle, A.; Eggermont, A.; Soria, J.-C.; Kroemer, G.; Zitvogel, L. Targeting the tumor microenvironment: Removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016, 27, 1482–1492. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.R.; Maute, R.L.; Dulken, B.W.; Hutter, G.; George, B.M.; McCracken, M.N.; Gupta, R.; Tsai, J.M.; Sinha, R.; Corey, D. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017, 545, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Lingel, H.; Brunner-Weinzierl, M.C. CTLA-4 (CD152): A versatile receptor for immune-based therapy. Semin. Immunol. 2019, 42, 101298. [Google Scholar] [CrossRef]
- Li, X.; Su, Y.; Zhang, J.; Zhu, Y.; Xu, Y.; Wu, G. LAPTM5 Plays a Key Role in the Diagnosis and Prognosis of Testicular Germ Cell Tumors. Int. J. Genom. 2021, 2021, 8816456. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, S.; Dai, W.; Xie, C.; Li, J.-C. A Comprehensive Prognostic and Immune Analysis of SLC41A3 in Pan-Cancer. Front. Oncol. 2021, 10. [Google Scholar] [CrossRef]
- Vermeulen, K.; Van Bockstaele, D.R.; Berneman, Z.N. The cell cycle: A review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003, 36, 131–149. [Google Scholar] [CrossRef] [PubMed]
- Solaki, M.; Ewald, J.C. Fueling the Cycle: CDKs in Carbon and Energy Metabolism. Front. Cell Dev. Biol 2018, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoodless, L.J.; Robb, C.T.; Felton, J.M.; Tucker, C.S.; Rossi, A.G. Models for the Study of the Cross Talk Between Inflammation and Cell Cycle. Methods Mol. Biol. 2016, 1336, 179–209. [Google Scholar] [CrossRef]
- Sundar, V.; Vimal, S.; Sai Mithlesh, M.s.; Dutta, A.; Tamizhselvi, R.; Manickam, V. Transcriptional cyclin-dependent kinases as the mediators of inflammation-a review. Gene 2021, 769, 145200. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-C.; Wu, A.T.H.; Chen, J.-H.; Huang, W.-Y.; Lawal, B.; Mokgautsi, N.; Huang, H.-S.; Ho, C.-L. HNC0014, a Multi-Targeted Small-Molecule, Inhibits Head and Neck Squamous Cell Carcinoma by Suppressing c-Met/STAT3/CD44/PD-L1 Oncoimmune Signature and Eliciting Antitumor Immune Responses. Cancers 2020, 12, 3759. [Google Scholar] [CrossRef]
- Ishibashi, K.; Koguchi, T.; Matsuoka, K.; Onagi, A.; Tanji, R.; Takinami-Honda, R.; Hoshi, S.; Onoda, M.; Kurimura, Y.; Hata, J. Interleukin-6 induces drug resistance in renal cell carcinoma. Fukushima J. Med Sci. 2018, 64, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Priego, N.; Zhu, L.; Monteiro, C.; Mulders, M.; Wasilewski, D.; Bindeman, W.; Doglio, L.; Martínez, L.; Martínez-Saez, E.; y Cajal, S.R. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis. Nat. Med. 2018, 24, 1024–1035. [Google Scholar] [CrossRef]
- De Bruin, R.A.; McDonald, W.H.; Kalashnikova, T.I.; Yates, J., III; Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 2004, 117, 887–898. [Google Scholar] [CrossRef] [Green Version]
- Goel, B.; Tripathi, N.; Bhardwaj, N.; Jain, S.K. Small Molecule CDK Inhibitors for the Therapeutic Management of Cancer. Curr. Top. Med. Chem. 2020, 20, 1535–1563. [Google Scholar] [CrossRef] [PubMed]
- Brown, V.D.; Phillips, R.A.; Gallie, B.L. Cumulative effect of phosphorylation of pRB on regulation of E2F activity. Mol. Cell. Biol. 1999, 19, 3246–3256. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Dynlacht, B.; Imai, T.; Hori, T.-a.; Harlow, E. Expression of NPAT, a novel substrate of cyclin E–CDK2, promotes S-phase entry. Genes Dev. 1998, 12, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wu, G.; Li, J.; Li, J.; Ruan, N.; Ma, L.; Han, X.; Wei, Y.; Li, L.; Zhang, H. Screening and Identification of Key Biomarkers for Bladder Cancer: A Study Based on TCGA and GEO Data. Biomed Res. Int. 2020. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Li, X.; Han, Y.; Wang, Z.; Han, C.; Ruan, N.; Li, J.; Yu, X.; Xia, Q.; Wu, G. A new prognostic risk model based on PPAR pathway-related genes in kidney renal clear cell carcinoma. PPAR Res. 2020. [Google Scholar] [CrossRef]
- Polo, A.; Crispo, A.; Cerino, P.; Falzone, L.; Candido, S.; Giudice, A.; De Petro, G.; Ciliberto, G.; Montella, M.; Budillon, A.; et al. Environment and bladder cancer: Molecular analysis by interaction networks. Oncotarget 2017, 8, 65240–65252. [Google Scholar] [CrossRef] [Green Version]
- Falzone, L.; Grimaldi, M.; Celentano, E.; Augustin, L.S.A.; Libra, M. Identification of Modulated MicroRNAs Associated with Breast Cancer, Diet, and Physical Activity. Cancers 2020, 12, 2555. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; He, T.; Liu, J.; Li, Z.; Xie, F.; Chen, C.; Xing, Y. Bioinformatics analysis of multi-omics data identifying molecular biomarker candidates and epigenetically regulatory targets associated with retinoblastoma. Medicine 2020, 99, e23314. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Shi, X.; Ding, K.; Lv, M.; Qian, Y.; Zhu, S.; Guo, C.; Zhang, Y. The Joint Analysis of Multi-Omics Data Revealed the Methylation-Expression Regulations in Atrial Fibrillation. Front. Bioeng. Biotechnol. 2020, 8. [Google Scholar] [CrossRef]
- Cazaly, E.; Saad, J.; Wang, W.; Heckman, C.; Ollikainen, M.; Tang, J. Making Sense of the Epigenome Using Data Integration Approaches. Front Pharm. 2019, 10, 126. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Jiang, P.; Gu, S.; Pan, D.; Fu, J.; Sahu, A.; Hu, X.; Li, Z.; Traugh, N.; Bu, X.; Li, B.; et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018, 24, 1550–1558. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef] [Green Version]
- Nathanson, T.; Ahuja, A.; Rubinsteyn, A.; Aksoy, B.A.; Hellmann, M.D.; Miao, D.; Van Allen, E.; Merghoub, T.; Wolchok, J.D.; Snyder, A.; et al. Somatic Mutations and Neoepitope Homology in Melanomas Treated with CTLA-4 Blockade. Cancer Immunol. Res. 2017, 5, 84–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Chen, A.X.; Gartrell, R.D.; Silverman, A.M.; Aparicio, L.; Chu, T.; Bordbar, D.; Shan, D.; Samanamud, J.; Mahajan, A.; et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 2019, 25, 462–469. [Google Scholar] [CrossRef]
- Ge, P.; Wang, W.; Li, L.; Zhang, G.; Gao, Z.; Tang, Z.; Dang, X.; Wu, Y. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of colorectal cancer. Biomed. Pharmacother. 2019, 118, 109228. [Google Scholar] [CrossRef] [PubMed]
- Rathnagiriswaran, S.; Wan, Y.-W.; Abraham, J.; Castranova, V.; Qian, Y.; Guo, N.L. A population-based gene signature is predictive of breast cancer survival and chemoresponse. Int. J. Oncol. 2010, 36, 607–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haslem, D.S.; Chakravarty, I.; Fulde, G.; Gilbert, H.; Tudor, B.P.; Lin, K.; Ford, J.M.; Nadauld, L.D. Precision oncology in advanced cancer patients improves overall survival with lower weekly healthcare costs. Oncotarget 2018, 9, 12316–12322. [Google Scholar] [CrossRef] [Green Version]
- Bharadwaj, M.; Vallurupalli, M.; Huang, F.W. Global Precision Oncology: A Call to Action on Expanding Access to Targeted Cancer Therapies. Oncologist 2021. [Google Scholar] [CrossRef]
- Falzone, L.; Salomone, S.; Libra, M. Evolution of Cancer Pharmacological Treatments at the Turn of the Third Millennium. Front Pharm. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, J.H.; Olver, I. Precision medicine-based drug treatment individualization in oncology. Br. J. Clin. Pharmacol. 2021, 87, 223–226. [Google Scholar] [CrossRef]
- Lawal, B.; Liu, Y.-L.; Mokgautsi, N.; Khedkar, H.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.-S. Pharmacoinformatics and Preclinical Studies of NSC765690 and NSC765599, Potential STAT3/CDK2/4/6 Inhibitors with Antitumor Activities against NCI60 Human Tumor Cell Lines. Biomedicines 2021, 9, 92. [Google Scholar] [CrossRef]
- Laraia, L.; McKenzie, G.; Spring, D.R.; Venkitaraman, A.R.; Huggins, D.J. Overcoming Chemical, Biological, and Computational Challenges in the Development of Inhibitors Targeting Protein-Protein Interactions. Chem. Biol. 2015, 22, 689–703. [Google Scholar] [CrossRef] [Green Version]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Semple, J.W.; Duncker, B.P. ORC-associated replication factors as biomarkers for cancer. Biotechnol. Adv. 2004, 22, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Niu, G.; Kortylewski, M.; Burdelya, L.; Shain, K.; Zhang, S.; Bhattacharya, R.; Gabrilovich, D.; Heller, R.; Coppola, D. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004, 10, 48–54. [Google Scholar] [CrossRef]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br J Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, N.; Yamazaki-Itoh, R.; Ino, Y.; Mizuguchi, Y.; Yamada, T.; Hirohashi, S.; Kanai, Y. CXCL17 and ICAM2 Are Associated With a Potential Anti-Tumor Immune Response in Early Intraepithelial Stages of Human Pancreatic Carcinogenesis. Gastroenterology 2011, 140, 310–321.e314. [Google Scholar] [CrossRef]
- Kortylewski, M.; Kujawski, M.; Wang, T.; Wei, S.; Zhang, S.; Pilon-Thomas, S.; Niu, G.; Kay, H.; Mulé, J.; Kerr, W.G. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005, 11, 1314–1321. [Google Scholar] [CrossRef]
- Villarino, A.V.; Kanno, Y.; O’Shea, J.J. Mechanisms and consequences of Jak–STAT signaling in the immune system. Nat. Immunol. 2017, 18, 374. [Google Scholar] [CrossRef]
- Spranger, S.; Gajewski, T.F. Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology 2016, 5, e1086862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaderbhaï, C.; Tharin, Z.; Ghiringhelli, F. The Role of Molecular Profiling to Predict the Response to Immune Checkpoint Inhibitors in Lung Cancer. Cancers 2019, 11, 201. [Google Scholar] [CrossRef] [Green Version]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- An, X.; Zhu, Y.; Zheng, T.; Wang, G.; Zhang, M.; Li, J.; Ji, H.; Li, S.; Yang, S.; Xu, D.; et al. An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol. Ther. Nucleic Acids 2019, 14, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Caccese, M.; Indraccolo, S.; Zagonel, V.; Lombardi, G. PD-1/PD-L1 immune-checkpoint inhibitors in glioblastoma: A concise review. Crit. Rev. Oncol. /Hematol. 2019, 135, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Tien, A.-C.; Li, J.; Bao, X.; Derogatis, A.; Kim, S.; Mehta, S.; Sanai, N. A Phase 0 Trial of Ribociclib in Recurrent Glioblastoma Patients Incorporating a Tumor Pharmacodynamic- and Pharmacokinetic-Guided Expansion Cohort. Clin. Cancer Res. 2019, 25, 5777–5786. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.W.; Parikh, M.; Phillips, J.J.; James, C.D.; Molinaro, A.M.; Butowski, N.A.; Clarke, J.L.; Oberheim-Bush, N.A.; Chang, S.M.; Berger, M.S.; et al. Phase-2 trial of palbociclib in adult patients with recurrent RB1-positive glioblastoma. J. Neurooncol. 2018, 140, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bu, X.; Wang, H.; Zhu, Y.; Geng, Y.; Nihira, N.T.; Tan, Y.; Ci, Y.; Wu, F.; Dai, X.; et al. Cyclin D–CDK4 kinase destabilizes PD-L1 via cullin 3–SPOP to control cancer immune surveillance. Nature 2018, 553, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Anurag, M.; Haricharan, S.; Ellis, M.J. CDK4/6 Inhibitor Biomarker Research: Are We Barking Up the Wrong Tree? Clin. Cancer Res. 2020, 26, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Pearce, O.M.; Delaine-Smith, R.M.; Maniati, E.; Nichols, S.; Wang, J.; Böhm, S.; Rajeeve, V.; Ullah, D.; Chakravarty, P.; Jones, R.R. Deconstruction of a metastatic tumor microenvironment reveals a common matrix response in human cancers. Cancer Discov. 2018, 8, 304–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell 2018, 174, 1293–1308.e1236. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Tong, Q.; Liu, B.; Huang, W.; Tian, Y.; Fu, X. Targeting STAT3 in Cancer Immunotherapy. Mol. Cancer 2020, 19, 145. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [Green Version]
- Harryvan, T.J.; Verdegaal, E.M.E.; Hardwick, J.C.H.; Hawinkels, L.J.A.C.; van der Burg, S.H. Targeting of the Cancer-Associated Fibroblast—T-Cell Axis in Solid Malignancies. J. Clin. Med. 2019, 8, 1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannoni, E.; Bianchini, F.; Masieri, L.; Serni, S.; Torre, E.; Calorini, L.; Chiarugi, P. Reciprocal Activation of Prostate Cancer Cells and Cancer-Associated Fibroblasts Stimulates Epithelial-Mesenchymal Transition and Cancer Stemness. Cancer Res. 2010, 70, 6945–6956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erez, N.; Truitt, M.; Olson, P.; Arron, S.T.; Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-kappaB-Dependent Manner. Cancer Cell 2010, 17, 135–147. [Google Scholar] [CrossRef] [Green Version]
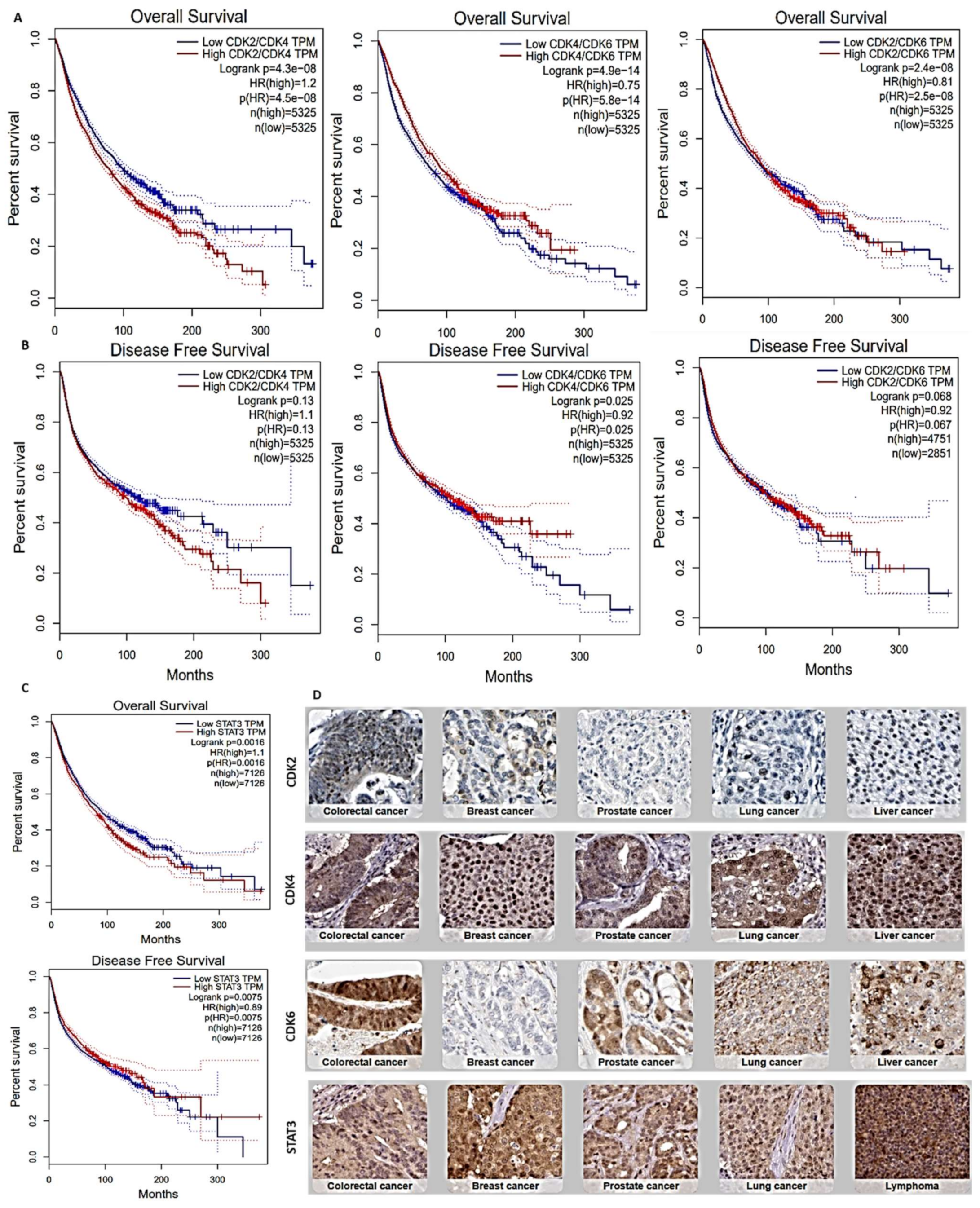




| Cancer Types | Sample in HPA | Patient Age | Patient Gender | Tumor-Histology | ||
|---|---|---|---|---|---|---|
| Total Sample | High Antibody Detected | Mean Age | Male n (%) | Female n (%) | Patient Tumor-Histology (%) | |
| CDK2 (Antibody: CAB013115) | ||||||
| Breast | 11 | 6 (54.54%) | 54.16 | - | 6 (100%) | DCN (33.33%) and LCN (66.66%) |
| Head and Neck | 4 | 4 (100 %) | 56.75 | 2 (50.00%) | 2 (50.00%) | HN-SCC (50.00%) and HN-ADC (50.00%) |
| Glioma | 11 | 7 (63.63%) | 58.14 | 5 (83.33%) | 3 (16.66%) | HGG (57.14%), LGG (42.85%) |
| Colorectal | 10 | 10 (100%) | 69.90 | 6 (60.00%) | 4 (40.00%) | C-ADC (70.00%), R-ADC (20.00%) |
| Prostate | 10 | 5 (50.00%) | 59.44 | 5 (100%) | - | HG_PA (60.00%) and LG_PA (40.00%) |
| Lung | 10 | 6 (60.00%) | 58.83 | 3 (50.00%) | 3 (50.00%) | L-SSC (66.66%) and L-AND (33.33%) |
| Liver | 11 | 4 (36.36%) | 63.50 | - | 4 (100.00%) | CCN (25.00%) and HCN (75.00%) |
| Pancreatic | 12 | 6 (50.00%) | 64.16 | 3 (50.00%) | 3 (50.00%) | PAC (100.00%) |
| CDK4 (Antibody: CAB013116) | ||||||
| Breast | 11 | 9 (81.81%) | 64.00 | - | 9 (100%) | DCN (55.55%) and LCN (44.44%) |
| Head and Neck | 4 | 4 (100%) | 71.5 | 3 (75.0%) | 1 (25.00%) | HN-SCC (75.00%) and HN-ADC (35.00%) |
| Glioma | 11 | 9 (81.81%) | 48.11 | 5 55.55%) | 4 (44.44%) | HGG (55.55%) and LGG (44.44%) |
| Colorectal | 12 | 12 (100%) | 79.50 | 6 (50.00%) | 6 (50.00%) | C-ADC (66.66%) and R-ADC (33.33%) |
| Prostate | 11 | 10 (90.90%) | 58.80 | 10 (100%) | - | HG_PA (70.00%) and LG_PA (30.00%) |
| Lung | 12 | 12 (100%) | 67.58 | 7 (58.33%) | 5 (41.66%) | L-SSC (58.33%) and L-AND (41.66%) |
| Liver | 12 | 8 (66.66%) | 63.25 | 5 62.55%) | 3 (37.5%) | CCN (25.00%) and 6 HCN (75.00%) |
| Pancreatic | 11 | 7 (63.63%) | 63.71 | 4 (57.15%) | 3 (42.85%) | PAC (100.00%) |
| CDK6 (Antibody: HPA002637) | ||||||
| Breast | 12 | 3 (25.00%) | 54.00 | - | 3 (100%) | DCN (75.00%) and LCN (25.00%) |
| Head and Neck | 4 | 4 (100.00%) | 58.25 | 1 (25%) | 3 (75%) | HN-SCC (50.00%) and HN-ADC (50.00%) |
| Glioma | 12 | 11 (91.66%) | 44.58 | 3 (100%) | - | HGG (63.63%) and LGG (36.36%) |
| Colorectal | 10 | 9 (90.00%) | 61.60 | 6 (66.6%) | 3 (33.33%) | C-ADC (66.66%) and R-ADC (6.33%) |
| Prostate | 10 | 2 (20.00%) | 66.00 | 2 (100%) | - | HG_PA (50.00%) and LG_PA (50.00%) |
| Lung | 11 | 4 (36.36%) | 61.00 | 2 (50%) | 2 (50%) | L-SSC (75.00%) and L-AND (25.00%) |
| Liver | 12 | 9 (75.00%) | 62.77 | 5 (55.55%) | 4 (44.5%) | CCN (66.66%) and HCN (33.33%) |
| Pancreatic | 11 | 7 (63.63%) | 62.00 | 3 (42.85%) | 4 (57.1%) | PAC (100.00%) |
| STAT3 (Antibody: HPA001671) | ||||||
| Breast | 11 | 11 (100%) | 63.18 | - | 11 (100%) | DCN (72.72%) and LCN (27.27%) |
| Head and Neck | 4 | 4 (100%) | 70.50 | 3 (75.00%) | 1 (25.00%) | HN-SCC (75.00%) and HN-ADC (25.00%) |
| Glioma | 12 | 5 (41.66%) | 45.60 | 2 (40.00%) | 3 (60.00%) | HGG (80.00%) and LGG (20.00%) |
| Colorectal | 12 | 12 (100%) | 64.83 | 4 (33.33%) | 8 (66.66%) | C-ADC (75.00%) and R-ADC (25.00%) |
| Prostate | 10 | 9 (90.00%) | 67.44 | 10 (100%) | - | HG_PA (88.88%) and LG_PA (11.11%) |
| Lung | 12 | 6 (50.00%) | 69.50 | 4 (66.66%) | 2 (33.33%) | L-SSC (50.00%) and L-AND (50.00%) |
| Liver | 11 | 4 (36.36%) | 57.75 | 2 (50.00%) | 2 (50.00%) | CCN (5000%) and 6 HCN (50.00%) |
| Pancreatic | 9 | 6 (66.66%) | 63.71 | 3 (50.00%) | 3 (50.00%) | PAC (100.00%) |
| Cancer Types | Variable | CDK2 | CDK4 | CDK6 | STAT3 | ||||
|---|---|---|---|---|---|---|---|---|---|
| rho-Value | p-Value | rho-Value | p-Value | rho-Value | p-Value | rho-Value | p-Value | ||
| BRCA | Purity | 0.173772 | 3.46 × 10−8 | 0.093476 | 0.003164 | −0.31833 | 7.21 × 10−25 | −0.10635 | 0.000779 |
| B Cell | 0.122448 | 0.000125 | 0.08921 | 0.005264 | 0.240495 | 2.54 × 10−14 | 0.066812 | 0.036797 | |
| CD8+ T Cell | 0.192216 | 1.41 × 10−9 | 0.032843 | 0.305355 | 0.391337 | 4.53 × 10−37 | 0.239179 | 3.65 × 10−14 | |
| CD4+ T Cell | 0.140028 | 1.31 × 10−5 | 0.042232 | 0.190619 | 0.320767 | 1.85 × 10−24 | 0.233229 | 2.38 × 10−13 | |
| Macrophage | 0.079207 | 0.012988 | −0.01619 | 0.612245 | 0.257793 | 2.19 × 10−16 | 0.25456 | 5.28 × 10−16 | |
| Neutrophil | 0.233463 | 3.07 × 10−13 | 0.096267 | 0.002962 | 0.391841 | 2.93 × 10−36 | 0.313132 | 4.44 × 10−23 | |
| Dendritic Cell | 0.169013 | 1.58 × 10−7 | 0.113947 | 0.00043 | 0.384255 | 8.01 × 10−35 | 0.202448 | 2.97 × 10−10 | |
| GBM | Purity | 0.286993 | 2.19 × 10−9 | 0.430653 | 2.39 × 10−20 | 0.192757 | 7.15 × 10−5 | −0.16588 | 0.000652 |
| B Cell | −0.05908 | 0.228103 | 0.01657 | 0.73552 | 0.02255 | 0.645725 | −0.00829 | 0.865847 | |
| CD8+ T Cell | −0.03756 | 0.443695 | −0.04642 | 0.343759 | 0.138459 | 0.004568 | −0.14558 | 0.002851 | |
| CD4+ T Cell | −0.05077 | 0.300396 | −0.02841 | 0.562404 | −0.0257 | 0.600353 | 0.277782 | 7.64 × 10−9 | |
| Macrophage | −0.01146 | 0.815223 | −0.02026 | 0.679627 | −0.03727 | 0.447329 | 0.056771 | 0.246809 | |
| Neutrophil | 0.089753 | 0.006772 | 0.052908 | 0.28049 | −0.16547 | 0.000683 | 0.175587 | 0.00031 | |
| Dendritic Cell | 0.200704 | 3.58 × 10−5 | 0.023584 | 0.630667 | −0.12206 | 0.012513 | 0.43801 | 5.06 × 10−21 | |
| HNSC | Purity | 0.229737 | 2.51 × 10−7 | 0.304761 | 4.69 × 10−12 | −0.06379 | 0.157308 | −0.01693 | 0.707708 |
| B Cell | 0.110468 | 0.015902 | 0.124189 | 0.00667 | −0.21932 | 1.36 × 10−6 | 0.230367 | 3.75 × 10−7 | |
| CD8+ T Cell | 0.092891 | 0.043236 | 0.07504 | 0.102739 | −0.25071 | 3.16 × 10−8 | 0.235486 | 2.14 × 10−7 | |
| CD4+ T Cell | 0.299532 | 2.09 × 10−11 | 0.140472 | 0.002036 | 0.189753 | 2.86 × 10−5 | 0.456441 | 4.47 × 10−26 | |
| Macrophage | 0.146554 | 0.001238 | 0.190636 | 2.47 × 10−5 | −0.013 | 0.775631 | 0.21852 | 1.24 × 10−6 | |
| Neutrophil | 0.215968 | 1.84 × 10−6 | 0.021853 | 0.6333 | 0.082199 | 0.072281 | 0.346725 | 5.62 × 10−15 | |
| Dendritic Cell | 0.253224 | 1.73 × 10−8 | 0.103755 | 0.00272 | 0.040238 | 0.378066 | 0.387553 | 1.01 × 10−18 | |
| LIHC | Purity | 0.181946 | 0.000672 | 0.069596 | 0.196552 | −0.11342 | 0.034946 | −0.23257 | 1.24 × 10−5 |
| B Cell | 0.397861 | 1.70 × 10−14 | 0.446746 | 2.80 × 10−18 | 0.077473 | 0.151618 | 0.167119 | 0.001869 | |
| CD8+ T Cell | 0.300309 | 1.47 × 10−8 | 0.327963 | 5.11 × 10−10 | 0.024258 | 0.654848 | 0.128993 | 0.016998 | |
| CD4+ T Cell | 0.423424 | 2.13 × 10−16 | 0.379031 | 3.39 × 10−13 | 0.062486 | 0.247738 | 0.348425 | 2.97 × 10−11 | |
| Macrophage | 0.476735 | 9.42 × 10−21 | 0.51059 | 4.90 × 10−24 | 0.097956 | 0.070829 | 0.359076 | 8.16 × 10−12 | |
| Neutrophil | 0.477554 | 4.69 × 10−21 | 0.368888 | 1.46 × 10−12 | 0.076032 | 0.158794 | 0.448825 | 1.67 × 10−18 | |
| Dendritic Cell | 0.480477 | 4.86 × 10−21 | 0.482455 | 3.18 × 10−21 | 0.052521 | 0.334277 | 0.285271 | 8.68 × 10−08 | |
| LUAD | Purity | 0.06579 | 0.144252 | 0.060096 | 0.182364 | −0.16182 | 0.000304 | 0.007492 | 0.868083 |
| B Cell | −0.04115 | 0.366308 | −0.10408 | 0.022022 | −0.03669 | 0.420564 | 0.119856 | 0.008302 | |
| CD8+ T Cell | 0.146119 | 0.001222 | 0.006762 | 0.881683 | 0.275158 | 6.57 × 10−10 | 0.128065 | 0.004647 | |
| CD4+ T Cell | 0.071922 | 0.001145 | −0.09706 | 0.032783 | 0.146028 | 0.001275 | 0.167818 | 0.000208 | |
| Macrophage | 0.03284 | 0.470574 | −0.01899 | 0.67652 | 0.207622 | 4.01 × 10−6 | 0.158864 | 0.000445 | |
| Neutrophil | 0.27285 | 1.08 × 10−9 | 0.078989 | 0.082887 | 0.345911 | 5.06 × 10−15 | 0.219087 | 1.16 × 10−6 | |
| Dendritic Cell | 0.134315 | 0.002949 | 0.021403 | 0.637169 | 0.208912 | 3.25 × 10−6 | 0.179555 | 6.64 × 10−5 | |
| SKCM | Purity | 0.134716 | 0.003873 | 0.33632 | 1.42 × 10−13 | 0.208989 | 6.48 × 10−6 | −0.09559 | 0.040865 |
| B Cell | −0.04378 | 0.355269 | 0.035404 | 0.454766 | 0.092088 | 0.051436 | 0.190704 | 4.85 × 10−5 | |
| CD8+ T Cell | −0.025 | 0.601752 | −0.09342 | 0.050716 | 0.273715 | 5.76 × 10−9 | 0.325874 | 2.70 × 10−12 | |
| CD4+ T Cell | −0.14917 | 0.001582 | −0.05216 | 0.271664 | 0.151926 | 0.00129 | 0.276917 | 2.71 × 10−9 | |
| Macrophage | −0.28478 | 6.72 × 10−10 | −0.06167 | 0.190155 | 0.252014 | 5.42 × 10−8 | 0.32638 | 1.05 × 10−12 | |
| Neutrophil | −0.18426 | 8.13 × 10−5 | −0.0556 | 0.238103 | 0.455928 | 1.39 × 10−24 | 0.500159 | 5.51 × 10−30 | |
| Dendritic Cell | −0.08306 | 0.079749 | −0.00475 | 0.920327 | 0.188403 | 6.24 × 10−5 | 0.373995 | 2.97 × 10−16 | |
| S/N | Genes ID | Cytoband | Altered Group | Unaltered Group | Log Ratio | p-Value | q-Value | Enriched in |
|---|---|---|---|---|---|---|---|---|
| Cyclin Dependent Kinase 2 | ||||||||
| 1 | FGD5 | 3p25.1 | 19 (15.32%) | 226 (2.19%) | 2.81 | 6.99 × 10−11 | 7.60 × 10−7 | Altered group |
| 2 | MMS22L | 6q16.1 | 17 (13.71%) | 178 (1.73%) | 2.99 | 1.23 × 10−10 | 7.60 × 10−7 | Altered group |
| 3 | LRP5 | 11q13.2 | 19 (15.32%) | 237 (2.30%) | 2.74 | 1.49 × 10−10 | 7.60 × 10−7 | Altered group |
| 4 | PALM2-AKAP2 | 9q31.3 | 17 (13.71%) | 181 (1.76%) | 2.97 | 1.56 × 10−10 | 7.60 × 10−7 | Altered group |
| 5 | GTF3C2 | 2p23.3 | 15 (12.10%) | 135 (1.31%) | 3.21 | 2.37 × 10−10 | 7.67 × 10−7 | Altered group |
| 6 | PAX8 | 2q14.1 | 12 (9.68%) | 72 (0.70%) | 3.79 | 2.53 × 10−10 | 7.67 × 10−7 | Altered group |
| 7 | MFHAS1 | 8p23.1 | 13 (10.48%) | 93 (0.90%) | 3.54 | 3.11 × 10−10 | 7.67 × 10−7 | Altered group |
| 8 | SENP5 | 3q29 | 13 (10.48%) | 94 (0.91%) | 3.52 | 3.51 × 10−10 | 7.67 × 10−7 | Altered group |
| 9 | PRDM9 | 5p14.2 | 25 (20.16%) | 461 (4.47%) | 2.17 | 3.86 × 10−10 | 7.67 × 10−7 | Altered group |
| 10 | LRRFIP2 | 3p22.2 | 13 (10.48%) | 95 (0.92%) | 3.51 | 3.95 × 10−10 | 7.67 × 10−7 | Altered group |
| Cyclin Dependent Kinase 4 | ||||||||
| 1 | EGFR | 7p11.2 | 39 (13.68%) | 357 (3.52%) | 1.96 | 2.25 × 10−12 | 4.38 × 10−8 | Altered group |
| 2 | ZNF19 | 16q22.2 | 18 (6.32%) | 73 (0.72%) | 3.13 | 3.28 × 10−11 | 3.19 × 10−7 | Altered group |
| 3 | NUP107 | 12q15 | 19 (6.67%) | 128 (1.26%) | 2.4 | 1.69 × 10−8 | 5.45 × 10−5 | Altered group |
| 4 | KLHL9 | 9p21.3 | 15 (5.26%) | 75 (0.74%) | 2.83 | 1.76× 10−8 | 5.45 × 10−5 | Altered group |
| 5 | ATP13A5 | 3q29 | 25 (8.77%) | 227 (2.24%) | 1.97 | 2.23× 10−8 | 5.45 × 10−5 | Altered group |
| 6 | TENM2 | 5q34 | 35 (12.28%) | 418 (4.12%) | 1.58 | 2.27× 10−8 | 5.45 × 10−5 | Altered group |
| 7 | MYPN | 10q21.3 | 26 (9.12%) | 247 (2.43%) | 1.91 | 2.68× 10−8 | 5.45 × 10−5 | Altered group |
| 8 | B4GALNT1 | 12q13.3 | 15 (5.26%) | 78 (0.77%) | 2.78 | 2.79× 10−08 | 5.45 × 10−05 | Altered group |
| 9 | PTPRH | 19q13.42 | 24 (8.42%) | 214 (2.11%) | 2 | 3.15× 10−08 | 5.45 × 10−05 | Altered group |
| 10 | NEMF | 14q21.3 | 18 (6.32%) | 120 (1.18%) | 2.42 | 3.53× 10−08 | 5.45 × 10−05 | Altered group |
| Cyclin Dependent Kinase 6 | ||||||||
| 1 | TP53 | 17p13.1 | 159 (62.6%) | 3680 (36.14%) | 0.79 | 2.75 × 10−17 | 5.35 × 10−13 | Altered group |
| 2 | CFAP47 | Xp21.1 | 43 (16.93%) | 399 (3.92%) | 2.11 | 3.41 × 10−15 | 3.31 × 10−11 | Altered group |
| 3 | CUBN | 10p13 | 51 (20.08%) | 625 (6.14%) | 1.71 | 2.03 × 10−13 | 1.31 × 10−9 | Altered group |
| 4 | KBTBD7 | 13q14.11 | 20 (7.87%) | 81 (0.80%) | 3.31 | 2.90 × 10−13 | 1.41 × 10−9 | Altered group |
| 5 | EYS | 6q12 | 39 (15.35%) | 385 (3.78%) | 2.02 | 4.73 × 10−13 | 1.84 × 10−9 | Altered group |
| 6 | FAT3 | 11q14.3 | 59 (23.23%) | 839 (8.24%) | 1.5 | 7.48 × 10−13 | 2.23 × 10−9 | Altered group |
| 7 | SPTBN4 | 19q13.2 | 34 (13.39%) | 298 (2.93%) | 2.19 | 8.39 × 10−13 | 2.23 × 10−9 | Altered group |
| 8 | TCERG1L | 10q26.3 | 21 (8.27%) | 99 (0.97%) | 3.09 | 9.29 × 10−13 | 2.23 × 10−9 | Altered group |
| 9 | ATP2B1 | 12q21.33 | 25 (9.84%) | 153 (1.50%) | 2.71 | 1.03 × 10−12 | 2.23 × 10−9 | Altered group |
| 10 | UBA6 | 4q13.2 | 24 (9.45%) | 141 (1.38%) | 2.77 | 1.38 × 10−12 | 2.68 × 10−9 | Altered group |
| Signal Transducer and Activator of Transcription 3 | ||||||||
| 1 | NEURL4 | 17p13.1 | 39 (17.89%) | 177 (1.73%) | 3.37 | 5.90 × 10−26 | 1.15 × 10−21 | Altered group |
| 2 | ARHGAP5 | 14q12 | 38 (17.43%) | 202 (1.98%) | 3.14 | 3.78 × 10−23 | 3.67 × 10−19 | Altered group |
| 3 | DSG1 | 18q12.1 | 38 (17.43%) | 208 (2.04%) | 3.1 | 9.42 × 10−23 | 6.10 × 10−19 | Altered group |
| 4 | PCDHGB6 | 5q31.3 | 34 (15.60%) | 157 (1.54%) | 3.34 | 1.81 × 10−22 | 8.80 × 10−19 | Altered group |
| 5 | CEP350 | 1q25.2 | 44 (20.18%) | 318 (3.11%) | 2.7 | 5.68 × 10−22 | 2.21 × 10−18 | Altered group |
| 6 | MED13 | 17q23.2 | 40 (18.35%) | 255 (2.50%) | 2.88 | 9.18 × 10−22 | 2.97 × 10−18 | Altered group |
| 7 | HMCN1 | 1q25.3 | 67 (30.73%) | 846 (8.28%) | 1.89 | 5.92 × 10−21 | 1.64 × 10−17 | Altered group |
| 8 | DOCK8 | 9p24.3 | 39 (17.89%) | 255 (2.50%) | 2.84 | 7.08 × 10−21 | 1.72 × 10−17 | Altered group |
| 9 | DNMBP | 10q24.2 | 34 (15.60%) | 180 (1.76%) | 3.15 | 8.24 × 10−21 | 1.78 × 10−17 | Altered group |
| 10 | MAP1B | 5q13.2 | 40 (18.35%) | 277 (2.71%) | 2.76 | 1.37 × 10−20 | 2.65 × 10−17 | Altered group |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lawal, B.; Lin, L.-C.; Lee, J.-C.; Chen, J.-H.; Bekaii-Saab, T.S.; Wu, A.T.H.; Ho, C.-L. Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies. Cancers 2021, 13, 954. https://doi.org/10.3390/cancers13050954
Lawal B, Lin L-C, Lee J-C, Chen J-H, Bekaii-Saab TS, Wu ATH, Ho C-L. Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies. Cancers. 2021; 13(5):954. https://doi.org/10.3390/cancers13050954
Chicago/Turabian StyleLawal, Bashir, Li-Ching Lin, Jih-Chin Lee, Jia-Hong Chen, Tanios S. Bekaii-Saab, Alexander T. H. Wu, and Ching-Liang Ho. 2021. "Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies" Cancers 13, no. 5: 954. https://doi.org/10.3390/cancers13050954
APA StyleLawal, B., Lin, L.-C., Lee, J.-C., Chen, J.-H., Bekaii-Saab, T. S., Wu, A. T. H., & Ho, C.-L. (2021). Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies. Cancers, 13(5), 954. https://doi.org/10.3390/cancers13050954







