Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
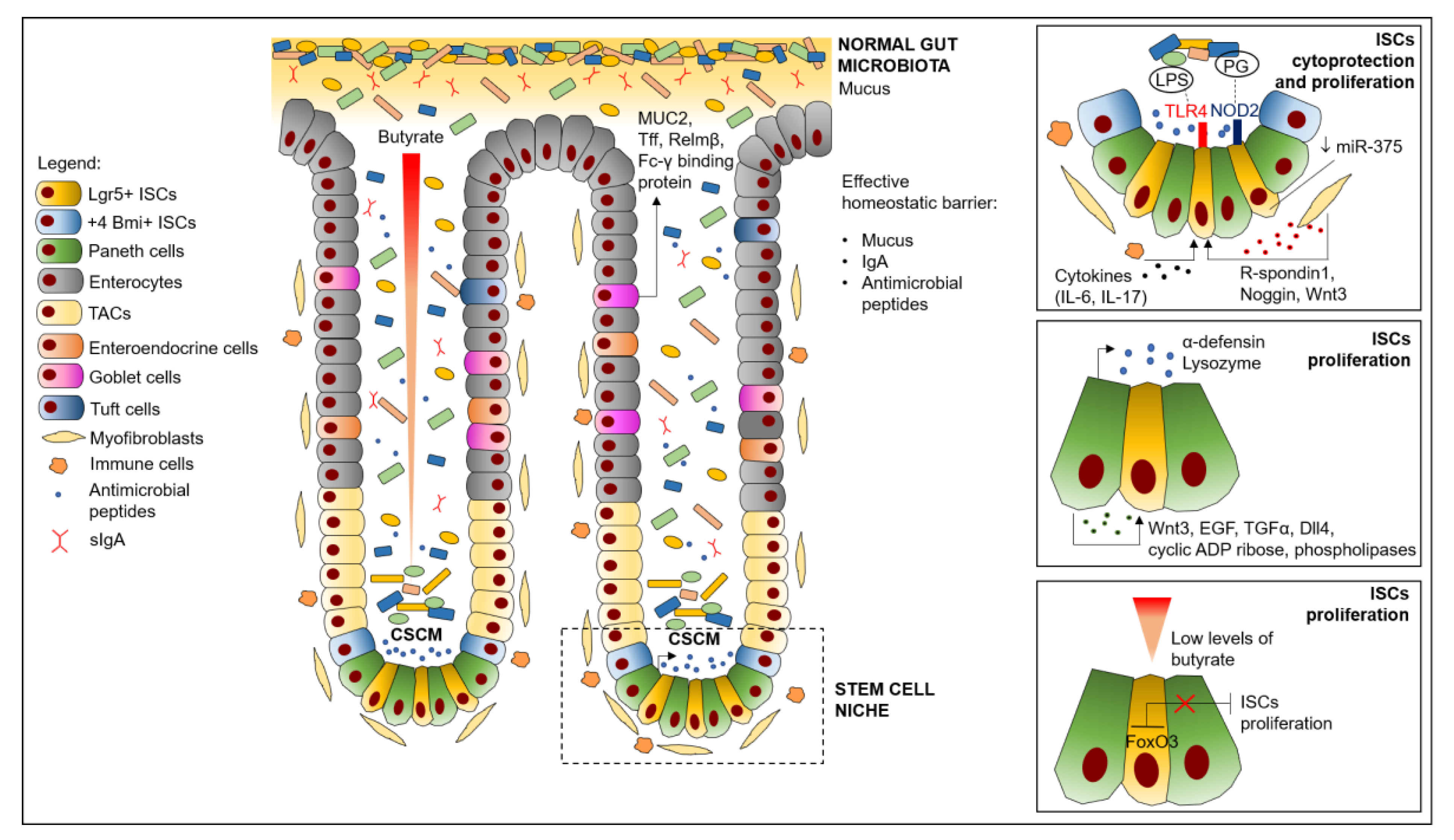
2. ISC Niche Structure and Functional Organization
3. Dysfunction of ISC Niche in CRC
4. Effects of Intestinal Microbiota on ISCs Homeostasis
4.1. ISCs Regulation Mediated by Microbiota Engagement of PRRs
4.2. ISCs Regulation Mediated by Microbiota Production of Reactive Oxygen Species (ROS)
4.3. ISCs Regulation Mediated by Microbiota-Derived Metabolites
4.4. ISCs Regulation due to Microbiota Effects on Paneth Cells
4.5. ISCs Regulation Mediated by Microbiota-Induced Production of microRNA
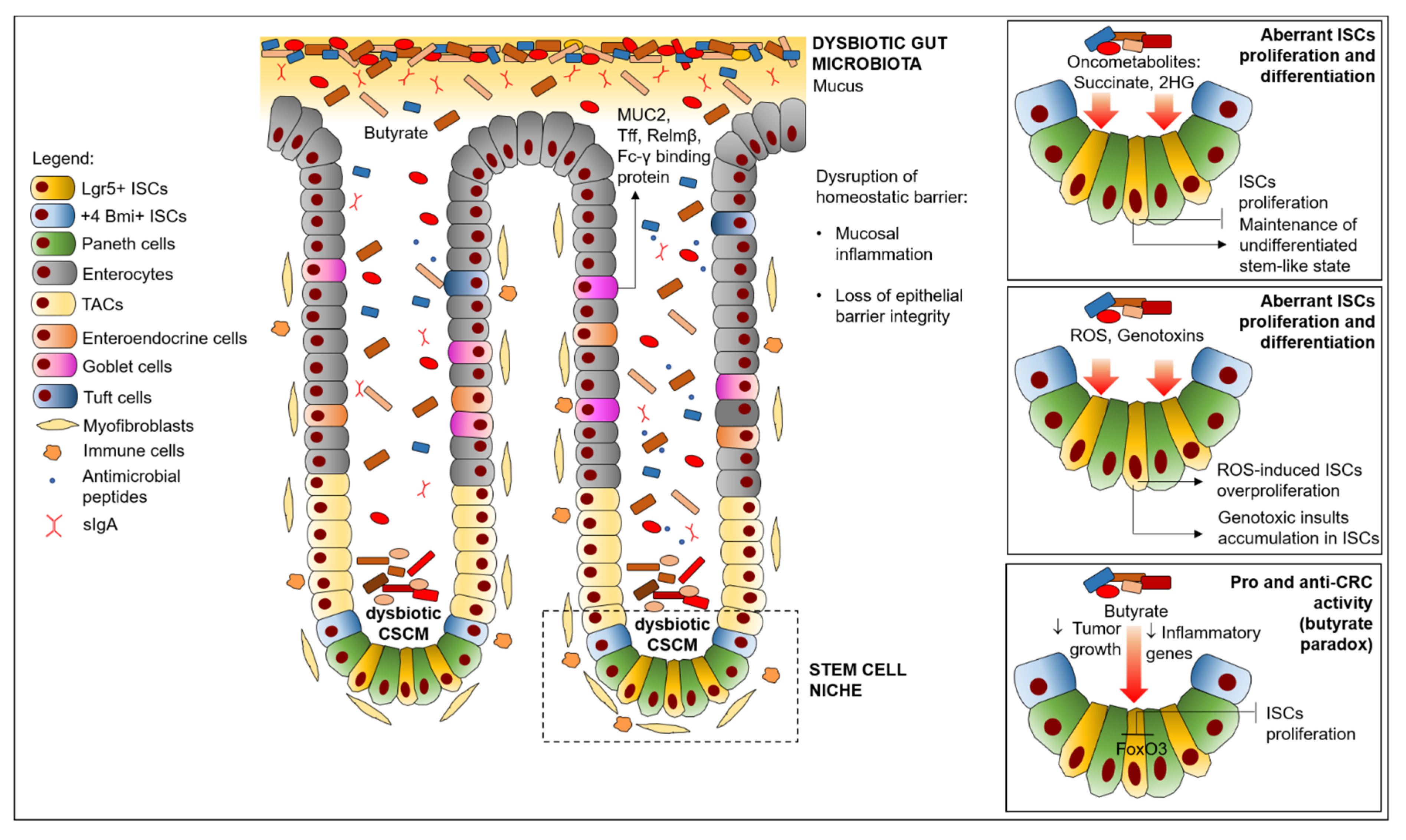
5. Effects of Dysbiosis on ISC Niche Impairment in CRC
5.1. Microbiota Promotion of Colon Inflammation, Leading to ISC Niche Impairment in CRC
5.2. Production of Microbiota Metabolites Affecting ISCs Proliferation/Differentiation in CRC
5.3. Invasive Microbes Inducing ISCs Impairment in CRC
6. OMICS Approaches at the Host-Microbiota Interface toward Novel Precision Medicine Strategies in CRC
6.1. Omics Approaches Potential in Deciphering Host-microbiota Interface at ISC Niche Level
6.2. Application of Different Multiomics to CRC Microbiota Research towards Precision Medicine
7. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thoo, L.; Noti, M.; Krebs, P. Keep Calm: The Intestinal Barrier at the Interface of Peace and War. Cell Death Dis. 2019, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Moossavi, S.; Zhang, H.; Sun, J.; Rezaei, N. Host–Microbiota Interaction and Intestinal Stem Cells in Chronic Inflammation and Colorectal Cancer. Expert Rev. Clin. Immunol. 2013, 9, 409–422. [Google Scholar] [CrossRef]
- Kreso, A.; Dick, J.E. Evolution of the Cancer Stem Cell Model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Fidelle, M.; Yonekura, S.; Picard, M.; Cogdill, A.; Hollebecque, A.; Roberti, M.P.; Zitvogel, L. Resolving the Paradox of Colon Cancer Through the Integration of Genetics, Immunology, and the Microbiota. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The Human Microbiome: Our Second Genome. Annu. Rev. Genom. Hum. Genet. 2012, 13, 151–170. [Google Scholar] [CrossRef] [PubMed]
- Kaiko, G.E.; Ryu, S.H.; Koues, O.I.; Collins, P.L.; Solnica-Krezel, L.; Pearce, E.J.; Pearce, E.L.; Oltz, E.M.; Stappenbeck, T.S. The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 2016, 165, 1708–1720. [Google Scholar] [CrossRef]
- O’Keefe, S.J.D. Diet, Microorganisms and Their Metabolites, and Colon Cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Saffarian, A.; Mulet, C.; Regnault, B.; Amiot, A.; Tran-Van-Nhieu, J.; Ravel, J.; Sobhani, I.; Sansonetti, P.J.; Pédron, T. Crypt- and Mucosa-Associated Core Microbiotas in Humans and Their Alteration in Colon Cancer Patients. mBio 2019, 10, e01315-19. [Google Scholar] [CrossRef]
- Allaire, J.M.; Crowley, S.M.; Law, H.T.; Chang, S.-Y.; Ko, H.-J.; Vallance, B.A. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018, 39, 677–696. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.S.; Chia, L.A.; Li, X.; Ootani, A.; Su, J.; Lee, J.Y.; Su, N.; Luo, Y.; Heilshorn, S.C.; Amieva, M.R.; et al. The Intestinal Stem Cell Markers Bmi1 and Lgr5 Identify Two Functionally Distinct Populations. Proc. Natl. Acad. Sci. USA 2012, 109, 466–471. [Google Scholar] [CrossRef]
- Clevers, H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef]
- Cheng, H.; Merzel, J.; Leblond, C.P. Renewal of Paneth Cells in the Small Intestine of the Mouse. Am. J. Anat. 1969, 126, 507–525. [Google Scholar] [CrossRef] [PubMed]
- De Lau, W.; Peng, W.C.; Gros, P.; Clevers, H. The R-Spondin/Lgr5/Rnf43 Module: Regulator of Wnt Signal Strength. Genes Dev. 2014, 28, 305–316. [Google Scholar] [CrossRef]
- Barker, N. Adult Intestinal Stem Cells: Critical Drivers of Epithelial Homeostasis and Regeneration. Nat. Rev. Mol. Cell Biol. 2014, 15, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth Cells Constitute the Niche for Lgr5 Stem Cells in Intestinal Crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Amaral, A.L.; Dias, A.; Mendes, N.; Muncan, V.; Silva, A.R.; Thibert, C.; Radu, A.G.; David, L.; Máximo, V.; et al. MEX3A Regulates Lgr5+ Stem Cell Maintenance in the Developing Intestinal Epithelium. EMBO Rep. 2020, 21, e48938. [Google Scholar] [CrossRef] [PubMed]
- Sangiorgi, E.; Capecchi, M.R. Bmi1 Is Expressed in Vivo in Intestinal Stem Cells. Nat. Genet. 2008, 40, 915–920. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.K.; Carlone, D.L.; Richmond, C.A.; Farilla, L.; Kranendonk, M.E.G.; Henderson, D.E.; Baffour-Awuah, N.Y.; Ambruzs, D.M.; Fogli, L.K.; Algra, S.; et al. Mouse Telomerase Reverse Transcriptase (MTert) Expression Marks Slowly Cycling Intestinal Stem Cells. Proc. Natl. Acad. Sci. USA 2011, 108, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.-K.; Yang, V.W.; Bialkowska, A.B. The Role of Intestinal Stem Cells in Epithelial Regeneration Following Radiation-Induced Gut Injury. Curr. Stem Cell Rep. 2017, 3, 320–332. [Google Scholar] [CrossRef]
- Farin, H.F.; Van Es, J.H.; Clevers, H. Redundant Sources of Wnt Regulate Intestinal Stem Cells and Promote Formation of Paneth Cells. Gastroenterology 2012, 143, 1518–1529.e7. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Sachs, N.; Wiebrands, K.; Ellenbroek, S.I.J.; Fumagalli, A.; Lyubimova, A.; Begthel, H.; van den Born, M.; van Es, J.H.; Karthaus, W.R.; et al. Reg4+ Deep Crypt Secretory Cells Function as Epithelial Niche for Lgr5+ Stem Cells in Colon. Proc. Natl. Acad. Sci. USA 2016, 113, E5399–E5407. [Google Scholar] [CrossRef]
- Schmitt, M.; Schewe, M.; Sacchetti, A.; Feijtel, D.; van de Geer, W.S.; Teeuwssen, M.; Sleddens, H.F.; Joosten, R.; van Royen, M.E.; van de Werken, H.J.G.; et al. Paneth Cells Respond to Inflammation and Contribute to Tissue Regeneration by Acquiring Stem-like Features through SCF/c-Kit Signaling. Cell Rep. 2018, 24, 2312–2328. [Google Scholar] [CrossRef]
- Nakanishi, Y.; Reina-Campos, M.; Nakanishi, N.; Llado, V.; Elmen, L.; Peterson, S.; Campos, A.; De, S.K.; Leitges, M.; Ikeuchi, H.; et al. Control of Paneth Cell Fate, Intestinal Inflammation and Tumorigenesis by PKCλ/ι. Cell Rep. 2016, 16, 3297–3310. [Google Scholar] [CrossRef] [PubMed]
- Lei, N.Y.; Jabaji, Z.; Wang, J.; Joshi, V.S.; Brinkley, G.J.; Khalil, H.; Wang, F.; Jaroszewicz, A.; Pellegrini, M.; Li, L.; et al. Intestinal Subepithelial Myofibroblasts Support the Growth of Intestinal Epithelial Stem Cells. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-Expressing Mesenchymal Cells Form the Essential Wnt-Secreting Niche for Colon Stem Cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef]
- Valenta, T.; Degirmenci, B.; Moor, A.E.; Herr, P.; Zimmerli, D.; Moor, M.B.; Hausmann, G.; Cantù, C.; Aguet, M.; Basler, K. Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep. 2016, 15, 911–918. [Google Scholar] [CrossRef]
- Haramis, A.-P.G. De Novo Crypt Formation and Juvenile Polyposis on BMP Inhibition in Mouse Intestine. Science 2004, 303, 1684–1686. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Davis, H.; Irshad, S.; Sandberg, T.; Worthley, D.; Leedham, S. Microenvironmental Control of Stem Cell Fate in Intestinal Homeostasis and Disease. J. Pathol. 2015, 237, 135–145. [Google Scholar] [CrossRef]
- Jeffery, V.; Goldson, A.J.; Dainty, J.R.; Chieppa, M.; Sobolewski, A. Interleukin-6 Signaling Regulates Small Intestinal Crypt Homeostasis. J. Immunol. Baltim. Md 1950 2017, 199, 304–311. [Google Scholar] [CrossRef]
- Takahashi, T.; Shiraishi, A. Stem Cell Signaling Pathways in the Small Intestine. Int. J. Mol. Sci. 2020, 21, 2032. [Google Scholar] [CrossRef]
- Adams, P.D.; Jasper, H.; Rudolph, K.L. Aging-Induced Stem Cell Mutations as Drivers for Disease and Cancer. Cell Stem Cell 2015, 16, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Zeuner, A.; Todaro, M.; Stassi, G.; De Maria, R. Colorectal Cancer Stem Cells: From the Crypt to the Clinic. Cell Stem Cell 2014, 15, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The Cancer Stem Cell: Premises, Promises and Challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Huels, D.J.; Sansom, O.J. Stem vs. Non-Stem Cell Origin of Colorectal Cancer. Br. J. Cancer 2015, 113, 1–5. [Google Scholar] [CrossRef]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; da Silva-Diz, V.; Muñoz, P.; et al. The Intestinal Stem Cell Signature Identifies Colorectal Cancer Stem Cells and Predicts Disease Relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef]
- De Robertis, M.; Poeta, M.L.; Signori, E.; Fazio, V.M. Current Understanding and Clinical Utility of MiRNAs Regulation of Colon Cancer Stem Cells. Semin. Cancer Biol. 2018, 53, 232–247. [Google Scholar] [CrossRef] [PubMed]
- Fearon, E.R.; Vogelstein, B. A Genetic Model for Colorectal Tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Vermeulen, L.; Morrissey, E.; van der Heijden, M.; Nicholson, A.M.; Sottoriva, A.; Buczacki, S.; Kemp, R.; Tavaré, S.; Winton, D.J. Defining Stem Cell Dynamics in Models of Intestinal Tumor Initiation. Science 2013, 342, 995. [Google Scholar] [CrossRef]
- Snippert, H.J.; Schepers, A.G.; van Es, J.H.; Simons, B.D.; Clevers, H. Biased Competition between Lgr5 Intestinal Stem Cells Driven by Oncogenic Mutation Induces Clonal Expansion. EMBO Rep. 2014, 15, 62–69. [Google Scholar] [CrossRef]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt Stem Cells as the Cells-of-Origin of Intestinal Cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.; Zimberlin, C.D.; Nicholson, A.M.; Colak, S.; Kemp, R.; Meijer, S.L.; Medema, J.P.; Greten, F.R.; Jansen, M.; Winton, D.J.; et al. Bcl-2 Is a Critical Mediator of Intestinal Transformation. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Van der Heijden, M.; Vermeulen, L. Stem Cells in Homeostasis and Cancer of the Gut. Mol. Cancer 2019, 18, 66. [Google Scholar] [CrossRef] [PubMed]
- Guarner, F.; Malagelada, J.-R. Gut Flora in Health and Disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Wang, M.; Ahrné, S.; Jeppsson, B.; Molin, G. Comparison of Bacterial Diversity along the Human Intestinal Tract by Direct Cloning and Sequencing of 16S RRNA Genes. FEMS Microbiol. Ecol. 2005, 54, 219–231. [Google Scholar] [CrossRef]
- Carroll, I.M.; Threadgill, D.W.; Threadgill, D.S. The Gastrointestinal Microbiome: A Malleable, Third Genome of Mammals. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2009, 20, 395–403. [Google Scholar] [CrossRef]
- Johansson, M.E.V.; Ambort, D.; Pelaseyed, T.; Schütte, A.; Gustafsson, J.K.; Ermund, A.; Subramani, D.B.; Holmén-Larsson, J.M.; Thomsson, K.A.; Bergström, J.H.; et al. Composition and Functional Role of the Mucus Layers in the Intestine. Cell. Mol. Life Sci. 2011, 68, 3635. [Google Scholar] [CrossRef]
- Spadoni, I.; Pietrelli, A.; Pesole, G.; Rescigno, M. Gene Expression Profile of Endothelial Cells during Perturbation of the Gut Vascular Barrier. Gut Microbes 2016, 7, 540–548. [Google Scholar] [CrossRef]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; et al. Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial Organization and Composition of the Mucosal Flora in Patients with Inflammatory Bowel Disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef]
- Elphick, D.A.; Mahida, Y.R. Paneth Cells: Their Role in Innate Immunity and Inflammatory Disease. Gut 2005, 54, 1802–1809. [Google Scholar] [CrossRef]
- Macpherson, A.J.; Uhr, T. Induction of Protective IgA by Intestinal Dendritic Cells Carrying Commensal Bacteria. Science 2004, 303, 1662. [Google Scholar] [CrossRef] [PubMed]
- Peterson, D.A.; McNulty, N.P.; Guruge, J.L.; Gordon, J.I. IgA Response to Symbiotic Bacteria as a Mediator of Gut Homeostasis. Cell Host Microbe 2007, 2, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Perruzza, L.; Gargari, G.; Proietti, M.; Fosso, B.; D’Erchia, A.M.; Faliti, C.E.; Rezzonico-Jost, T.; Scribano, D.; Mauri, L.; Colombo, D.; et al. T Follicular Helper Cells Promote a Beneficial Gut Ecosystem for Host Metabolic Homeostasis by Sensing Microbiota-Derived Extracellular ATP. Cell Rep. 2017, 18, 2566–2575. [Google Scholar] [CrossRef]
- Perruzza, L.; Strati, F.; Gargari, G.; D’Erchia, A.M.; Fosso, B.; Pesole, G.; Guglielmetti, S.; Grassi, F. Enrichment of Intestinal Lactobacillus by Enhanced Secretory IgA Coating Alters Glucose Homeostasis in P2rx7−/− Mice. Sci. Rep. 2019, 9, 9315. [Google Scholar] [CrossRef]
- Johansen, F.-E.; Kaetzel, C. Regulation of the Polymeric Immunoglobulin Receptor and IgA Transport: New Advances in Environmental Factors That Stimulate PIgR Expression and Its Role in Mucosal Immunity. Mucosal Immunol. 2011, 4, 598–602. [Google Scholar] [CrossRef] [PubMed]
- Hooper, L.V.; Macpherson, A.J. Immune Adaptations That Maintain Homeostasis with the Intestinal Microbiota. Nat. Rev. Immunol. 2010, 10, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Fransen, F.; Zagato, E.; Mazzini, E.; Fosso, B.; Manzari, C.; El Aidy, S.; Chiavelli, A.; D’Erchia, A.M.; Sethi, M.K.; Pabst, O.; et al. BALB/c and C57BL/6 Mice Differ in Polyreactive IgA Abundance, Which Impacts the Generation of Antigen-Specific IgA and Microbiota Diversity. Immunity 2015, 43, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Macpherson, A.J.; Harris, N.L. Interactions between Commensal Intestinal Bacteria and the Immune System. Nat. Rev. Immunol. 2004, 4, 478–485. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Shima, T.; Imaoka, A.; Kuwahara, T.; Momose, Y.; Cheng, G.; Yamasaki, S.; Saito, T.; Ohba, Y.; et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science 2011, 331, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Rigoni, R.; Fontana, E.; Guglielmetti, S.; Fosso, B.; D’Erchia, A.M.; Maina, V.; Taverniti, V.; Castiello, M.C.; Mantero, S.; Pacchiana, G.; et al. Intestinal Microbiota Sustains Inflammation and Autoimmunity Induced by Hypomorphic RAG Defects. J. Exp. Med. 2016, 213, 355–375. [Google Scholar] [CrossRef] [PubMed]
- Pédron, T.; Mulet, C.; Dauga, C.; Frangeul, L.; Chervaux, C.; Grompone, G.; Sansonetti, P.J. A Crypt-Specific Core Microbiota Resides in the Mouse Colon. mBio 2012, 3. [Google Scholar] [CrossRef]
- Neish, A.S. Microbes in Gastrointestinal Health and Disease. Gastroenterology 2009, 136, 65–80. [Google Scholar] [CrossRef]
- Fukata, M.; Vamadevan, A.S.; Abreu, M.T. Toll-like Receptors (TLRs) and Nod-like Receptors (NLRs) in Inflammatory Disorders. Semin. Immunol. 2009, 21, 242–253. [Google Scholar] [CrossRef]
- Neal, M.D.; Sodhi, C.P.; Jia, H.; Dyer, M.; Egan, C.E.; Yazji, I.; Good, M.; Afrazi, A.; Marino, R.; Slagle, D.; et al. Toll-like Receptor 4 Is Expressed on Intestinal Stem Cells and Regulates Their Proliferation and Apoptosis via the P53 Up-Regulated Modulator of Apoptosis. J. Biol. Chem. 2012, 287, 37296–37308. [Google Scholar] [CrossRef] [PubMed]
- Neal, M.D.; Sodhi, C.P.; Yazji, I.; Russo, A.M.; Siggers, R.; Jia, H.; Prindle, T.; Branca, M.; Hackam, D.J. Toll-like Receptor 4 (TLR4) Is Expressed on Intestinal Stem Cells and Inhibits Stem Cell Proliferation in the Pathogenesis of Intestinal Inflammation. J. Am. Coll. Surg. 2011, 213, S85. [Google Scholar] [CrossRef]
- Naito, T.; Mulet, C.; De Castro, C.; Molinaro, A.; Saffarian, A.; Nigro, G.; Bérard, M.; Clerc, M.; Pedersen, A.B.; Sansonetti, P.J.; et al. Lipopolysaccharide from Crypt-Specific Core Microbiota Modulates the Colonic Epithelial Proliferation-to-Differentiation Balance. mBio 2017, 8, e01680-17. [Google Scholar] [CrossRef]
- Yi, H.; Patel, A.K.; Sodhi, C.P.; Hackam, D.J.; Hackam, A.S. Novel Role for the Innate Immune Receptor Toll-Like Receptor 4 (TLR4) in the Regulation of the Wnt Signaling Pathway and Photoreceptor Apoptosis. PLoS ONE 2012, 7, e36560. [Google Scholar] [CrossRef] [PubMed]
- Sodhi, C.P.; Shi, X.; Richardson, W.M.; Grant, Z.S.; Shapiro, R.A.; Prindle, T.; Branca, M.; Russo, A.; Gribar, S.C.; Ma, C.; et al. Toll-like-Receptor-4 Inhibits Enterocyte Proliferation via Impaired β-Catenin Signaling in Necrotizing Enterocolitis. Gastroenterology 2010, 138, 185. [Google Scholar] [CrossRef]
- Nigro, G.; Rossi, R.; Commere, P.-H.; Jay, P.; Sansonetti, P.J. The Cytosolic Bacterial Peptidoglycan Sensor Nod2 Affords Stem Cell Protection and Links Microbes to Gut Epithelial Regeneration. Cell Host Microbe 2014, 15, 792–798. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.M.; Neish, A.S. Redox Signaling Mediated by the Gut Microbiota. Free Radic. Biol. Med. 2017, 105, 41–47. [Google Scholar] [CrossRef]
- Ren, F.; Wang, K.; Zhang, T.; Jiang, J.; Nice, E.C.; Huang, C. New Insights into Redox Regulation of Stem Cell Self-Renewal and Differentiation. Biochim. Biophys. Acta BBA-Gen. Subj. 2015, 1850, 1518–1526. [Google Scholar] [CrossRef]
- Jones, R.M.; Luo, L.; Ardita, C.S.; Richardson, A.N.; Kwon, Y.M.; Mercante, J.W.; Alam, A.; Gates, C.L.; Wu, H.; Swanson, P.A.; et al. Symbiotic Lactobacilli Stimulate Gut Epithelial Proliferation via Nox-Mediated Generation of Reactive Oxygen Species. EMBO J. 2013, 32, 3017–3028. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.K.; Harrington, H.A.; Shepherd, S.; Brennan, K.; Dale, T.; Osborne, J.M.; Gavaghan, D.J.; Byrne, H.M. The Role of the Hes1 Crosstalk Hub in Notch-Wnt Interactions of the Intestinal Crypt. PLoS Comput. Biol. 2017, 13, e1005400. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Prasad, P.D.; Singh, N. Benefits of Short-Chain Fatty Acids and Their Receptors in Inflammation and Carcinogenesis. Pharmacol. Ther. 2016, 164, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Iván, J.; Major, E.; Sipos, A.; Kovács, K.; Horváth, D.; Tamás, I.; Bay, P.; Dombrádi, V.; Lontay, B. The Short-Chain Fatty Acid Propionate Inhibits Adipogenic Differentiation of Human Chorion-Derived Mesenchymal Stem Cells Through the Free Fatty Acid Receptor 2. Stem Cells Dev. 2017, 26, 1724–1733. [Google Scholar] [CrossRef] [PubMed]
- Jocken, J.W.E.; González Hernández, M.A.; Hoebers, N.T.H.; van der Beek, C.M.; Essers, Y.P.G.; Blaak, E.E.; Canfora, E.E. Short-Chain Fatty Acids Differentially Affect Intracellular Lipolysis in a Human White Adipocyte Model. Front. Endocrinol. 2018, 8. [Google Scholar] [CrossRef]
- Farin, H.F.; Karthaus, W.R.; Kujala, P.; Rakhshandehroo, M.; Schwank, G.; Vries, R.G.J.; Kalkhoven, E.; Nieuwenhuis, E.E.S.; Clevers, H. Paneth Cell Extrusion and Release of Antimicrobial Products Is Directly Controlled by Immune Cell–Derived IFN-γ. J. Exp. Med. 2014, 211, 1393–1405. [Google Scholar] [CrossRef]
- Vaishnava, S.; Behrendt, C.L.; Ismail, A.S.; Eckmann, L.; Hooper, L.V. Paneth Cells Directly Sense Gut Commensals and Maintain Homeostasis at the Intestinal Host-Microbial Interface. Proc. Natl. Acad. Sci. USA 2008, 105, 20858–20863. [Google Scholar] [CrossRef]
- Martinez Rodriguez, N.R.; Eloi, M.D.; Huynh, A.; Dominguez, T.; Lam, A.H.C.; Carcamo-Molina, D.; Naser, Z.; Desharnais, R.; Salzman, N.H.; Porter, E. Expansion of Paneth Cell Population in Response to Enteric Salmonella Enterica Serovar Typhimurium Infection. Infect. Immun. 2012, 80, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Hirao, L.A.; Grishina, I.; Bourry, O.; Hu, W.K.; Somrit, M.; Sankaran-Walters, S.; Gaulke, C.A.; Fenton, A.N.; Li, J.A.; Crawford, R.W.; et al. Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef]
- Lee, Y.-S.; Kim, T.-Y.; Kim, Y.; Lee, S.-H.; Kim, S.; Kang, S.W.; Yang, J.-Y.; Baek, I.-J.; Sung, Y.H.; Park, Y.-Y.; et al. Microbiota-Derived Lactate Accelerates Intestinal Stem-Cell-Mediated Epithelial Development. Cell Host Microbe 2018, 24, 833–846.e6. [Google Scholar] [CrossRef]
- De Robertis, M.; Mazza, T.; Fusilli, C.; Loiacono, L.; Poeta, M.L.; Sanchez, M.; Massi, E.; Lamorte, G.; Diodoro, M.G.; Pescarmona, E.; et al. EphB2 Stem-Related and EphA2 Progression-Related MiRNA-Based Networks in Progressive Stages of CRC Evolution: Clinical Significance and Potential MiRNA Drivers. Mol. Cancer 2018, 17, 169. [Google Scholar] [CrossRef] [PubMed]
- De Robertis, M.; Loiacono, L.; Fusilli, C.; Poeta, M.L.; Mazza, T.; Sanchez, M.; Marchionni, L.; Signori, E.; Lamorte, G.; Vescovi, A.L.; et al. Dysregulation of EGFR Pathway in EphA2 Cell Subpopulation Significantly Associates with Poor Prognosis in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 159. [Google Scholar] [CrossRef] [PubMed]
- Archambaud, C.; Sismeiro, O.; Toedling, J.; Soubigou, G.; Bécavin, C.; Lechat, P.; Lebreton, A.; Ciaudo, C.; Cossart, P. The Intestinal Microbiota Interferes with the MicroRNA Response upon Oral Listeria Infection. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Peck, B.C.E.; Mah, A.T.; Pitman, W.A.; Ding, S.; Lund, P.K.; Sethupathy, P. Functional Transcriptomics in Diverse Intestinal Epithelial Cell Types Reveals Robust MicroRNA Sensitivity in Intestinal Stem Cells to Microbial Status. J. Biol. Chem. 2017, 292, 2586–2600. [Google Scholar] [CrossRef] [PubMed]
- Shanahan, M.T.; Kanke, M.; Singh, A.P.; Villanueva, J.W.; McNairn, A.J.; Oyesola, O.O.; Bonfini, A.; Hung, Y.-H.; Sheahan, B.; Bloom, J.C.; et al. Single Cell Analysis Reveals Multi-Faceted MiR-375 Regulation of the Intestinal Crypt. bioRxiv 2020. [Google Scholar] [CrossRef]
- Peck, B.C.E.; Sincavage, J.; Feinstein, S.; Mah, A.T.; Simmons, J.G.; Lund, P.K.; Sethupathy, P. MiR-30 Family Controls Proliferation and Differentiation of Intestinal Epithelial Cell Models by Directing a Broad Gene Expression Program That Includes SOX9 and the Ubiquitin Ligase Pathway. J. Biol. Chem. 2016, 291, 15975–15984. [Google Scholar] [CrossRef]
- Niel, G.V.; Raposo, G.; Candalh, C.; Boussac, M.; Hershberg, R.; Cerf–Bensussan, N.; Heyman, M. Intestinal Epithelial Cells Secrete Exosome–like Vesicles. Gastroenterology 2001, 121, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; da Cunha, A.P.; Rezende, R.M.; Cialic, R.; Wei, Z.; Bry, L.; Comstock, L.E.; Gandhi, R.; Weiner, H.L. The Host Shapes the Gut Microbiota via Fecal MicroRNA. Cell Host Microbe 2016, 19, 32–43. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A Bacterial Driver–Passenger Model for Colorectal Cancer: Beyond the Usual Suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Ternes, D.; Karta, J.; Tsenkova, M.; Wilmes, P.; Haan, S.; Letellier, E. Microbiome in Colorectal Cancer: How to Get from Meta-Omics to Mechanism? Trends Microbiol. 2020, 28, 401–423. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L.; et al. Fusobacterium Nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor Immune Microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Chung, L.; Orberg, E.T.; Geis, A.L.; Chan, J.L.; Fu, K.; DeStefano Shields, C.E.; Dejea, C.M.; Fathi, P.; Chen, J.; Finard, B.B.; et al. Bacteroides Fragilis Toxin Coordinates a Pro-Carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe 2018, 23, 203–214.e5. [Google Scholar] [CrossRef]
- Zagato, E.; Pozzi, C.; Bertocchi, A.; Schioppa, T.; Saccheri, F.; Guglietta, S.; Fosso, B.; Melocchi, L.; Nizzoli, G.; Troisi, J.; et al. Endogenous Murine Microbiota Member Faecalibaculum Rodentium and Its Human Homologue Protect from Intestinal Tumour Growth. Nat. Microbiol. 2020, 5, 511–524. [Google Scholar] [CrossRef]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and Colon Cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Wang, L.; Zhang, C. Interplay between Inflammatory Tumor Microenvironment and Cancer Stem Cells. Oncol. Lett. 2018, 16, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Bollrath, J.; Phesse, T.J.; von Burstin, V.A.; Putoczki, T.; Bennecke, M.; Bateman, T.; Nebelsiek, T.; Lundgren-May, T.; Canli, Ö.; Schwitalla, S.; et al. Gp130-Mediated Stat3 Activation in Enterocytes Regulates Cell Survival and Cell-Cycle Progression during Colitis-Associated Tumorigenesis. Cancer Cell 2009, 15, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Keenan, J.I.; Aitchison, A.; Purcell, R.V.; Greenlees, R.; Pearson, J.F.; Frizelle, F.A. Screening for Enterotoxigenic Bacteroides Fragilis in Stool Samples. Anaerobe 2016, 40, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with Familial Adenomatous Polyposis Harbor Colonic Biofilms Containing Tumorigenic Bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef]
- Lee, Y.K.; Mehrabian, P.; Boyajian, S.; Wu, W.-L.; Selicha, J.; Vonderfecht, S.; Mazmanian, S.K. The Protective Role of Bacteroides Fragilis in a Murine Model of Colitis-Associated Colorectal Cancer. mSphere 2018, 3. [Google Scholar] [CrossRef]
- Ruiz, P.A.; Shkoda, A.; Kim, S.C.; Sartor, R.B.; Haller, D. IL-10 Gene-Deficient Mice Lack TGF-Beta/Smad-Mediated TLR2 Degradation and Fail to Inhibit Proinflammatory Gene Expression in Intestinal Epithelial Cells under Conditions of Chronic Inflammation. Ann. N. Y. Acad. Sci. 2006, 1072, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Kikushige, Y.; Miyawaki, K.; Kunisaki, Y.; Mizuno, S.; Takenaka, K.; Tamura, S.; Okumura, Y.; Ito, M.; Ariyama, H.; et al. Dedifferentiation Process Driven by TGF-Beta Signaling Enhances Stem Cell Properties in Human Colorectal Cancer. Oncogene 2019, 38, 780–793. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, Y.; Huycke, M.M. Commensal Bacteria Drive Endogenous Transformation and Tumour Stem Cell Marker Expression through a Bystander Effect. Gut 2015, 64, 459–468. [Google Scholar] [CrossRef]
- Shin, S.C.; Kim, S.-H.; You, H.; Kim, B.; Kim, A.C.; Lee, K.-A.; Yoon, J.-H.; Ryu, J.-H.; Lee, W.-J. Drosophila Microbiome Modulates Host Developmental and Metabolic Homeostasis via Insulin Signaling. Science 2011, 334, 670. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.R.J.; Patel, K.; Putnam, W.C.; Kapur, P.; Rakheja, D. Oncometabolites: A New Paradigm for Oncology, Metabolism, and the Clinical Laboratory. Clin. Chem. 2017, 63, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, J.; Rao, B.; Deng, L. Gut Flora Profiling and Fecal Metabolite Composition of Colorectal Cancer Patients and Healthy Individuals. Exp. Ther. Med. 2017, 13, 2848–2854. [Google Scholar] [CrossRef]
- Fernández-Veledo, S.; Vendrell, J. Gut Microbiota-Derived Succinate: Friend or Foe in Human Metabolic Diseases? Rev. Endocr. Metab. Disord. 2019, 20, 439–447. [Google Scholar] [CrossRef]
- Connors, J.; Dawe, N.; Van Limbergen, J. The Role of Succinate in the Regulation of Intestinal Inflammation. Nutrients 2019, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Littlewood-Evans, A.; Sarret, S.; Apfel, V.; Loesle, P.; Dawson, J.; Zhang, J.; Muller, A.; Tigani, B.; Kneuer, R.; Patel, S.; et al. GPR91 Senses Extracellular Succinate Released from Inflammatory Macrophages and Exacerbates Rheumatoid Arthritis. J. Exp. Med. 2016, 213, 1655–1662. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.M.; Hu, Z.; Klimko, C.; Narayanan, S.; Deberardinis, R.; Sperandio, V. The Gut Commensal Bacteroides Thetaiotaomicron Exacerbates Enteric Infection through Modification of the Metabolic Landscape. Cell Host Microbe 2014, 16, 759–769. [Google Scholar] [CrossRef] [PubMed]
- In, J.; Foulke-Abel, J.; Zachos, N.C.; Hansen, A.-M.; Kaper, J.B.; Bernstein, H.D.; Halushka, M.; Blutt, S.; Estes, M.K.; Donowitz, M.; et al. Enterohemorrhagic Escherichia Coli Reduces Mucus and Intermicrovillar Bridges in Human Stem Cell-Derived Colonoids. Cell. Mol. Gastroenterol. Hepatol. 2015, 2, 48–62.e3. [Google Scholar] [CrossRef]
- Inagaki, A.; Ichikawa, H.; Sakata, T. Inhibitory Effect of Succinic Acid on Epithelial Cell Proliferation of Colonic Mucosa in Rats. J. Nutr. Sci. Vitaminol. (Tokyo) 2007, 53, 377–379. [Google Scholar] [CrossRef]
- Ariake, K.; Ohkusa, T.; Sakurazawa, T.; Kumagai, J.; Eishi, Y.; Hoshi, S.; Yajima, T. Roles of Mucosal Bacteria and Succinic Acid in Colitis Caused by Dextran Sulfate Sodium in Mice. J. Med. Dent. Sci. 2000, 47, 233–241. [Google Scholar] [PubMed]
- Chowdhury, R.; Yeoh, K.K.; Tian, Y.-M.; Hillringhaus, L.; Bagg, E.A.; Rose, N.R.; Leung, I.K.H.; Li, X.S.; Woon, E.C.Y.; Yang, M.; et al. The Oncometabolite 2-Hydroxyglutarate Inhibits Histone Lysine Demethylases. EMBO Rep. 2011, 12, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Ježek, P. 2-Hydroxyglutarate in Cancer Cells. Antioxid. Redox Signal. 2020, 33, 903–926. [Google Scholar] [CrossRef]
- Li, H.; Chawla, G.; Hurlburt, A.J.; Sterrett, M.C.; Zaslaver, O.; Cox, J.; Karty, J.A.; Rosebrock, A.P.; Caudy, A.A.; Tennessen, J.M. Drosophila Larvae Synthesize the Putative Oncometabolite L-2-Hydroxyglutarate during Normal Developmental Growth. Proc. Natl. Acad. Sci. USA 2017, 114, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz-López, K.G.; Castro-Muñoz, L.J.; Reyes-Hernández, D.O.; García-Carrancá, A.; Manzo-Merino, J. Lactate in the Regulation of Tumor Microenvironment and Therapeutic Approaches. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Tazoe, H.; Otomo, Y.; Kaji, I.; Tanaka, R.; Karaki, S.-I.; Kuwahara, A. Roles of Short-Chain Fatty Acids Receptors, GPR41 and GPR43 on Colonic Functions. J. Physiol. Pharmacol. Off. J. Pol. Physiol. Soc. 2008, 59 (Suppl. 2), 251–262. [Google Scholar]
- Bigarella, C.L.; Liang, R.; Ghaffari, S. Stem Cells and the Impact of ROS Signaling. Dev. Camb. Engl. 2014, 141, 4206–4218. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the Effects of Diet on Bacterial Metabolism in the Large Intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg Effect Dictates the Mechanism of Butyrate Mediated Histone Acetylation and Cell Proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Hu, S.; Dong, T.S.; Dalal, S.R.; Wu, F.; Bissonnette, M.; Kwon, J.H.; Chang, E.B. The Microbe-Derived Short Chain Fatty Acid Butyrate Targets MiRNA-Dependent P21 Gene Expression in Human Colon Cancer. PLoS ONE 2011, 6, e16221. [Google Scholar] [CrossRef]
- Hu, S.; Liu, L.; Chang, E.B.; Wang, J.-Y.; Raufman, J.-P. Butyrate Inhibits Pro-Proliferative MiR-92a by Diminishing c-Myc-Induced MiR-17-92a Cluster Transcription in Human Colon Cancer Cells. Mol. Cancer 2015, 14. [Google Scholar] [CrossRef]
- Scheppach, W. Effects of Short Chain Fatty Acids on Gut Morphology and Function. Gut 1994, 35, S35–S38. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K.; Bultman, S.J. The Microbiome and Butyrate Regulate Energy Metabolism and Autophagy in the Mammalian Colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef]
- Yin, X.; Farin, H.F.; van Es, J.H.; Clevers, H.; Langer, R.; Karp, J.M. Niche-Independent High-Purity Cultures of Lgr5+ Intestinal Stem Cells and Their Progeny. Nat. Methods 2014, 11, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Burgess, D.J. Warburg behind the Butyrate Paradox? Nat. Rev. Cancer 2012, 12, 798–799. [Google Scholar] [CrossRef] [PubMed]
- Wrzosek, L.; Miquel, S.; Noordine, M.-L.; Bouet, S.; Chevalier-Curt, M.J.; Robert, V.; Philippe, C.; Bridonneau, C.; Cherbuy, C.; Robbe-Masselot, C.; et al. Bacteroides Thetaiotaomicron and Faecalibacterium Prausnitzii Influence the Production of Mucus Glycans and the Development of Goblet Cells in the Colonic Epithelium of a Gnotobiotic Model Rodent. BMC Biol. 2013, 11, 61. [Google Scholar] [CrossRef] [PubMed]
- Priyamvada, S.; Anbazhagan, A.; Chatterjee, I.; Alrefai, W.; Dudeja, P.; Borthakur, A. Gut Bacterial Metabolite Propionate Upregulates Intestinal Epithelial Kruppel-like Factor 4 Expression via a PPAR-γ-Dependent Mechanism. FASEB J. 2015, 29, 854.4. [Google Scholar] [CrossRef]
- Corrêa-Oliveira, R.; Fachi, J.L.; Vieira, A.; Sato, F.T.; Vinolo, M.A.R. Regulation of Immune Cell Function by Short-Chain Fatty Acids. Clin. Transl. Immunol. 2016, 5, e73. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Islam, K.B.M.S.; Fukiya, S.; Hagio, M.; Fujii, N.; Ishizuka, S.; Ooka, T.; Ogura, Y.; Hayashi, T.; Yokota, A. Bile Acid Is a Host Factor That Regulates the Composition of the Cecal Microbiota in Rats. Gastroenterology 2011, 141, 1773–1781. [Google Scholar] [CrossRef]
- Martínez-Augustin, O.; de Medina, F.S. Intestinal Bile Acid Physiology and Pathophysiology. World J. Gastroenterol. WJG 2008, 14, 5630–5640. [Google Scholar] [CrossRef] [PubMed]
- Farhana, L.; Nangia-Makker, P.; Arbit, E.; Shango, K.; Sarkar, S.; Mahmud, H.; Hadden, T.; Yu, Y.; Majumdar, A.P.N. Bile Acid: A Potential Inducer of Colon Cancer Stem Cells. Stem Cell Res. Ther. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, H.; Bernstein, C.; Payne, C.M.; Dvorak, K. Bile Acids as Endogenous Etiologic Agents in Gastrointestinal Cancer. World J. Gastroenterol. WJG 2009, 15, 3329–3340. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Y.; Moore, D.R.; Nimmo, S.L.; Lightfoot, S.A.; Huycke, M.M. 4-Hydroxy-2-Nonenal Mediates Genotoxicity and Bystander Effects Caused by Enterococcus Faecalis-Infected Macrophages. Gastroenterology 2012, 142, 543–551.e7. [Google Scholar] [CrossRef]
- Goodwin, A.C.; Shields, C.E.D.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine Catabolism Contributes to Enterotoxigenic Bacteroides Fragilis-Induced Colon Tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Tsoi, H.; Chu, E.S.H.; Zhang, X.; Sheng, J.; Nakatsu, G.; Ng, S.C.; Chan, A.W.H.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Peptostreptococcus Anaerobius Induces Intracellular Cholesterol Biosynthesis in Colon Cells to Induce Proliferation and Causes Dysplasia in Mice. Gastroenterology 2017, 152, 1419–1433.e5. [Google Scholar] [CrossRef]
- Kwong, T.N.Y.; Wang, X.; Nakatsu, G.; Chow, T.C.; Tipoe, T.; Dai, R.Z.W.; Tsoi, K.K.K.; Wong, M.C.S.; Tse, G.; Chan, M.T.V.; et al. Association Between Bacteremia From Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology 2018, 155, 383–390.e8. [Google Scholar] [CrossRef]
- Myant, K.B.; Cammareri, P.; McGhee, E.J.; Ridgway, R.A.; Huels, D.J.; Cordero, J.B.; Schwitalla, S.; Kalna, G.; Ogg, E.-L.; Athineos, D.; et al. ROS Production and NF-ΚB Activation Triggered by RAC1 Facilitate WNT-Driven Intestinal Stem Cell Proliferation and Colorectal Cancer Initiation. Cell Stem Cell 2013, 12, 761–773. [Google Scholar] [CrossRef]
- Balskus, E.P. Colibactin: Understanding an Elusive Gut Bacterial Genotoxin. Nat. Prod. Rep. 2015, 32, 1534–1540. [Google Scholar] [CrossRef]
- Wassenaar, T.M. E. Coli and Colorectal Cancer: A Complex Relationship That Deserves a Critical Mindset. Crit. Rev. Microbiol. 2018, 44, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Aung, K.M.; Uhlin, B.E.; Wai, S.N. Reversible Senescence of Human Colon Cancer Cells after Blockage of Mitosis/Cytokinesis Caused by the CNF1 Cyclomodulin from Escherichia Coli. Sci. Rep. 2018, 8, 17780. [Google Scholar] [CrossRef]
- Tomkovich, S.; Yang, Y.; Winglee, K.; Gauthier, J.; Mühlbauer, M.; Sun, X.; Mohamadzadeh, M.; Liu, X.; Martin, P.; Wang, G.P.; et al. Locoregional Effects of Microbiota in a Preclinical Model of Colon Carcinogenesis. Cancer Res. 2017, 77, 2620–2632. [Google Scholar] [CrossRef]
- Wlodarska, M.; Willing, B.; Keeney, K.M.; Menendez, A.; Bergstrom, K.S.; Gill, N.; Russell, S.L.; Vallance, B.A.; Finlay, B.B. Antibiotic Treatment Alters the Colonic Mucus Layer and Predisposes the Host to Exacerbated Citrobacter Rodentium-Induced Colitis. Infect. Immun. 2011, 79, 1536–1545. [Google Scholar] [CrossRef] [PubMed]
- Leatham, M.P.; Banerjee, S.; Autieri, S.M.; Mercado-Lubo, R.; Conway, T.; Cohen, P.S. Precolonized Human Commensal Escherichia Coli Strains Serve as a Barrier to E. Coli O157:H7 Growth in the Streptomycin-Treated Mouse Intestine. Infect. Immun. 2009, 77, 2876–2886. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Blaser, M.J. The Human Microbiome: At the Interface of Health and Disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. Inflammatory Responses and Inflammation-Associated Diseases in Organs. Oncotarget 2017, 9, 7204–7218. [Google Scholar] [CrossRef] [PubMed]
- Garrett, W.S. Cancer and the Microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Afify, S.M.; Seno, M. Conversion of Stem Cells to Cancer Stem Cells: Undercurrent of Cancer Initiation. Cancers 2019, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Peuker, K.; Muff, S.; Wang, J.; Künzel, S.; Bosse, E.; Zeissig, Y.; Luzzi, G.; Basic, M.; Strigli, A.; Ulbricht, A.; et al. Epithelial Calcineurin Controls Microbiota-Dependent Intestinal Tumor Development. Nat. Med. 2016, 22, 506–515. [Google Scholar] [CrossRef]
- Sahu, U.; Choudhury, A.; Parvez, S.; Biswas, S.; Kar, S. Induction of Intestinal Stemness and Tumorigenicity by Aberrant Internalization of Commensal Non-Pathogenic E. Coli. Cell Death Dis. 2017, 8, e2667. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota Organization Is a Distinct Feature of Proximal Colorectal Cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and Its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Konstantinov, S.R.; Smits, R.; Peppelenbosch, M.P. Bacterial Biofilms in Colorectal Cancer Initiation and Progression. Trends Mol. Med. 2017, 23, 18–30. [Google Scholar] [CrossRef]
- Raskov, H.; Kragh, K.N.; Bjarnsholt, T.; Alamili, M.; Gögenur, I. Bacterial Biofilm Formation inside Colonic Crypts May Accelerate Colorectal Carcinogenesis. Clin. Transl. Med. 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Hold, G.L.; Allen-Vercoe, E. Gut Microbial Biofilm Composition and Organisation Holds the Key to CRC. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 329–330. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.Y.; Koh, E.; Wong, A.; March, J.C.; Bentley, W.E.; Lee, Y.S.; Chang, M.W. Engineered Probiotic Escherichia Coli Can Eliminate and Prevent Pseudomonas Aeruginosa Gut Infection in Animal Models. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Kaur, S.; Sharma, P.; Kalia, N.; Singh, J.; Kaur, S. Anti-Biofilm Properties of the Fecal Probiotic Lactobacilli Against Vibrio Spp. Front. Cell. Infect. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.R.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-Associated and Non-Tumour-Associated Microbiota in Colorectal Cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic Analysis of Faecal Microbiome as a Tool towards Targeted Non-Invasive Biomarkers for Colorectal Cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.T.; Ruffin, M.T.; Rogers, M.A.M.; Schloss, P.D. Microbiota-Based Model Improves the Sensitivity of Fecal Immunochemical Test for Detecting Colonic Lesions. Genome Med. 2016, 8, 37. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.M.; Ruffin, M.T.; Schloss, P.D. The Human Gut Microbiome as a Screening Tool for Colorectal Cancer. Cancer Prev. Res. Phila. Pa 2014, 7, 1112–1121. [Google Scholar] [CrossRef]
- Nakatsu, G.; Li, X.; Zhou, H.; Sheng, J.; Wong, S.H.; Wu, W.K.K.; Ng, S.C.; Tsoi, H.; Dong, Y.; Zhang, N.; et al. Gut Mucosal Microbiome across Stages of Colorectal Carcinogenesis. Nat. Commun. 2015, 6, 8727. [Google Scholar] [CrossRef] [PubMed]
- Kinross, J.; Mirnezami, R.; Alexander, J.; Brown, R.; Scott, A.; Galea, D.; Veselkov, K.; Goldin, R.; Darzi, A.; Nicholson, J.; et al. A Prospective Analysis of Mucosal Microbiome-Metabonome Interactions in Colorectal Cancer Using a Combined MAS 1HNMR and Metataxonomic Strategy. Sci. Rep. 2017, 7, 8979. [Google Scholar] [CrossRef]
- Chistoserdova, L. Recent Progress and New Challenges in Metagenomics for Biotechnology. Biotechnol. Lett. 2010, 32, 1351–1359. [Google Scholar] [CrossRef]
- Stringer, A.M.; Al-Dasooqi, N.; Bowen, J.M.; Tan, T.H.; Radzuan, M.; Logan, R.M.; Mayo, B.; Keefe, D.M.K.; Gibson, R.J. Biomarkers of Chemotherapy-Induced Diarrhoea: A Clinical Study of Intestinal Microbiome Alterations, Inflammation and Circulating Matrix Metalloproteinases. Support. Care Cancer 2013, 21, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.Y.; Park, H.T.; Dinh, N.T.H.; Choi, S.J.; Lee, J.; Kim, J.H.; Lee, S.-W.; Gho, Y.S. Bacterial Outer Membrane Vesicles Suppress Tumor by Interferon-γ-Mediated Antitumor Response. Nat. Commun. 2017, 8, 626. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Van Nuenen, M.H.M.C.; Venema, K.; van der Woude, J.C.J.; Kuipers, E.J. The Metabolic Activity of Fecal Microbiota from Healthy Individuals and Patients with Inflammatory Bowel Disease. Dig. Dis. Sci. 2004, 49, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Wilson, I.D. Gut Microorganisms, Mammalian Metabolism and Personalized Health Care. Nat. Rev. Microbiol. 2005, 3, 431–438. [Google Scholar] [CrossRef]
- Lin, X.B.; Dieleman, L.A.; Ketabi, A.; Bibova, I.; Sawyer, M.B.; Xue, H.; Field, C.J.; Baracos, V.E.; Gänzle, M.G. Irinotecan (CPT-11) Chemotherapy Alters Intestinal Microbiota in Tumour Bearing Rats. PLoS ONE 2012, 7, e39764. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.B.; Farhangfar, A.; Valcheva, R.; Sawyer, M.B.; Dieleman, L.; Schieber, A.; Gänzle, M.G.; Baracos, V. The Role of Intestinal Microbiota in Development of Irinotecan Toxicity and in Toxicity Reduction through Dietary Fibres in Rats. PLoS ONE 2014, 9, e83644. [Google Scholar] [CrossRef] [PubMed]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut Microbiota Modulation of Chemotherapy Efficacy and Toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356–365. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-Cell Genome Sequencing: Current State of the Science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A Single-Cell Survey of the Small Intestinal Epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Campbell, J.H.; O’Donoghue, P.; Campbell, A.G.; Schwientek, P.; Sczyrba, A.; Woyke, T.; Söll, D.; Podar, M. UGA Is an Additional Glycine Codon in Uncultured SR1 Bacteria from the Human Microbiota. Proc. Natl. Acad. Sci. USA 2013, 110, 5540–5545. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.; Stecher, B.; Schintlmeister, A.; Reichert, J.; Brugiroux, S.; Wild, B.; Wanek, W.; Richter, A.; Rauch, I.; Decker, T.; et al. Host-Compound Foraging by Intestinal Microbiota Revealed by Single-Cell Stable Isotope Probing. Proc. Natl. Acad. Sci. USA 2013, 110, 4720. [Google Scholar] [CrossRef] [PubMed]
- Props, R.; Kerckhof, F.-M.; Rubbens, P.; De Vrieze, J.; Hernandez Sanabria, E.; Waegeman, W.; Monsieurs, P.; Hammes, F.; Boon, N. Absolute Quantification of Microbial Taxon Abundances. ISME J. 2017, 11, 584–587. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Kim, J.K.; Tsang, J.C.H.; Ilicic, T.; Henriksson, J.; Natarajan, K.N.; Tuck, A.C.; Gao, X.; Bühler, M.; Liu, P.; et al. Single Cell RNA-Sequencing of Pluripotent States Unlocks Modular Transcriptional Variation. Cell Stem Cell 2015, 17, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.V.; Thaiss, C.A. Host-Microbiome Interactions in the Era of Single-Cell Biology. Front. Cell. Infect. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liao, B.; Zhang, T.; Xu, Y. Editorial: Bioinformatics Analysis of Single Cell Sequencing Data and Applications in Precision Medicine. Front. Genet. 2020, 10. [Google Scholar] [CrossRef]
- Yoon, H.S.; Price, D.C.; Stepanauskas, R.; Rajah, V.D.; Sieracki, M.E.; Wilson, W.H.; Yang, E.C.; Duffy, S.; Bhattacharya, D. Single-Cell Genomics Reveals Organismal Interactions in Uncultivated Marine Protists. Science 2011, 332, 714. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, M.; Brazel, D.; Poulton, N.J.; Swan, B.K.; Gomez, M.L.; Masland, D.; Sieracki, M.E.; Stepanauskas, R. Unveiling in Situ Interactions between Marine Protists and Bacteria through Single Cell Sequencing. ISME J. 2012, 6, 703–707. [Google Scholar] [CrossRef]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifesta, M.; Viappiani, A.; Lugli, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering Bifidobacterial-Mediated Metabolic Interactions and Their Impact on Gut Microbiota by a Multi-Omics Approach. ISME J. 2016, 10, 1656–1668. [Google Scholar] [CrossRef]
- Sattin, E.; Andreani, N.A.; Carraro, L.; Lucchini, R.; Fasolato, L.; Telatin, A.; Balzan, S.; Novelli, E.; Simionati, B.; Cardazzo, B. A Multi-Omics Approach to Evaluate the Quality of Milk Whey Used in Ricotta Cheese Production. Front. Microbiol. 2016, 7, 1272. [Google Scholar] [CrossRef]
- Habowski, A.N.; Flesher, J.L.; Bates, J.M.; Tsai, C.-F.; Martin, K.; Zhao, R.; Ganesan, A.K.; Edwards, R.A.; Shi, T.; Wiley, H.S.; et al. Transcriptomic and Proteomic Signatures of Stemness and Differentiation in the Colon Crypt. Commun. Biol. 2020, 3, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Lindeboom, R.G.; van Voorthuijsen, L.; Oost, K.C.; Rodríguez-Colman, M.J.; Luna-Velez, M.V.; Furlan, C.; Baraille, F.; Jansen, P.W.; Ribeiro, A.; Burgering, B.M.; et al. Integrative Multi-omics Analysis of Intestinal Organoid Differentiation. Mol. Syst. Biol. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Emara, S.; Amer, S.; Ali, A.; Abouleila, Y.; Oga, A.; Masujima, T. Single-Cell Metabolomics. In Metabolomics: From Fundamentals to Clinical Applications; Advances in Experimental Medicine and Biology; Sussulini, A., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 323–343. ISBN 978-3-319-47656-8. [Google Scholar]
- Brazovskaja, A.; Treutlein, B.; Camp, J.G. High-Throughput Single-Cell Transcriptomics on Organoids. Curr. Opin. Biotechnol. 2019, 55, 167–171. [Google Scholar] [CrossRef]
- Jiang, D.; Armour, C.R.; Hu, C.; Mei, M.; Tian, C.; Sharpton, T.J.; Jiang, Y. Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Front. Genet. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-Omics of the Gut Microbial Ecosystem in Inflammatory Bowel Diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Andersen, J.N.; Futreal, P.A. Cancer Genomics: From Discovery Science to Personalized Medicine. Nat. Med. 2011, 17, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Sadanandam, A.; Lyssiotis, C.A.; Homicsko, K.; Collisson, E.A.; Gibb, W.J.; Wullschleger, S.; Ostos, L.C.G.; Lannon, W.A.; Grotzinger, C.; Del Rio, M.; et al. A Colorectal Cancer Classification System That Associates Cellular Phenotype and Responses to Therapy. Nat. Med. 2013, 19, 619–625. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut Microbiome Influences Efficacy of PD-1–Based Immunotherapy against Epithelial Tumors. Science 2018, 359, 91. [Google Scholar] [CrossRef] [PubMed]
- García-González, A.P.; Ritter, A.D.; Shrestha, S.; Andersen, E.C.; Yilmaz, L.S.; Walhout, A.J.M. Bacterial Metabolism Affects the C. Elegans Response to Cancer Chemotherapeutics. Cell 2017, 169, 431–441.e8. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Maier, L.; Pruteanu, M.; Kuhn, M.; Zeller, G.; Telzerow, A.; Anderson, E.E.; Brochado, A.R.; Fernandez, K.C.; Dose, H.; Mori, H.; et al. Extensive Impact of Non-Antibiotic Drugs on Human Gut Bacteria. Nature 2018, 555, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Gough, E.; Shaikh, H.; Manges, A.R. Systematic Review of Intestinal Microbiota Transplantation (Fecal Bacteriotherapy) for Recurrent Clostridium Difficile Infection. Clin. Infect. Dis. 2011, 53, 994–1002. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marzano, M.; Fosso, B.; Piancone, E.; Defazio, G.; Pesole, G.; De Robertis, M. Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers 2021, 13, 996. https://doi.org/10.3390/cancers13050996
Marzano M, Fosso B, Piancone E, Defazio G, Pesole G, De Robertis M. Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers. 2021; 13(5):996. https://doi.org/10.3390/cancers13050996
Chicago/Turabian StyleMarzano, Marinella, Bruno Fosso, Elisabetta Piancone, Giuseppe Defazio, Graziano Pesole, and Mariangela De Robertis. 2021. "Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer" Cancers 13, no. 5: 996. https://doi.org/10.3390/cancers13050996
APA StyleMarzano, M., Fosso, B., Piancone, E., Defazio, G., Pesole, G., & De Robertis, M. (2021). Stem Cell Impairment at the Host-Microbiota Interface in Colorectal Cancer. Cancers, 13(5), 996. https://doi.org/10.3390/cancers13050996








