V-Domain Ig Suppressor of T Cell Activation (VISTA) Expression Is an Independent Prognostic Factor in Multiple Myeloma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
2.1. Transcriptomics of Bone Marrow Samples Identifies VISTA as an Independent Prognostic Factor for MM
2.2. MM Bone Marrow CD11b+ Cells, but Not Plasma Cells, CD4, or CD8 T-Cell Subsets nor CD163+ Cells, Express VISTA Protein
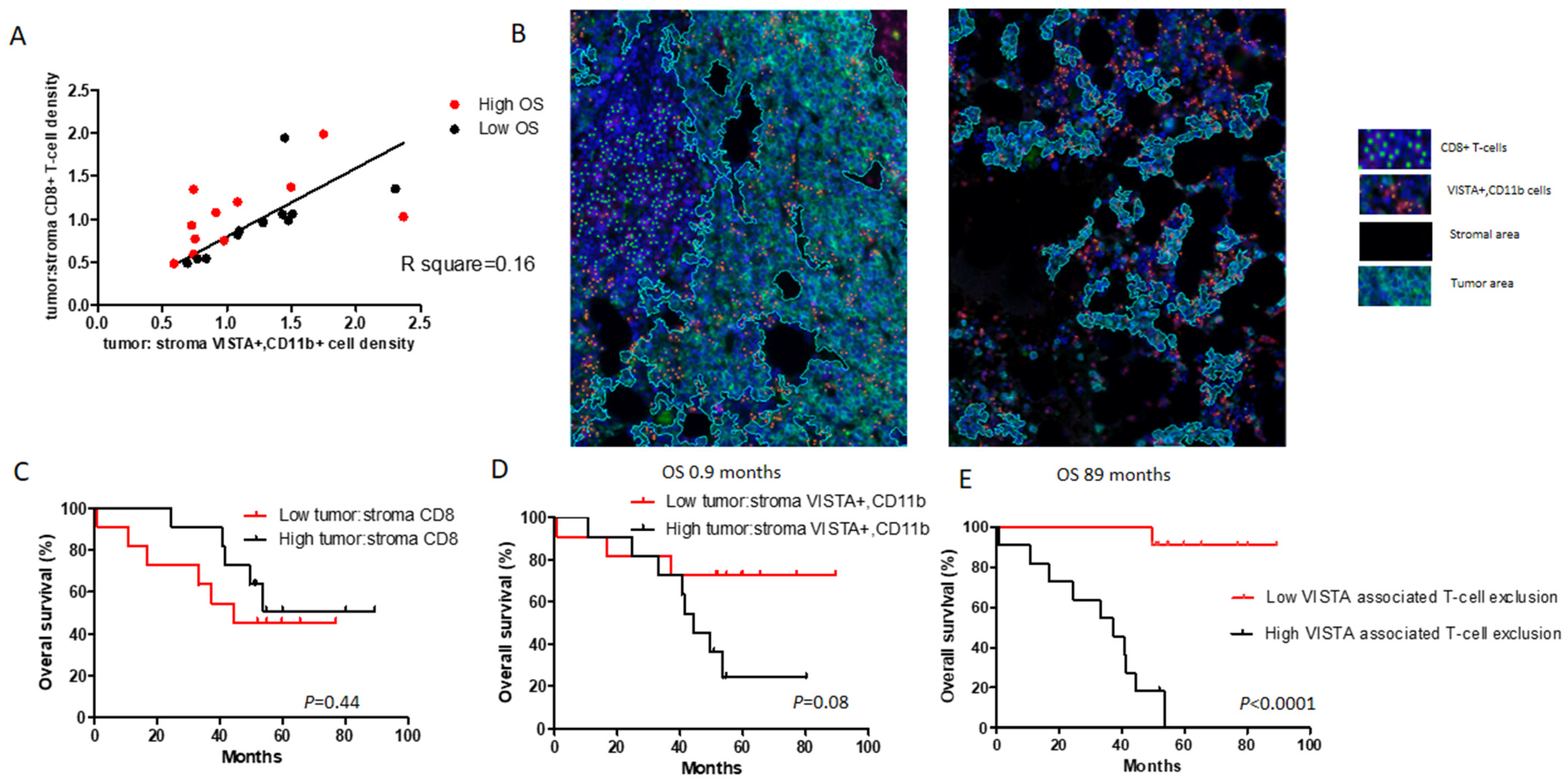
2.3. Densities of VISTA+, CD11b+ Cells or Distances between VISTA+, CD11b+, and CD8+ T-Cells in Different Tissue Compartments Do Not Correlate to OS in MM
2.4. High Density of VISTA+, CD11b+ Cells in Tumor over Stroma Combined with Low Density of CD8+ T-Cells in Tumor over Stroma Associates with Short OS in MM
3. Discussion
4. Materials and Methods
4.1. Patients and Assessment of Clinical Responses
4.2. Bone Marrow Aspirates and Gene Expression Analysis
4.3. Frequencies of Immune Cell Populations
4.4. Multiplexed Immunofluorescence (IF)
4.5. Analysis of Multiplexed Immunofluorescence Images
4.6. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dhodapkar, M.V.; Krasovsky, J.; Osman, K.; Geller, M.D. Vigorous Premalignancy-specific Effector T Cell Response in the Bone Marrow of Patients with Monoclonal Gammopathy. J. Exp. Med. 2003, 198, 1753–1757. [Google Scholar] [CrossRef] [Green Version]
- Suen, H.; Brown, R.; Yang, S.; Weatherburn, C.; Ho, P.J.; Woodland, N.; Nassif, N.; Barbaro, P.; Bryant, C.; Hart, D.; et al. Multiple myeloma causes clonal T-cell immunosenescence: Identification of potential novel targets for promoting tumour immunity and implications for checkpoint blockade. Leukemia 2016, 30, 1716–1724. [Google Scholar] [CrossRef]
- Debets, R.; Donnadieu, E.; Chouaib, S.; Coukos, G. TCR-engineered T cells to treat tumors: Seeing but not touching? Semin. Immunol. 2016, 28, 10–21. [Google Scholar] [CrossRef]
- Hammerl, D.; Rieder, D.; Martens, J.W.; Trajanoski, Z.; Debets, R. Adoptive T Cell Therapy: New Avenues Leading to Safe Targets and Powerful Allies. Trends Immunol. 2018, 39, 921–936. [Google Scholar] [CrossRef]
- Giannopoulos, K.; Kaminska, W.; Hus, I.; Dmoszynska, A. The frequency of T regulatory cells modulates the survival of multiple myeloma patients: Detailed characterisation of immune status in multiple myeloma. Br. J. Cancer 2012, 106, 546–552. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, I.R.; Martner, A.; Pisklakova, A.; Condamine, T.; Chase, T.; Vogl, T.; Roth, J.; Gabrilovich, D.; Nefedova, Y. Myeloid-Derived Suppressor Cells Regulate Growth of Multiple Myeloma by Inhibiting T Cells in Bone Marrow. J. Immunol. 2013, 190, 3815–3823. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zhang, L.; Wang, H.; Xiong, S.; Li, Y.; Tao, Q.; Xiao, W.; Qin, H.; Wang, Y.; Zhai, Z. Tumor-induced CD14+HLA-DR−/low myeloid-derived suppressor cells correlate with tumor progression and outcome of therapy in multiple myeloma patients. Cancer Immunol. Immunother. 2015, 64, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Paiva, B.S.R.; Azpilikueta, A.; Puig, N.; Ocio, E.M.; Sharma, R.K.; Oyajobi, B.O.; Labiano, S.; San-Segundo, L.; Rodriguez, A.C.; Airesmejia, I.; et al. PD-L1/PD-1 presence in the tumor microenvironment and activity of PD-1 blockade in multiple myeloma. Leuk. 2015, 29, 2110–2113. [Google Scholar] [CrossRef]
- Bae, J.; Nguyen, B.; Tai, Y.-T.; Hideshima, T.; Chauhan, D.; Munshi, N.C.; Anderson, K.C. Function and expression of checkpoint inhibitors and immune agonists on immune cells in monoclonal gammopathy of undetermined significance (MGUS), smoldering multiple myeloma (SMM) and MM and tumor-specific T lymphocytes. J. Clin. Oncol. 2017, 35, 11577. [Google Scholar] [CrossRef]
- Lesokhin, A.M.; Ansell, S.M.; Armand, P.; Scott, E.C.; Halwani, A.; Gutierrez, M.; Millenson, M.M.; Cohen, A.D.; Schuster, S.J.; Lebovic, D.; et al. Nivolumab in Patients With Relapsed or Refractory Hematologic Malignancy: Preliminary Results of a Phase Ib Study. J. Clin. Oncol. 2016, 34, 2698–2704. [Google Scholar] [CrossRef] [Green Version]
- Badros, A.; Hyjek, E.; Ma, N.; Lesokhin, A.; Dogan, A.; Rapoport, A.P.; Kocoglu, M.; Lederer, E.; Philip, S.; Milliron, T.; et al. Pembrolizumab, pomalidomide, and low-dose dexamethasone for relapsed/refractory multiple myeloma. Blood 2017, 130, 1189–1197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mateos, M.; Orlowski, R.Z.; Ocio, E.M.; Rodríguez-Otero, P.; Reece, D.; Moreau, P.; Munshi, N.; Avigan, D.E.; Siegel, D.S.; Ghori, R.; et al. Pembrolizumab combined with lenalidomide and low-dose dexamethasone for relapsed or refractory multiple myeloma: Phase I KEYNOTE-023 study. Br. J. Haematol. 2019, 186, e117–e121. [Google Scholar] [CrossRef] [Green Version]
- Gormley, N.J.; Pazdur, R. Immunotherapy Combinations in Multiple Myeloma—Known Unknowns. N. Engl. J. Med. 2018, 379, 1791–1795. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [Green Version]
- Hammerl, D.; Massink, M.P.G.; Smid, M.; van Deurzen, C.H.M.; Meijers-Heijboer, H.E.J.; Waisfisz, Q.; Debets, R.; Martens, J.W.M. Clonality, Antigen Recognition, and Suppression of CD8+ T Cells Differentially Affect Prognosis of Breast Cancer Subtypes. Clin. Cancer Res. 2019, 26, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wu, G.; Manick, B.; Hernandez, V.; Renelt, M.; Erickson, C.; Guan, J.; Singh, R.; Rollins, S.; Solorz, A.; et al. VSIG-3 as a ligand of VISTA inhibits human T-cell function. Immunology 2019, 156, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Johnston, R.J.; Su, L.J.; Pinckney, J.; Critton, D.; Boyer, E.; Krishnakumar, A.; Corbett, M.; Rankin, A.L.; Dibella, R.; Campbell, L.; et al. VISTA is an acidic pH-selective ligand for PSGL-1. Nature 2019, 574, 565–570. [Google Scholar] [CrossRef]
- Tagliamento, M.; Bironzo, P.; Novello, S. New emerging targets in cancer immunotherapy: The role of VISTA. ESMO Open 2019, 4, e000683. [Google Scholar] [CrossRef]
- Wang, L.; Rubinstein, R.; Lines, J.L.; Wasiuk, A.; Ahonen, C.; Guo, Y.; Lu, L.-F.; Gondek, D.; Wang, Y.; Fava, R.A.; et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J. Exp. Med. 2011, 208, 577–592. [Google Scholar] [CrossRef] [PubMed]
- Lines, J.L.; Pantazi, E.; Mak, J.; Sempere, L.F.; Wang, L.; O’Connell, S.; Ceeraz, S.; Suriawinata, A.A.; Yan, S.; Ernstoff, M.S.; et al. VISTA Is an Immune Checkpoint Molecule for Human T Cells. Cancer Res. 2014, 74, 1924–1932. [Google Scholar] [CrossRef] [Green Version]
- Le Mercier, I.; Chen, W.; Lines, J.L.; Day, M.; Li, J.; Sergent, P.; Noelle, R.J.; Wang, L. VISTA Regulates the Development of Protective Antitumor Immunity. Cancer Res. 2014, 74, 1933–1944. [Google Scholar] [CrossRef] [Green Version]
- Sharpe, A.H.; Pauken, K.E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 2018, 18, 153–167. [Google Scholar] [CrossRef]
- Gooden, M.J.M.; De Bock, G.H.; Leffers, N.; Daemen, T.; Nijman, H.W. The prognostic influence of tumour-infiltrating lymphocytes in cancer: A systematic review with meta-analysis. Br. J. Cancer 2011, 105, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Lines, J.L.; Sempere, L.F.; Broughton, T.; Wang, L.; Noelle, R. VISTA Is a Novel Broad-Spectrum Negative Checkpoint Regulator for Cancer Immunotherapy. Cancer Immunol. Res. 2014, 2, 510–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Pang, H.-J.; Zhao, W.; Li, Y.-F.; Yan, L.-X.; Dong, Z.-Y.; He, X.-F. VISTA expression associated with CD8 confers a favorable immune microenvironment and better overall survival in hepatocellular carcinoma. BMC Cancer 2018, 18, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Deng, W.-W.; Huang, C.-F.; Bu, L.-L.; Yu, G.-T.; Mao, L.; Zhang, W.-F.; Liu, B.; Sun, Z.-J. Expression of VISTA correlated with immunosuppression and synergized with CD8 to predict survival in human oral squamous cell carcinoma. Cancer Immunol. Immunother. 2017, 66, 627–636. [Google Scholar] [CrossRef]
- Chaudhary, B.; Elkord, E. Regulatory T Cells in the Tumor Microenvironment and Cancer Progression: Role and Therapeutic Targeting. Vaccines 2016, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Spranger, S.; Spaapen, R.M.; Zha, Y.; Williams, J.; Meng, Y.; Ha, T.T.; Gajewski, T.F. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 2013, 5, 200ra116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef] [Green Version]
- Holm, S. A Simple Sequentially Rejective Multiple Test Procedure. Scand. J. Stat. 1979, 6, 65–70. [Google Scholar]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Discovery n = 1045 | n | Type | Treatment | Analysis Platform |
|---|---|---|---|---|
| HOVON65/GMMG-HD4 | 327 | NDMM 1 | PAD/VAD 3 | Affymetrix Gene chip Plus 2.0 |
| UAMS-TT2 | 345 | NDMM | TD/VincristineDex 4 | Affymetrix Gene chip Plus 2.0 |
| MRC-IX non-IC | 109 | NDMM | CTDa/MP 5 | Affymetrix Gene chip Plus 2.0 |
| APEX | 264 | RRMM 2 | BOR/DEX 6 | Affymetrix Gene chip Plus A + B |
| Validation n = 609 | ||||
| HOVON87/NMSG18 | 178 | NDMM | MPT-T/MPR-R 7 | Affymetrix Gene chip Plus 2.0 |
| UAMS-TT3 | 238 | NDMM | VTD 8 | Affymetrix Gene chip Plus 2.0 |
| MRC-IX intensive | 138 | NDMM | CTD/CVAD 9 | Affymetrix Gene chip Plus 2.0 |
| UAMS-TT6 | 55 | RRMM | VTD | Affymetrix Gene chip Plus 2.0 |
| 2nd validation cohort n = 1000 | ||||
| CoMMpass | 718 | NDMM/RRMM | Multiple 1st, 2nd and 3rd regimens | RNAseq |
| Categories of Immune Evasion | p Value Discovery | p Value Validation | ||||
|---|---|---|---|---|---|---|
| Multivariate | Multivariate Holm | Passed | Multivariate | Multivariate Holm | Passed | |
| Antigen presentation (n = 14) | 0.92 × 10−3 | 9.20 × 10−4 | Yes | 1.50 × 10−1 | 1.50 × 10−1 | No |
| Immune cells (n = 1) | 2.78 × 10−7 | 5.57 × 10−7 | Yes | 6.00 × 10−2 | 1.20 × 10−1 | No |
| Cell death (n = 51) | 3.85 × 10−20 | 2.31 × 10−19 | Yes | 2.73 × 10−7 | 1.92 × 10−6 | Yes |
| Immune checkpoints (n = 9) | 1.27 × 10−9 | 3.81 × 10−9 | Yes | 2.76 × 10−7 | 1.92 × 10−6 | Yes |
| Metabolic checkpoints (n = 77) | 1.02 × 10−20 | 7.19 × 10−20 | Yes | 4.46 × 10−6 | 2.23 × 10−5 | Yes |
| Oncogenic signaling pathways (n = 38) | 4.18 × 10−16 | 2.09 × 10−15 | Yes | 1.71 × 10−5 | 6.23 × 10−5 | Yes |
| TME (n = 20) | 1.22 × 10−15 | 4.91 × 10−15 | Yes | 1.53 × 10−5 | 6.22 × 10−5 | Yes |
| Categories of Immune Evasion | Gene 1 | HR | 95% CI | p Value |
|---|---|---|---|---|
| Cell death | MAP1LC3A | 0.80 | 0.71–0.90 | 0.001 |
| Cell death | DRAM1 | 0.79 | 0.70–0.90 | 0.017 |
| Immune checkpoints | VISTA | 0.76 | 0.67–0.86 | 0.001 |
| Metabolic checkpoints | MTX2 | 1.30 | 1.14–1.48 | 0.003 |
| Oncogenic signaling pathways | IFI16 | 1.37 | 1.21–1.55 | 0.0005 |
| TME | PECAM1 | 0.73 | 0.64–0.82 | 0.0001 |
| Categories of Immune Evasion | Gene | HR | 95% CI | p Value |
|---|---|---|---|---|
| Cell death | MAP1LC3A | 0.87 | 0.69–1.1 | 0.25 |
| Cell death | DRAM1 | 0.86 | 0.69–1.1 | 0.19 |
| Cell death | BCL2 | 0.87 | 0.7–1.1 | 0.19 |
| Immune checkpoints | CD40 | 0.81 | 0.64–1.0 | 0.07 |
| Immune checkpoints | VISTA | 0.75 | 0.62–0.92 | 0.005 |
| Metabolic checkpoints | MTX2 | 0.88 | 1.7–3.4 | 0.22 |
| Oncogenic signaling pathways | IFI16 | 1.1 | 0.88–1.3 | 0.48 |
| TME | N/A | N/A | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mutsaers, P.; Balcioglu, H.E.; Kuiper, R.; Hammerl, D.; Wijers, R.; van Duin, M.; van der Holt, B.; Broijl, A.; Gregory, W.; Zweegman, S.; et al. V-Domain Ig Suppressor of T Cell Activation (VISTA) Expression Is an Independent Prognostic Factor in Multiple Myeloma. Cancers 2021, 13, 2219. https://doi.org/10.3390/cancers13092219
Mutsaers P, Balcioglu HE, Kuiper R, Hammerl D, Wijers R, van Duin M, van der Holt B, Broijl A, Gregory W, Zweegman S, et al. V-Domain Ig Suppressor of T Cell Activation (VISTA) Expression Is an Independent Prognostic Factor in Multiple Myeloma. Cancers. 2021; 13(9):2219. https://doi.org/10.3390/cancers13092219
Chicago/Turabian StyleMutsaers, Pim, Hayri E. Balcioglu, Rowan Kuiper, Dora Hammerl, Rebecca Wijers, Mark van Duin, Bronno van der Holt, Annemiek Broijl, Walter Gregory, Sonja Zweegman, and et al. 2021. "V-Domain Ig Suppressor of T Cell Activation (VISTA) Expression Is an Independent Prognostic Factor in Multiple Myeloma" Cancers 13, no. 9: 2219. https://doi.org/10.3390/cancers13092219
APA StyleMutsaers, P., Balcioglu, H. E., Kuiper, R., Hammerl, D., Wijers, R., van Duin, M., van der Holt, B., Broijl, A., Gregory, W., Zweegman, S., Sonneveld, P., & Debets, R. (2021). V-Domain Ig Suppressor of T Cell Activation (VISTA) Expression Is an Independent Prognostic Factor in Multiple Myeloma. Cancers, 13(9), 2219. https://doi.org/10.3390/cancers13092219







