Tumor-Associated Regulatory T Cell Expression of LAIR2 Is Prognostic in Lung Adenocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Gene Expression Data Analyses
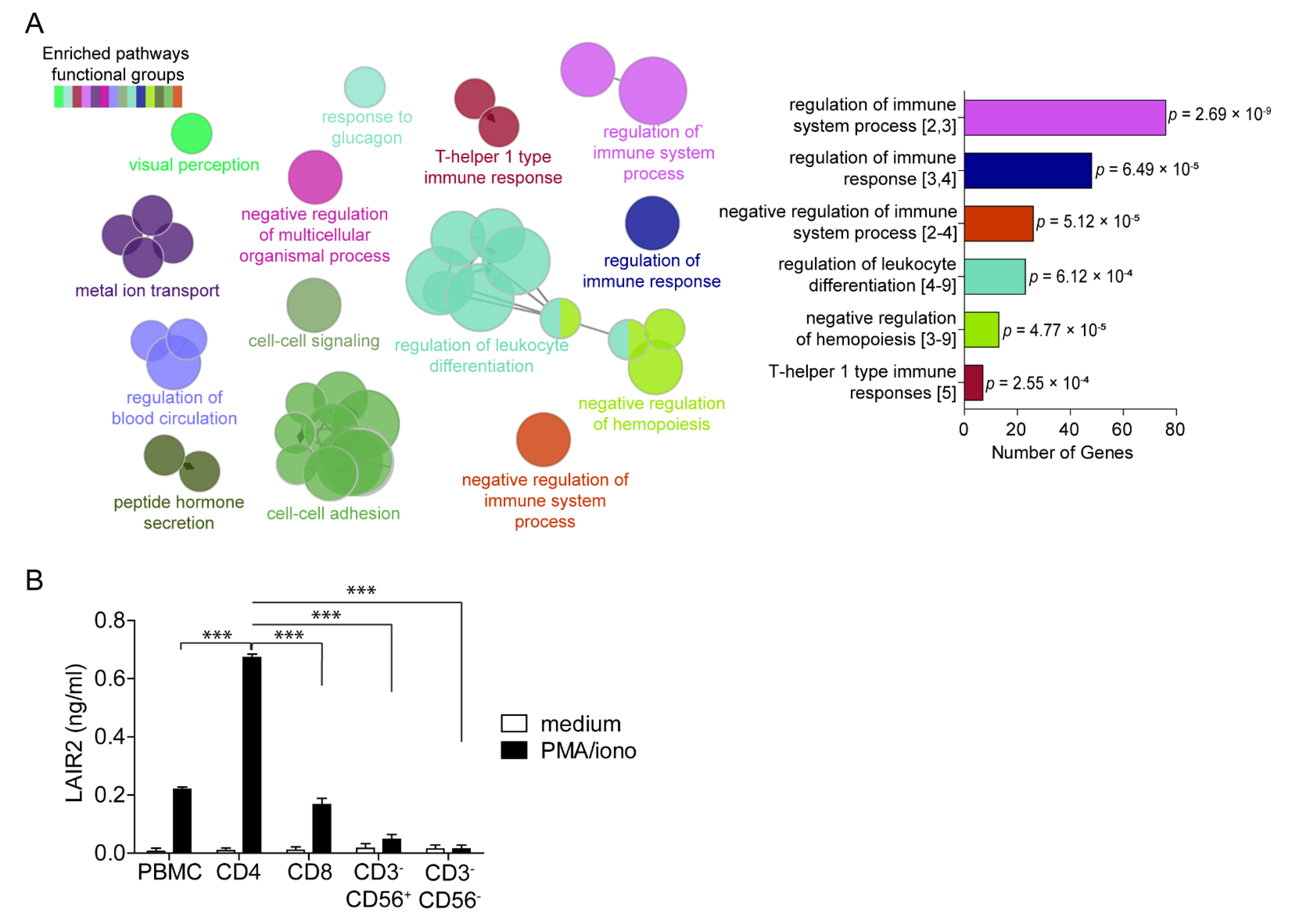
2.2. Pathway Enrichment, Gene Set Enrichment Analysis and Immune Estimation
2.3. Cell Isolation, Expansion and ELISA
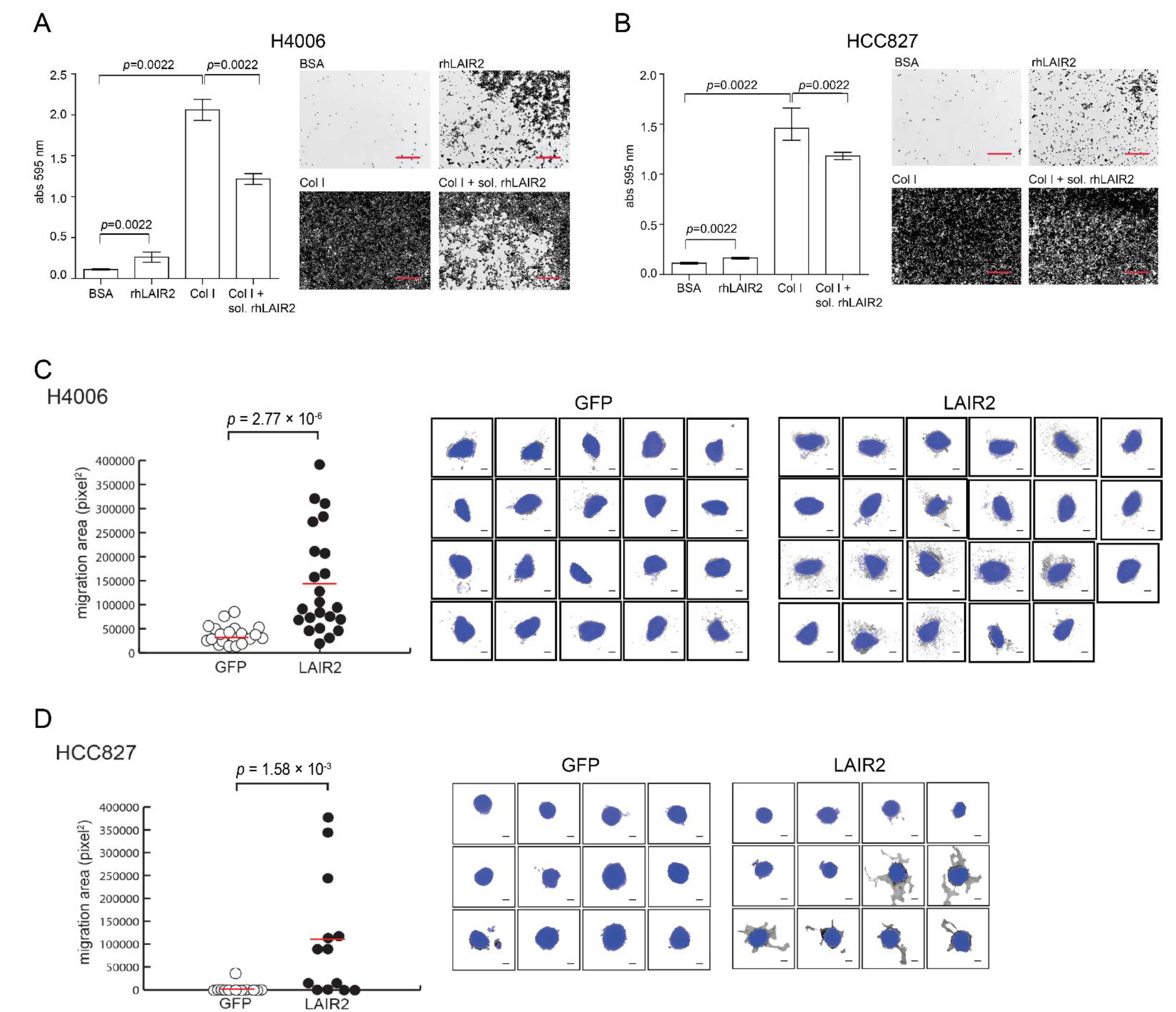
2.4. Cell Adhesion and Invasion Assay
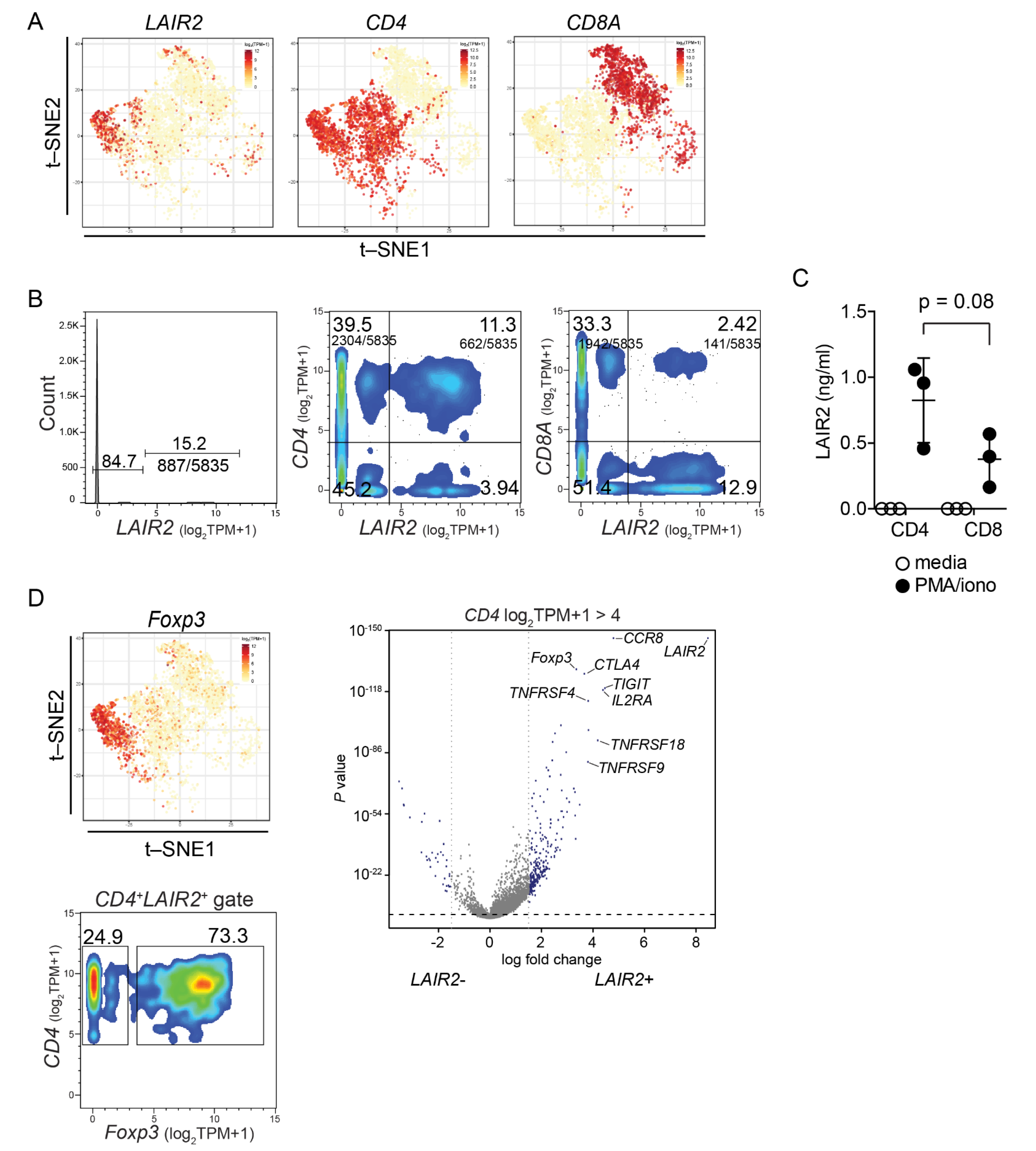
2.5. Single Cell RNASeq Analysis
2.6. Statistical Analysis
3. Results
3.1. High LAIR2 Expression Is Associated with Poor LUAD Patient Survival
3.2. CD4+ T Cells Are a Major Source of Soluble LAIR2
3.3. T Cell Derived LAIR2 Enhances Tumor Cell Invasion
3.4. LAIR2 Is Expressed by Tumor-Associated Treg Cells
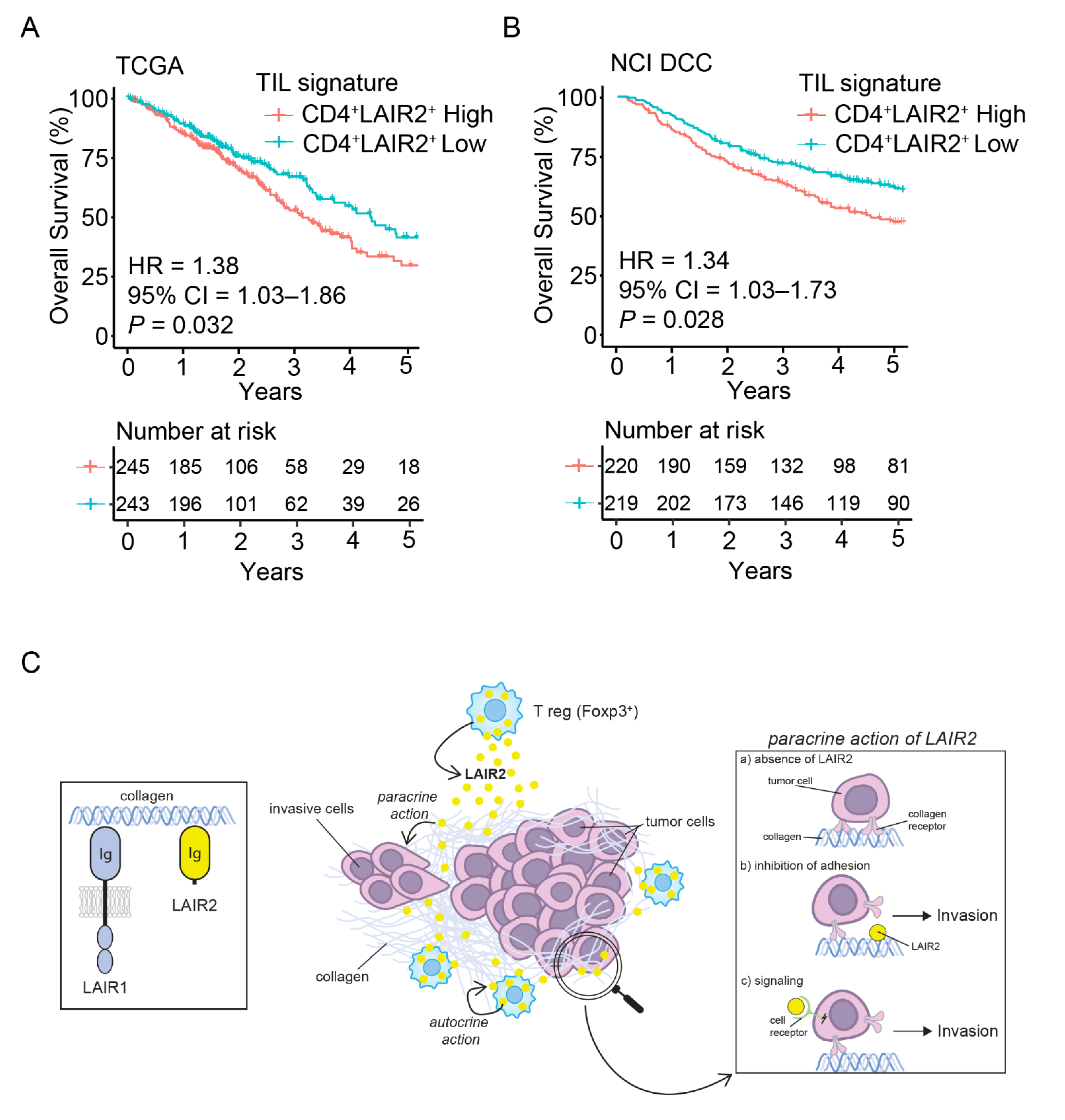
3.5. CD4+ LAIR2+ Tumor-Associated Treg Gene Signature Is Prognostic in LUAD
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus Docetaxel for Previously Treated, PD-L1-Positive, Advanced Non-Small-Cell Lung Cancer (KEYNOTE-010): A Randomised Controlled Trial. Lancet 2016, 387, 1540–1550. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the Tumor Immune Microenvironment (TIME) for Effective Therapy. Nat. Med. 2018, 24, 541. [Google Scholar] [CrossRef]
- Wei, S.C.; Duffy, C.R.; Allison, J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018, 8, 1069–1086. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Song, E. Turning Foes to Friends: Targeting Cancer-Associated Fibroblasts. Nat. Rev. Drug. Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef]
- Joyce, J.A.; Pollard, J.W. Microenvironmental Regulation of Metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Bremnes, R.M.; Busund, L.-T.; Kilvær, T.L.; Andersen, S.; Richardsen, E.; Paulsen, E.E.; Hald, S.; Khanehkenari, M.R.; Cooper, W.A.; Kao, S.C.; et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2016, 11, 789–800. [Google Scholar] [CrossRef] [Green Version]
- Remark, R.; Becker, C.; Gomez, J.E.; Damotte, D.; Dieu-Nosjean, M.-C.; Sautès-Fridman, C.; Fridman, W.-H.; Powell, C.A.; Altorki, N.K.; Merad, M.; et al. The Non-Small Cell Lung Cancer Immune Contexture. A Major Determinant of Tumor Characteristics and Patient Outcome. Am. J. Respir. Crit. Care Med. 2015, 191, 377–390. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Sakaguchi, S. Regulatory T Cells in Cancer Immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Chao, J.L.; Savage, P.A. Unlocking the Complexities of Tumor-Associated Regulatory T Cells. J. Immunol. 2018, 200, 415–421. [Google Scholar] [CrossRef] [Green Version]
- Scott, E.N.; Gocher, A.M.; Workman, C.J.; Vignali, D.A.A. Regulatory T Cells: Barriers of Immune Infiltration Into the Tumor Microenvironment. Front. Immunol. 2021, 12, 702726. [Google Scholar] [CrossRef]
- Shang, B.; Liu, Y.; Jiang, S.; Liu, Y. Prognostic Value of Tumor-Infiltrating FoxP3+ Regulatory T Cells in Cancers: A Systematic Review and Meta-Analysis. Sci. Rep. 2015, 5, 15179. [Google Scholar] [CrossRef] [Green Version]
- De Leeuw, R.J.; Kost, S.E.; Kakal, J.A.; Nelson, B.H. The Prognostic Value of FoxP3+ Tumor-Infiltrating Lymphocytes in Cancer: A Critical Review of the Literature. Clin. Cancer Res. 2012, 18, 3022–3029. [Google Scholar] [CrossRef] [Green Version]
- Saleh, R.; Elkord, E. FoxP3+ T Regulatory Cells in Cancer: Prognostic Biomarkers and Therapeutic Targets. Cancer Lett. 2020, 490, 174–185. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Zheng, L.; Zheng, C.; Song, J.; Zhang, Q.; Kang, B.; Liu, Z.; Jin, L.; Xing, R.; et al. Global Characterization of T Cells in Non-Small-Cell Lung Cancer by Single-Cell Sequencing. Nat. Med. 2018, 24, 978–985. [Google Scholar] [CrossRef]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Zheng, L.; Yoo, J.-K.; Guo, H.; Zhang, Y.; Guo, X.; Kang, B.; Hu, R.; Huang, J.Y.; Zhang, Q.; et al. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell 2017, 169, 1342–1356.e16. [Google Scholar] [CrossRef] [Green Version]
- Miragaia, R.J.; Gomes, T.; Chomka, A.; Jardine, L.; Riedel, A.; Hegazy, A.N.; Whibley, N.; Tucci, A.; Chen, X.; Lindeman, I.; et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 2019, 50, 493–504.e7. [Google Scholar] [CrossRef] [Green Version]
- De Simone, M.; Arrigoni, A.; Rossetti, G.; Gruarin, P.; Ranzani, V.; Politano, C.; Bonnal, R.J.P.; Provasi, E.; Sarnicola, M.L.; Panzeri, I.; et al. Transcriptional Landscape of Human Tissue Lymphocytes Unveils Uniqueness of Tumor-Infiltrating T Regulatory Cells. Immunity 2016, 45, 1135–1147. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yu, X.; Zheng, L.; Zhang, Y.; Li, Y.; Fang, Q.; Gao, R.; Kang, B.; Zhang, Q.; Huang, J.Y.; et al. Lineage Tracking Reveals Dynamic Relationships of T Cells in Colorectal Cancer. Nature 2018, 564, 268–272. [Google Scholar] [CrossRef]
- Peng, D.H.; Rodriguez, B.L.; Diao, L.; Chen, L.; Wang, J.; Byers, L.A.; Wei, Y.; Chapman, H.A.; Yamauchi, M.; Behrens, C.; et al. Collagen Promotes Anti-PD-1/PD-L1 Resistance in Cancer through LAIR1-Dependent CD8+ T Cell Exhaustion. Nat. Commun. 2020, 11, 4520. [Google Scholar] [CrossRef]
- Meyaard, L. The Inhibitory Collagen Receptor LAIR-1 (CD305). J. Leukoc. Biol. 2008, 83, 799–803. [Google Scholar] [CrossRef]
- Lebbink, R.J.; van den Berg, M.C.; de Ruiter, T.; Raynal, N.; van Roon, J.A.; Lenting, P.J.; Jin, B.; Meyaard, L. The Soluble Leukocyte-Associated Ig-like Receptor (LAIR)-2 Antagonizes the Collagen/LAIR-1 Inhibitory Immune Interaction. J. Immunol. 2008, 180, 1662–1669. [Google Scholar] [CrossRef] [Green Version]
- Lebbink, R.J.; Raynal, N.; de Ruiter, T.; Bihan, D.G.; Farndale, R.W.; Meyaard, L. Identification of Multiple Potent Binding Sites for Human Leukocyte Associated Ig-like Receptor LAIR on Collagens II and III. Matrix Biol. 2009, 28, 202–210. [Google Scholar] [CrossRef]
- Meyaard, L.; Adema, G.J.; Chang, C.; Woollatt, E.; Sutherland, G.R.; Lanier, L.L.; Phillips, J.H. LAIR-1, a Novel Inhibitory Receptor Expressed on Human Mononuclear Leukocytes. Immunity 1997, 7, 283–290. [Google Scholar] [CrossRef]
- Son, M.; Santiago-Schwarz, F.; Al-Abed, Y.; Diamond, B. C1q Limits Dendritic Cell Differentiation and Activation by Engaging LAIR-1. Proc. Natl. Acad. Sci. USA 2012, 109, E3160–E3167. [Google Scholar] [CrossRef] [Green Version]
- Olde Nordkamp, M.J.; Boross, P.; Yildiz, C.; Jansen, J.H.; Leusen, J.H.; Wouters, D.; Urbanus, R.T.; Hack, C.E.; Meyaard, L. Inhibition of the Classical and Lectin Pathway of the Complement System by Recombinant LAIR-2. J. Innate Immun. 2014, 6, 284–292. [Google Scholar] [CrossRef]
- Olde Nordkamp, M.J.; van Roon, J.A.; Douwes, M.; de Ruiter, T.; Urbanus, R.T.; Meyaard, L. Enhanced Secretion of Leukocyte-Associated Immunoglobulin-like Receptor 2 (LAIR-2) and Soluble LAIR-1 in Rheumatoid Arthritis: LAIR-2 Is a More Efficient Antagonist of the LAIR-1-Collagen Inhibitory Interaction than Is Soluble LAIR-1. Arthritis Rheum. 2011, 63, 3749–3757. [Google Scholar] [CrossRef]
- Simone, R.; Pesce, G.; Antola, P.; Merlo, D.F.; Bagnasco, M.; Saverino, D. Serum LAIR-2 Is Increased in Autoimmune Thyroid Diseases. PLoS ONE 2013, 8, e63282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Der, S.D.; Sykes, J.; Pintilie, M.; Zhu, C.Q.; Strumpf, D.; Liu, N.; Jurisica, I.; Shepherd, F.A.; Tsao, M.S. Validation of a Histology-Independent Prognostic Gene Signature for Early-Stage, Non-Small-Cell Lung Cancer Including Stage IA Patients. J. Thorac. Oncol. 2014, 9, 59–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganesan, A.-P.; Clarke, J.; Wood, O.; Garrido-Martin, E.M.; Chee, S.J.; Mellows, T.; Samaniego-Castruita, D.; Singh, D.; Seumois, G.; Alzetani, A.; et al. Tissue-Resident Memory Features Are Linked to the Magnitude of Cytotoxic T Cell Responses in Human Lung Cancer. Nat. Immunol. 2017, 18, 940–950. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. XCell: Digitally Portraying the Tissue Cellular Heterogeneity Landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef] [Green Version]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcicoma; Shedden, K.; Taylor, J.M.; Enkemann, S.A.; Tsao, M.S.; Yeatman, T.J.; Gerald, W.L.; Eschrich, S.; Jurisica, I.; Giordano, T.J.; et al. Gene Expression-Based Survival Prediction in Lung Adenocarcinoma: A Multi-Site, Blinded Validation Study. Nat. Med. 2008, 14, 822–827. [Google Scholar] [CrossRef]
- Viechtbauer, W. Conducting Meta-Analyses in R with the Metafor Package. J. Stat. Softw. 2010, 36, 48. [Google Scholar] [CrossRef] [Green Version]
- Fang, M.; Yuan, J.; Peng, C.; Li, Y. Collagen as a Double-Edged Sword in Tumor Progression. Tumour Biol. 2014, 35, 2871–2882. [Google Scholar] [CrossRef] [Green Version]
- Walker, C.; Mojares, E.; Del Río Hernández, A. Role of Extracellular Matrix in Development and Cancer Progression. Int. J. Mol. Sci. 2018, 19, 3028. [Google Scholar] [CrossRef] [Green Version]
- Egeblad, M.; Rasch, M.G.; Weaver, V.M. Dynamic Interplay between the Collagen Scaffold and Tumor Evolution. Curr. Opin. Cell Biol. 2010, 22, 697–706. [Google Scholar] [CrossRef] [Green Version]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The Role of the Extracellular Matrix and the Heparan Sulfate Proteoglycan Perlecan. Front. Oncol. 2020, 9, 1482. [Google Scholar] [CrossRef] [Green Version]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Fan, J.; He, Y.; Xiong, A.; Yu, J.; Li, Y.; Zhang, Y.; Zhao, W.; Zhou, F.; Li, W.; et al. Single-Cell Profiling of Tumor Heterogeneity and the Microenvironment in Advanced Non-Small Cell Lung Cancer. Nat. Commun. 2021, 12, 2540. [Google Scholar] [CrossRef]
- Liu, X.; Shang, X.; Li, J.; Zhang, S. The Prognosis and Immune Checkpoint Blockade Efficacy Prediction of Tumor-Infiltrating Immune Cells in Lung Cancer. Front. Cell Dev. Biol. 2021, 9, 707143. [Google Scholar] [CrossRef]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [Green Version]
- Kuczek, D.E.; Larsen, A.M.H.; Thorseth, M.-L.; Carretta, M.; Kalvisa, A.; Siersbæk, M.S.; Simões, A.M.C.; Roslind, A.; Engelholm, L.H.; Noessner, E.; et al. Collagen Density Regulates the Activity of Tumor-Infiltrating T Cells. J. Immunother. Cancer 2019, 7, 68. [Google Scholar] [CrossRef] [Green Version]
- Principe, D.R.; Chiec, L.; Mohindra, N.A.; Munshi, H.G. Regulatory T-Cells as an Emerging Barrier to Immune Checkpoint Inhibition in Lung Cancer. Front. Oncol. 2021, 11, 684098. [Google Scholar] [CrossRef]
- Kumagai, S.; Togashi, Y.; Kamada, T.; Sugiyama, E.; Nishinakamura, H.; Takeuchi, Y.; Vitaly, K.; Itahashi, K.; Maeda, Y.; Matsui, S.; et al. The PD-1 Expression Balance between Effector and Regulatory T Cells Predicts the Clinical Efficacy of PD-1 Blockade Therapies. Nat. Immunol. 2020, 21, 1346–1358. [Google Scholar] [CrossRef]
- Wu, S.-P.; Liao, R.-Q.; Tu, H.-Y.; Wang, W.-J.; Dong, Z.-Y.; Huang, S.-M.; Guo, W.-B.; Gou, L.-Y.; Sun, H.-W.; Zhang, Q.; et al. Stromal PD-L1-Positive Regulatory T Cells and PD-1-Positive CD8-Positive T Cells Define the Response of Different Subsets of Non-Small Cell Lung Cancer to PD-1/PD-L1 Blockade Immunotherapy. J. Thorac. Oncol. 2018, 13, 521–532. [Google Scholar] [CrossRef] [Green Version]
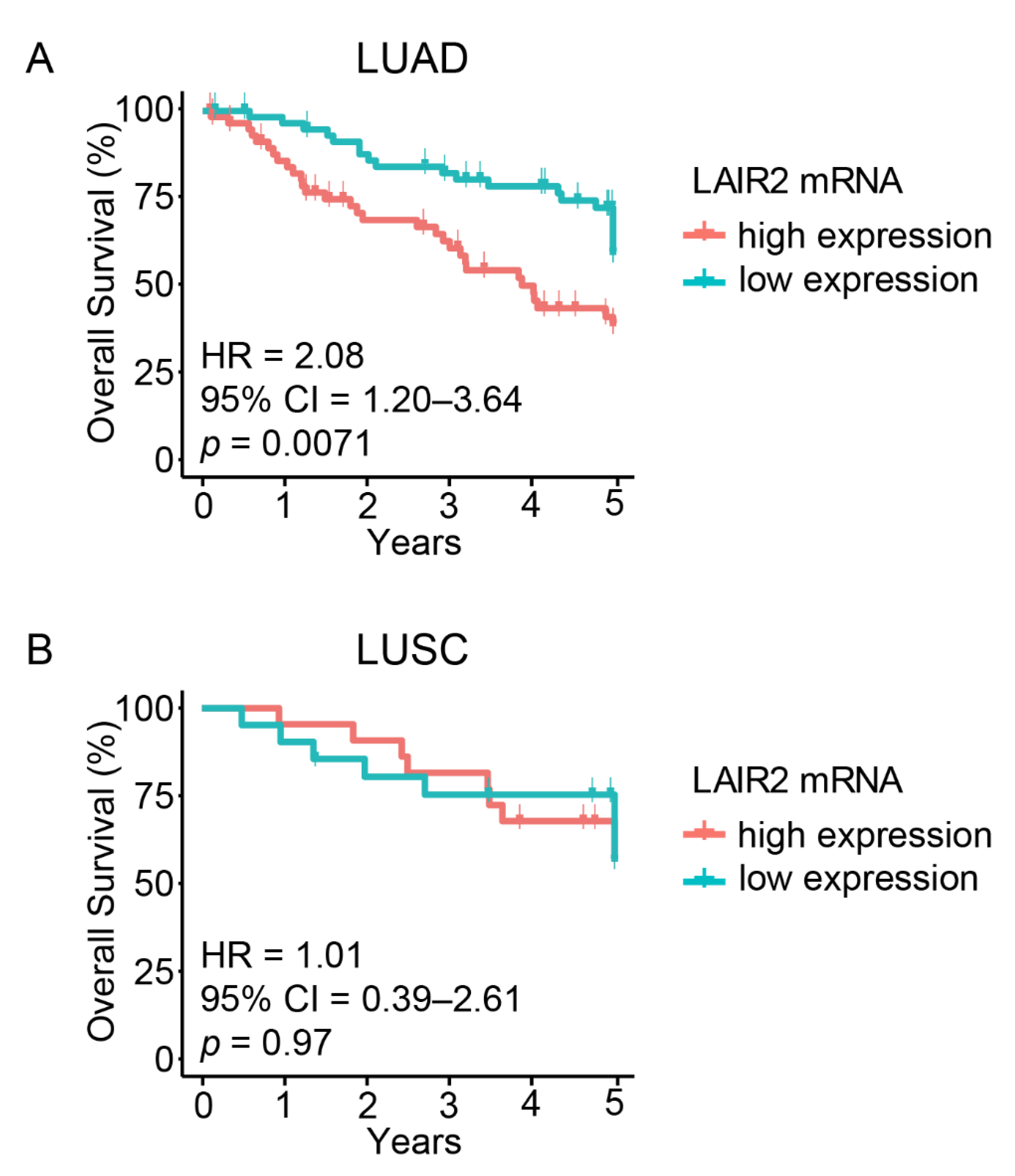
| Parameter | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-Value | HR | 95% CI | p-Value | |
| LAIR2 (high vs. low) | 2.08 | 1.20–3.64 | 0.0071 | 1.96 | 1.05–3.66 | 0.03 |
| LAIR1 (high vs. low) | 1.62 | 0.94–2.80 | 0.083 | 1.16 | 0.62–2.15 | 0.64 |
| Age (older vs. younger) | 1.00 | 0.99–1.1 | 0.12 | 1.02 | 0.99–1.05 | 0.13 |
| Sex (female vs. male) | 0.76 | 0.44–1.3 | 0.33 | 0.75 | 0.42–1.36 | 0.35 |
| Smoking (no vs. yes) | 0.96 | 0.62–1.5 | 0.87 | 0.99 | 0.62–1.59 | 0.97 |
| Stage (Stage II vs. I) | 2.4 | 1.4–4.2 | 0.002 | 2.21 | 1.22–3.98 | 0.008 |
| Histological Pattern (micropapillary/solid vs. acinar/papillar/lepidic) | 0.95 | 0.63–1.4 | 0.79 | 0.92 | 0.61–1.4 | 0.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ly, D.; Li, Q.; Navab, R.; Zeltz, C.; Fang, L.; Cabanero, M.; Zhu, C.-Q.; Tsao, M.-S.; Zhang, L. Tumor-Associated Regulatory T Cell Expression of LAIR2 Is Prognostic in Lung Adenocarcinoma. Cancers 2022, 14, 205. https://doi.org/10.3390/cancers14010205
Ly D, Li Q, Navab R, Zeltz C, Fang L, Cabanero M, Zhu C-Q, Tsao M-S, Zhang L. Tumor-Associated Regulatory T Cell Expression of LAIR2 Is Prognostic in Lung Adenocarcinoma. Cancers. 2022; 14(1):205. https://doi.org/10.3390/cancers14010205
Chicago/Turabian StyleLy, Dalam, Quan Li, Roya Navab, Cédric Zeltz, Linan Fang, Michael Cabanero, Chang-Qi Zhu, Ming-Sound Tsao, and Li Zhang. 2022. "Tumor-Associated Regulatory T Cell Expression of LAIR2 Is Prognostic in Lung Adenocarcinoma" Cancers 14, no. 1: 205. https://doi.org/10.3390/cancers14010205






