Infrared Spectroscopy of Urine for the Non-Invasive Detection of Endometrial Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Study Design
2.2. Sample Acquisition, Processing and Storage
2.3. Histopathological Assessment
2.4. Spectral Acquisition
2.5. Computational Analysis
2.6. Statistical Analysis
3. Results
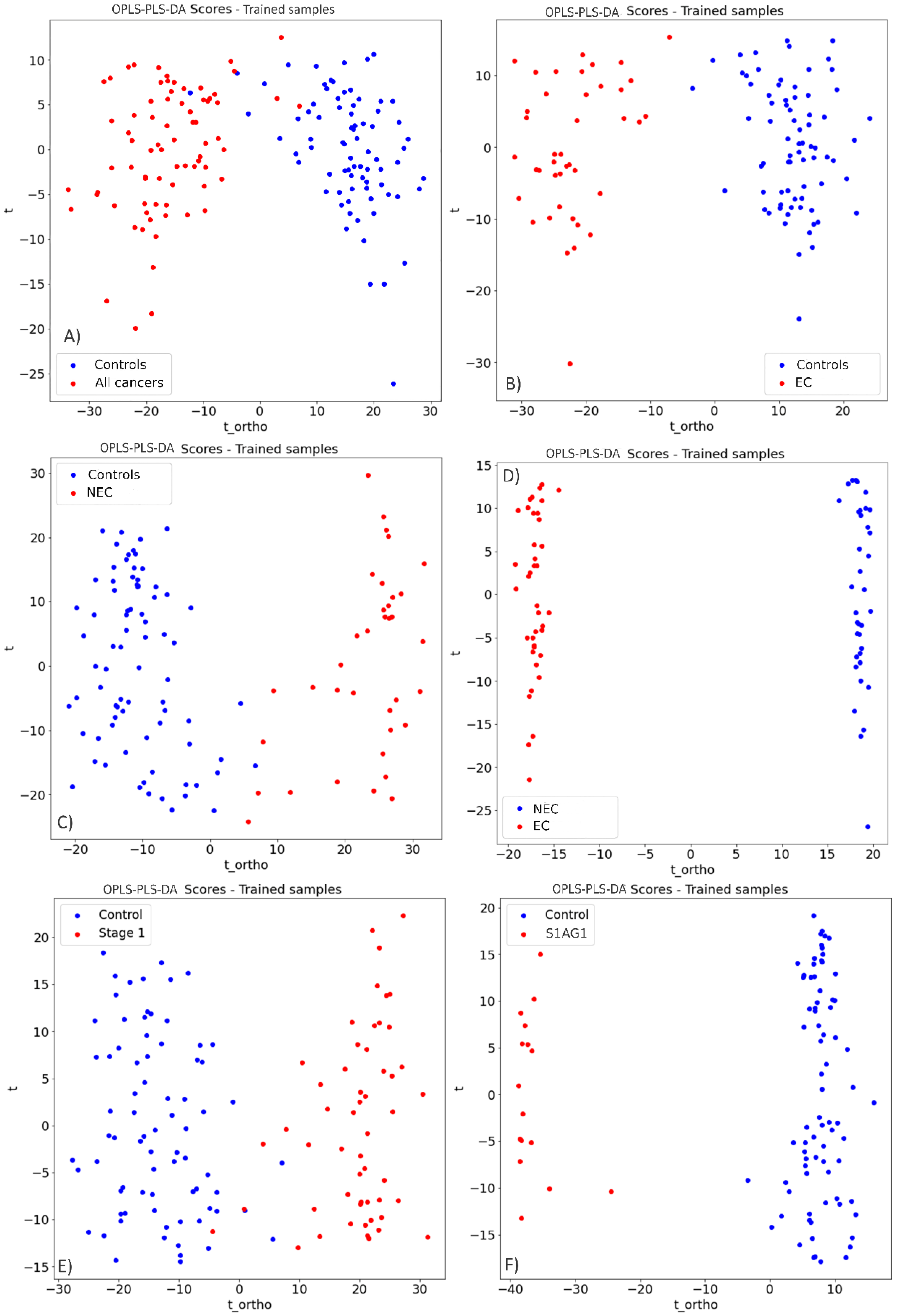
3.1. Endometrial Cancer Detection
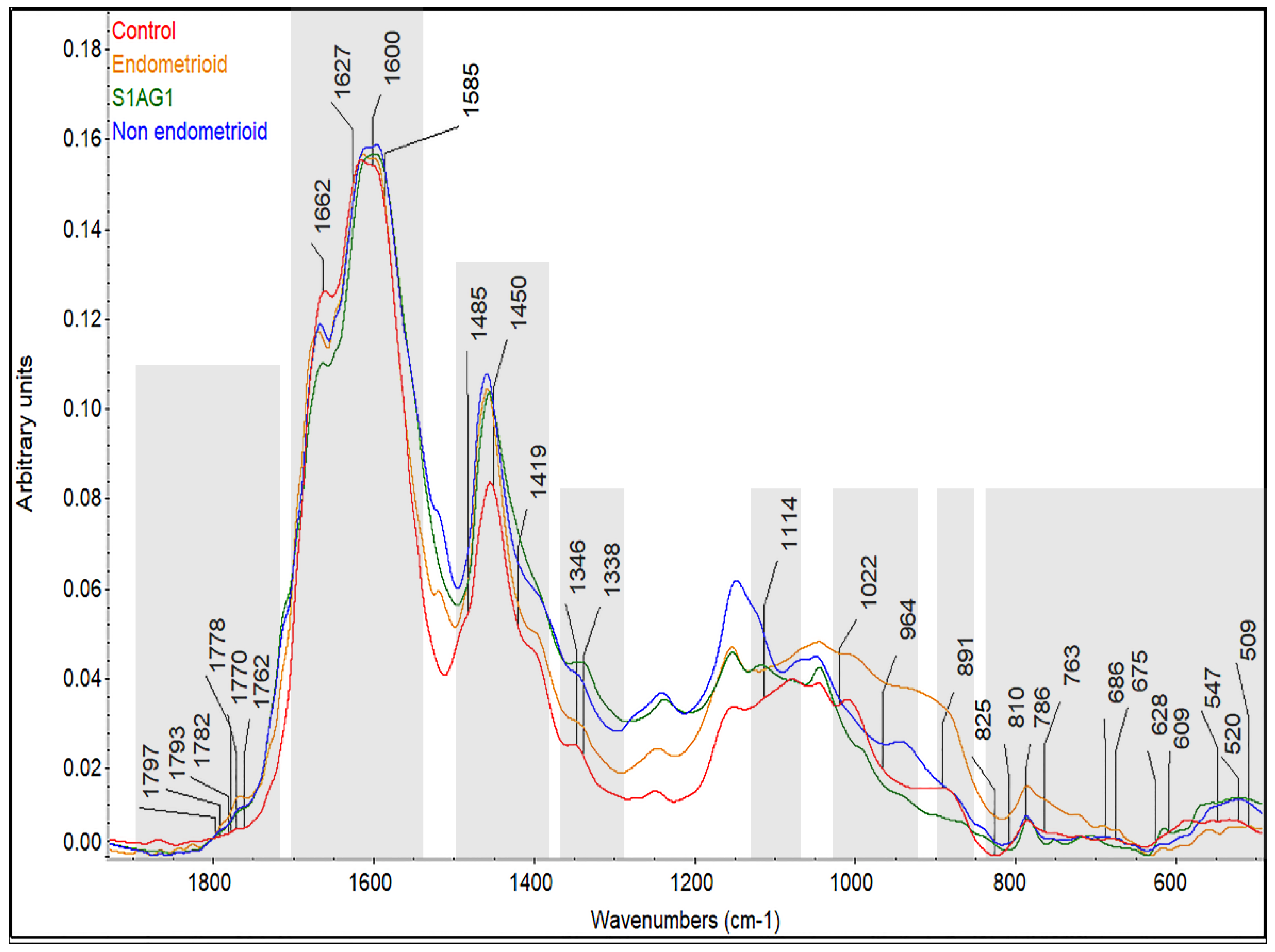
3.2. Prospective Spectral Biomarkers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Lortet-Tieulent, J.; Ferlay, J.; Bray, F.; Jemal, A. International Patterns and Trends in Endometrial Cancer Incidence, 1978–2013. JNCI J. Natl. Cancer Inst. 2018, 110, 354–361. [Google Scholar] [CrossRef]
- Crosbie, E.J.; Kitson, S.J.; McAlpine, J.N.; Mukhopadhyay, A.; Powell, M.E.; Singh, N. Endometrial cancer. Lancet 2022, 399, 1412–1428. [Google Scholar] [CrossRef]
- Cancer Research UK. Uterine Cancer. 2010. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/uterine-cancer#heading-Zero (accessed on 15 November 2021).
- Funston, G.; O’Flynn, H.; Ryan, N.A.J.; Hamilton, W.; Crosbie, E.J. Recognizing Gynecological Cancer in Primary Care: Risk Factors, Red Flags, and Referrals. Adv. Ther. 2018, 35, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.R.; O’Flynn, H.; Njoku, K.; Crosbie, E.J. Detecting endometrial cancer. Obstet. Gynaecol. 2021, 23, 103–112. [Google Scholar] [CrossRef]
- Long, B.; Clarke, M.A.; Morillo, A.D.M.; Wentzensen, N.; Bakkum-Gamez, J.N. Ultrasound detection of endometrial cancer in women with postmenopausal bleeding: Systematic review and meta-analysis. Gynecol. Oncol. 2020, 157, 624–633. [Google Scholar] [CrossRef] [PubMed]
- van Hanegem, N.; Prins, M.M.; Bongers, M.Y.; Opmeer, B.C.; Sahota, D.S.; Mol, B.W.; Timmermans, A. The accuracy of endometrial sampling in women with postmenopausal bleeding: A systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 2016, 197, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.L. Shining a new light into molecular workings. Nat. Methods 2011, 8, 385–387. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.G.; Trevisan, J.; Scott, A.D.; Carmichael, P.L.; Pollock, H.M.; Martin-Hirsch, P.L.; Martin, F.L. Biospectroscopy to metabolically profile biomolecular structure: A multistage approach linking computational analysis with biomarkers. J. Proteome Res. 2011, 10, 1437–1448. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Sitnikova, V.E.; Kotkova, M.A.; Nosenko, T.N.; Kotkova, T.N.; Martynova, D.M.; Uspenskaya, M.V. Breast cancer detection by ATR-FTIR spectroscopy of blood serum and multivariate data-analysis. Talanta 2020, 214, 120857. [Google Scholar] [CrossRef] [PubMed]
- Butler, H.J.; Brennan, P.M.; Cameron, J.M.; Finlayson, D.; Hegarty, M.G.; Jenkinson, M.D.; Palmer, D.S.; Smith, B.R.; Baker, M.J. Development of high-throughput ATR-FTIR technology for rapid triage of brain cancer. Nat. Commun. 2019, 10, 4501. [Google Scholar] [CrossRef] [PubMed]
- Maitra, I.; Morais, C.L.M.; Lima, K.M.G.; Ashton, K.M.; Date, R.S.; Martin, F.L. Attenuated total reflection Fourier-transform infrared spectral discrimination in human bodily fluids of oesophageal transformation to adenocarcinoma. Analyst 2019, 144, 7447–7456. [Google Scholar] [CrossRef] [PubMed]
- Gajjar, K.; Trevisan, J.; Owens, G.; Keating, P.J.; Wood, N.J.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Fourier-transform infrared spectroscopy coupled with a classification machine for the analysis of blood plasma or serum: A novel diagnostic approach for ovarian cancer. Analyst 2013, 138, 3917–3926. [Google Scholar] [CrossRef] [PubMed]
- Medipally, D.K.R.; Cullen, D.; Untereiner, V.; Sockalingum, G.D.; Maguire, A.; Nguyen, T.N.Q.; Bryant, J.; Noone, E.; Bradshaw, S.; Finn, M.; et al. Vibrational spectroscopy of liquid biopsies for prostate cancer diagnosis. Ther. Adv. Med. Oncol. 2020, 12, 175883592091849. [Google Scholar] [CrossRef] [PubMed]
- Paraskevaidi, M.; Morais, C.L.M.; Ashton, K.M.; Stringfellow, H.F.; McVey, R.J.; Ryan, N.A.J.; O’Flynn, H.; Sivalingam, V.N.; Kitson, S.J.; Mackintosh, M.L.; et al. Detecting Endometrial Cancer by Blood Spectroscopy: A Diagnostic Cross-Sectional Study. Cancers 2020, 12, 1256. [Google Scholar] [CrossRef] [PubMed]
- Paraskevaidi, M.; Morais, C.L.M.; Lima, K.M.G.; Ashton, K.M.; Stringfellow, H.F.; Martin-Hirsch, P.L.; Martin, F.L. Potential of mid-infrared spectroscopy as a non-invasive diagnostic test in urine for endometrial or ovarian cancer. Analyst 2018, 143, 3156–3163. [Google Scholar] [CrossRef]
- Mabwa, D.; Gajjar, K.; Furniss, D.; Schiemer, R.; Crane, R.; Fallaize, C.; Martin-Hirsch, P.L.; Martin, F.L.; Kypraios, T.; Seddon, A.B.; et al. Mid-infrared spectral classification of endometrial cancer compared to benign controls in serum or plasma samples. Analyst 2021, 146, 5631–5642. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.J.; Hussain, S.R.; Lovergne, L.; Untereiner, V.; Hughes, C.; Lukaszewski, R.A.; Thiéfin, G.; Sockalingum, G.D. Developing and understanding biofluid vibrational spectroscopy: A critical review. Chem. Soc. Rev. 2016, 45, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Paraskevaidi, M.; Baker, M.J.; Butler, J.H.; Hugh, J.H.; Chakkumpulakkal, T.P.V.; Loren, C.; Crean, S.; Gardener, P.; Gassner, C.S.; Kazarian, G.; et al. Clinical applications of infrared and Raman spectroscopy in the fields of cancer and infectious diseases. Appl. Spectrosc. Rev. 2021, 56, 804–868. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Feng, S.; Zheng, Z.; Xu, Y.; Lin, J.; Chen, G.; Weng, C.; Lin, D.; Qiu, S.; Cheng, M.; Huang, Z.; et al. A noninvasive cancer detection strategy based on gold nanoparticle surface-enhanced raman spectroscopy of urinary modified nucleosides isolated by affinity chromatography. Biosens. Bioelectron. 2020, 91, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Maitra, I.; Morais, C.L.M.; Lima, K.M.G.; Ashton, K.M.; Date, R.S.; Martin, F.L. Raman spectral discrimination in human liquid biopsies of oesophageal transformation to adenocarcinoma. J. Biophotonics 2019, 13, e201960132. [Google Scholar] [CrossRef] [PubMed]
- Brindha, E.; Rajasekaran, R.; Aruna, P.; Koteeswaran, D.; Ganesan, S. High wavenumber Raman spectroscopy in the characterization of urinary metabolites of normal subjects, oral premalignant and malignant patients. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 171, 52–59. [Google Scholar] [CrossRef]
- Pecorelli, S. Revised FIGO staging for carcinoma of the vulva, cervix, and endometrium. Int. J. Gynecol. Obstet. 2018, 105, 103–104. [Google Scholar] [CrossRef]
- Mbatani, N.; Olawaiye, A.B.; Prat, J. Uterine sarcomas. Int. J. Gynecol. Obstet. 2018, 143, 51–58. [Google Scholar] [CrossRef]
- Shi, C.; Wei, B.; Wei, S.; Wang, W.; Liu, H.; Liu, J. A quantitative discriminant method of elbow point for the optimal number of clusters in clustering algorithm. EURASIP J. Wirel. Commun. Netw. 2021, 2021, 31. [Google Scholar] [CrossRef]
- BiRG at Wright State University. Orhtogonal Projection to Latent Structures. 2020. Available online: https://github.com/BiRG/pyopls.2020 (accessed on 5 November 2021).
- Worley, B.; Powers, R. PCA as a Practical Indicator of OPLS-DA Model Reliability. Curr. Metab. 2016, 4, 97–103. [Google Scholar] [CrossRef]
- Fordellone, M.; Bellincontro, A.; Mencarelli, F. Partial least squares discriminant analysis: A dimensionality reduction method to classify hyperspectral data. arXiv 2018, arXiv:1806.09347. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Bloondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Machine Learning in Python. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Xu, Q.; Ding, C.; Liu, J.; Luo, B. PCA-guided search for K-means. Pattern Recognit. Lett. 2015, 54, 50–55. [Google Scholar] [CrossRef]
- Ramirez, C.A.M.; Greenop, M.; Ashton, L.; ur Rehman, I. Applications of machine learning in spectroscopy. Appl. Spectrosc. Rev. 2021, 56, 733–763. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; ur Rehman, I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Kumar, J.K.; Prasad, A.D. Fourier transform infrared spectroscopy an advanced technique for identification of biomolecules. Drug Invent. Today 2012, 14, 616–628. [Google Scholar]
- Perlich, C. Learning Curves in Machine Learning. In Encyclopedia of Machine Learning; Springer: Boston, MA, USA, 2011. [Google Scholar]
- Abdallah, F.F.; Darwish, H.W.; Darwish, I.A.; Naguib, I.A. Orthogonal projection to latent structures and first derivative for manipulation of PLSR and SVR chemometric models’ prediction: A case study. PLoS ONE 2019, 14, e0222197. [Google Scholar] [CrossRef]
- Stenlund, H.; Gorzsás, A.; Persson, P.; Sundberg, B.; Trygg, J. Orthogonal Projections to Latent Structures Discriminant Analysis Modeling on in Situ FT-IR Spectral Imaging of Liver Tissue for Identifying Sources of Variability. Anal. Chem. 2013, 80, 6898–6906. [Google Scholar] [CrossRef]
- Talari, A.C.S.; Evans, C.A.; Holen, I.; Coleman, R.E.; ur Rehman, I. Raman spectroscopic analysis differentiates between breast cancer cell lines. J. Raman Spectrosc. 2015, 46, 421–427. [Google Scholar] [CrossRef]
- Yeramian, A.; Moreno-Bueno, G.; Dolcet, X.; Catasus, L.; Abal, M.; Colas, E.; Reventos, J.; Palacios, J.; Prat, J.; Matias-Guiu, X. Endometrial carcinoma: Molecular alterations involved in tumor development and progression. Oncogene 2013, 32, 403–413. [Google Scholar] [CrossRef]
- Njoku, K.; Chiasserini, D.; Jones, E.R.; Barr, C.E.; O’Flynn, H.; Whetton, A.D.; Crosbie, E.J. Urinary Biomarkers and Their Potential for the Non-Invasive Detection of Endometrial Cancer. Front. Oncol. 2020, 10, 559016. [Google Scholar] [CrossRef]
- Mattila, A.L.; Saario, I.; Viinikka, L.; Ylikorkala, O.; Perheentupa, J. Urinary epidermal growth factor concentrations in various human malignancies. Br. J. Cancer 1988, 57, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Grayson, K.; Gregory, E.; Khan, G.; Guinn, B.-A. Guinn, Urine Biomarkers for the Early Detection of Ovarian Cancer—Are We There Yet? Biomark. Cancer 2019, 11, 1179299X1983097. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Lorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Jirásko, R.; Idkowiak, J.; Wolrab, D.; Kvasnička, A.; Friedecký, D.; Polański, K.; Študentová, H.; Študent, V.; Melichar, B.; Holčapek, M. Altered plasma, urine, and tissue profiles of sulfatides and sphingomyelins in patients with renal cell carcinoma. Cancers 2022, 44, 4622. [Google Scholar] [CrossRef]
- Ruskin, R.; Srivastava, A.; McMeekin, D.; Ramesh, R.; Moxley, K. Liquid Gold: Urinary exosomes as a potential source of biomarkers in endometrial cancer. Gynecol. Oncol. 2016, 143, 207. [Google Scholar] [CrossRef]
- Langenbeck, U.; Baum, F.; Mench-Hoinowski, A.; Luthe, H.; Behbehani, A.W. Predicting the phenylalanine blood concentration from urine analyses. An approach to noninvasive monitoring of patients with phenylketonuria. J. Inherit. Metab. Dis. 2005, 28, 855–861. [Google Scholar] [CrossRef]
- Kido, J.; Mitsubuchi, H.; Itoh, F.; Yoshida, T.; Matsumoto, S.; Sakamoto, R.; Endo, F.; Nakamura, K. Advanced Endometrial Cancer in Phenylketonuria. Med. Sci. Case Rep. 2016, 3, 108–111. [Google Scholar] [CrossRef]
- Petitclerc, E.; Deschesnes, R.G.; Côté, M.F.; Marquis, C.; Janvier, R.; Lacroix, J.; Miot-Noirault, E.; Legault, J.; Mounetou, E.; Madelmont, J.C.; et al. Antiangiogenic and Antitumoral Activity of Phenyl-3-(2-Chloroethyl) Ureas: A Class of Soft Alkylating Agents Disrupting Microtubules That Are Unaffected by Cell Adhesion-Mediated Drug Resistance. Cancer Res. 2004, 64, 4654–4663. [Google Scholar] [CrossRef]
- Liew, S.K.; Malagobadan, S.; Arshad, N.M.; Nagoor, N.H. A Review of the Structure–Activity Relationship of Natural and Synthetic Antimetastatic Compounds. Biomolecules 2020, 10, 138. [Google Scholar] [CrossRef]
- O’Flynn, H.; Ryan, N.A.J.; Narine, N.; Shelton, D.; Rana, D.; Crosbie, E.J. Diagnostic accuracy of cytology for the detection of endometrial cancer in urine and vaginal samples. Nat. Commun. 2021, 12, 952. [Google Scholar] [CrossRef]



| Patient Characteristics | Endometrial Cancers (n = 109) | Benign Controls (n = 110) |
|---|---|---|
| Age, years | ||
| 67 (38–88) | 56 (27–89) | |
| <60 yrs | 29/109 (26.6%) | 60/110 (55%) |
| ≥60 yrs | 80/109 (73.4%) | 50/110 (45%) |
| BMI (kg/m2) | ||
| 31.9 (18.4–65.5) | 27.8 (18.3–49.8) | |
| <30 BMI | 51/109 (46.8%) | 79/110 (71.8%) |
| ≥30 BMI | 58/109 (53.2%) | 31/110 (28.2%) |
| Type 2 Diabetes | ||
| Yes, Diabetes | 19/109 (17.4%) | 9/110 (8.2%) |
| No, Diabetes | 90/109 (82.6%) | 101/110 (91.8%) |
| Uterine Cancers | Benign Controls | ||
|---|---|---|---|
| Endometrioid | n = 57 | Ovarian cysts (non-endometriomas) | 43 |
| Grade1 | 32 | Uterine fibroids and/or adenomyosis | 27 |
| Grade 2 | 17 | Endometriosis (inc. endometriomas) | 15 |
| Grade 3 | 8 | Endometrial polyps | 4 |
| Non-endometrioid | 45 | Uterine prolapse | 3 |
| Mucinous | 1 | Pelvic inflammatory disease | 1 |
| Clear cell | 7 | Endometrial hyperplasia | 1 |
| Serous | 14 | Cervical intraepithelial neoplasia (CIN) | 1 |
| Carcinosarcoma | 14 | Normal (no pathology identified) | 15 |
| Leiomyosarcoma | 3 | ||
| Adenosarcoma | 3 | ||
| Endometrial stromal sarcoma | 2 | ||
| Sex cord tumour | 1 | ||
| Mixed tumours | 7 | ||
| Mixed endometrioid-serous | 4 | ||
| Mixed endometrioid-clear cell | 2 | ||
| Mixed endometrioid-serous-clear cell | 1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meza Ramirez, C.A.; Stringfellow, H.; Naik, R.; Crosbie, E.J.; Paraskevaidi, M.; Rehman, I.U.; Martin-Hirsch, P. Infrared Spectroscopy of Urine for the Non-Invasive Detection of Endometrial Cancer. Cancers 2022, 14, 5015. https://doi.org/10.3390/cancers14205015
Meza Ramirez CA, Stringfellow H, Naik R, Crosbie EJ, Paraskevaidi M, Rehman IU, Martin-Hirsch P. Infrared Spectroscopy of Urine for the Non-Invasive Detection of Endometrial Cancer. Cancers. 2022; 14(20):5015. https://doi.org/10.3390/cancers14205015
Chicago/Turabian StyleMeza Ramirez, Carlos A., Helen Stringfellow, Raj Naik, Emma J. Crosbie, Maria Paraskevaidi, Ihtesham U. Rehman, and Pierre Martin-Hirsch. 2022. "Infrared Spectroscopy of Urine for the Non-Invasive Detection of Endometrial Cancer" Cancers 14, no. 20: 5015. https://doi.org/10.3390/cancers14205015
APA StyleMeza Ramirez, C. A., Stringfellow, H., Naik, R., Crosbie, E. J., Paraskevaidi, M., Rehman, I. U., & Martin-Hirsch, P. (2022). Infrared Spectroscopy of Urine for the Non-Invasive Detection of Endometrial Cancer. Cancers, 14(20), 5015. https://doi.org/10.3390/cancers14205015








