JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Reagents and Plasmids
2.3. Cell Transfection and Reporter Assays
2.4. RNA Isolation, RT-PCR and qPCR Analysis
2.5. Western Blot Analysis
2.6. Next-Generation Sequencing (NGS) High-Throughput RNA-Seq Analysis
2.7. Generation of JICD-Overexpressing Adenovirus (AdJICD)
2.8. Development of Stable JICD-Expressing CRPC (CWR22Rv1 and C4-2) Cells
2.9. Cell Proliferation Assays
2.10. Colony Formation Assay
2.11. Cell Mobility Assays
2.12. Three-Dimensional Cell Culture Assays
2.13. Xenograft Animal Model
2.14. Quantification and Statistical Analysis
3. Results
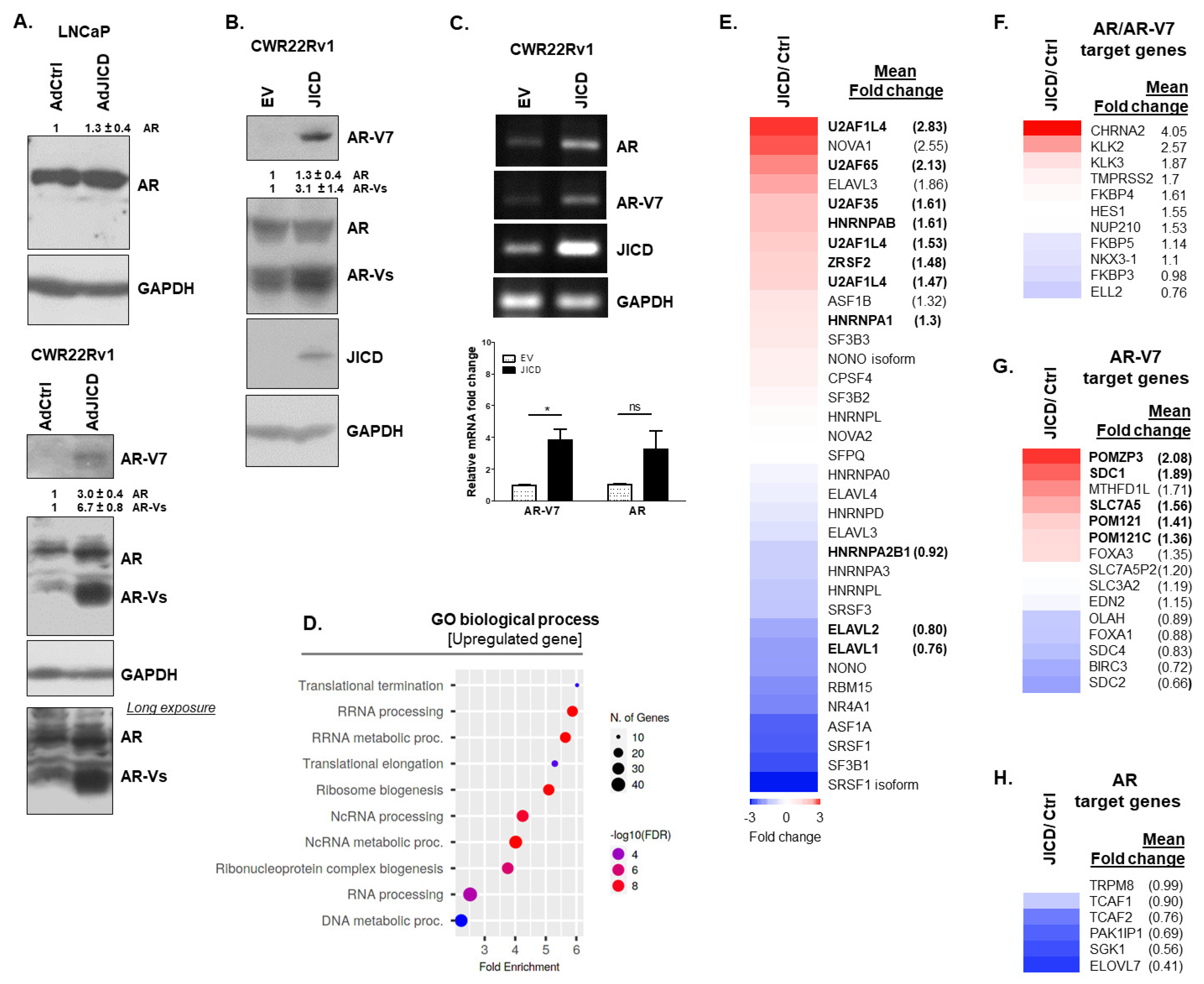
3.1. JAG1 Regulates the Expression of AR and AR-Vs in PC Cells
3.2. JICD Upregulates the Expression of AR-Vs in PC Cells
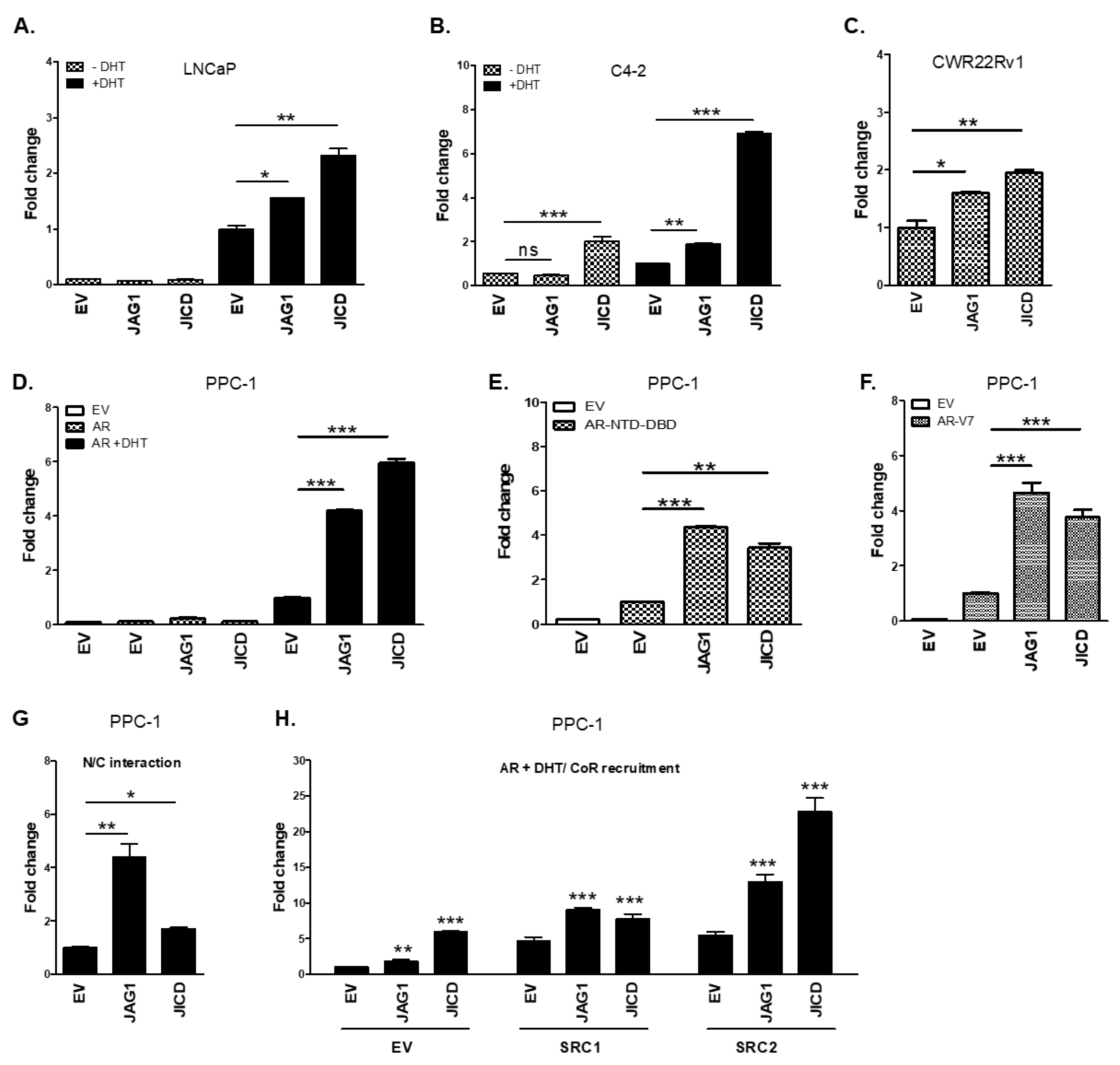
3.3. JICD Enhances Androgen-Dependent and Androgen-Independent Transactivation of ARs
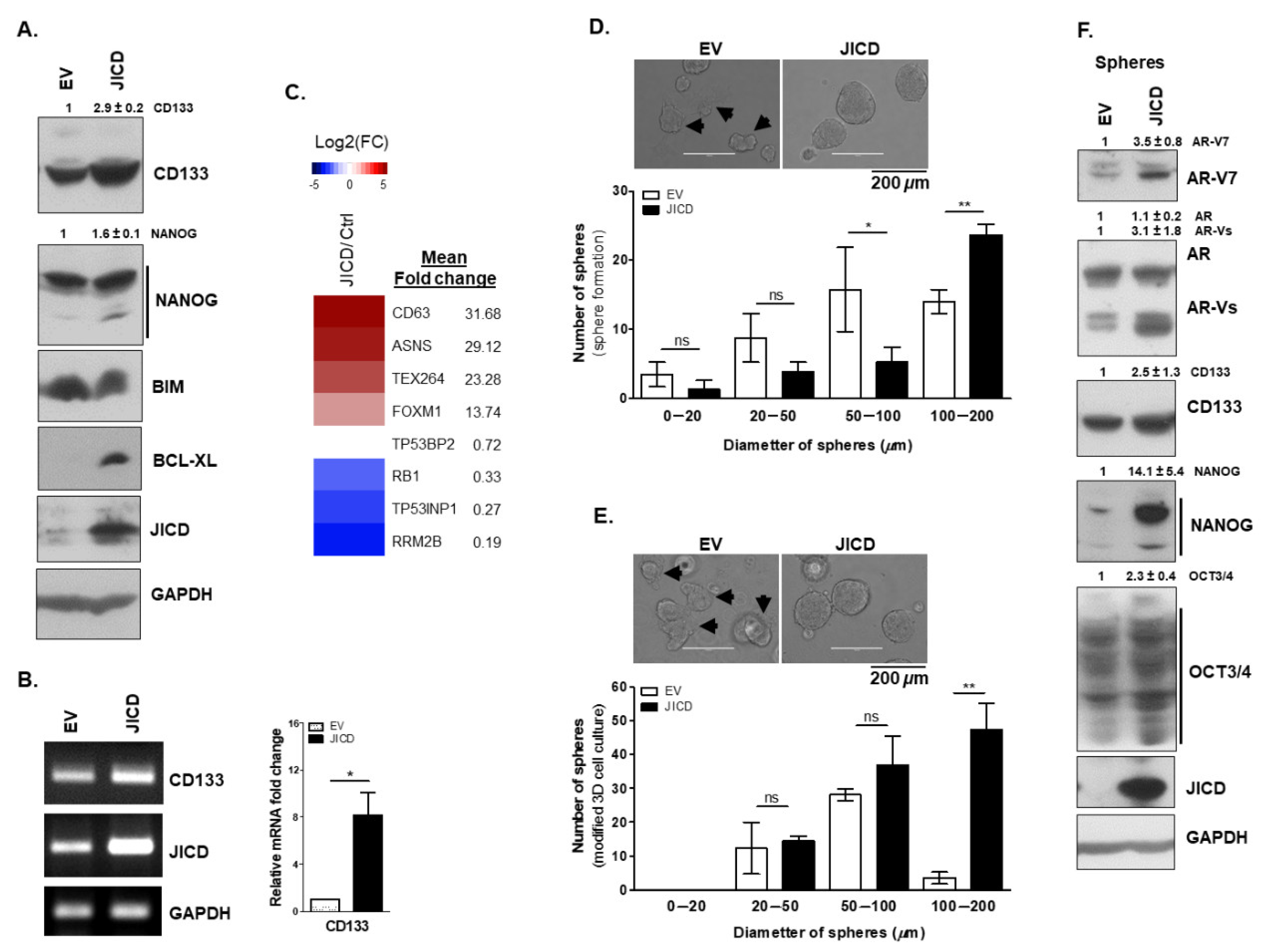
3.4. JICD Promotes Stem-Like Cell Properties in PC Cells
3.5. JICD Increases PC Cell Mobility and In Vivo Tumorigenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AR | androgen receptor |
| ARE | androgen-response element |
| AR-Vs | androgen receptor variants |
| AR-FL | full-length AR |
| AR-NTD-DBD | AR N-terminal and DNA-binding domain |
| CE3 | cryptic exon 3 |
| CRPC | castration-resistant prostate cancer |
| DBD | DNA-binding domain |
| LDB | ligand-binding domain |
| NTD | NH2-terminal transactivation domain |
| JAG1 | JAGGED1 |
| JICD | JAG1 intracellular domain |
| PCSC | prostate cancer stem cell |
References
- Tan, M.H.; Li, J.; Xu, H.E.; Melcher, K.; Yong, E.L. Androgen receptor: Structure, role in prostate cancer and drug discovery. Acta Pharmacol. Sin. 2015, 36, 3–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharifi, N. Mechanisms of androgen receptor activation in castration-resistant prostate cancer. Endocrinology 2013, 154, 4010–4017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, C.; Heemers, H.; Sharifi, N. Androgen Signaling in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030452. [Google Scholar] [CrossRef] [Green Version]
- Wadosky, K.M.; Koochekpour, S. Androgen receptor splice variants and prostate cancer: From bench to bedside. Oncotarget 2017, 8, 18550–18576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprenger, C.C.; Plymate, S.R. The link between androgen receptor splice variants and castration-resistant prostate cancer. Horm. Cancer 2014, 5, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.L.; Xie, N.; Sun, S.; Plymate, S.; Mostaghel, E.; Dong, X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene 2014, 33, 3140–3150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldman, B.J.; Feldman, D. The development of androgen-independent prostate cancer. Nat. Rev. Cancer 2001, 1, 34–45. [Google Scholar] [CrossRef]
- Basu, S.; Dong, Y.; Kumar, R.; Jeter, C.; Tang, D.G. Slow-cycling (dormant) cancer cells in therapy resistance, cancer relapse and metastasis. Semin. Cancer Biol. 2022, 78, 90–103. [Google Scholar] [CrossRef]
- Pienta, K.J.; Bradley, D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin. Cancer Res. 2006, 12, 1665–1671. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Liang, D.; Liu, J.; Axcrona, K.; Kvalheim, G.; Stokke, T.; Nesland, J.M.; Suo, Z. Prostate cancer cell lines under hypoxia exhibit greater stem-like properties. PLoS ONE 2011, 6, e29170. [Google Scholar] [CrossRef]
- Bae, K.M.; Su, Z.; Frye, C.; McClellan, S.; Allan, R.W.; Andrejewski, J.T.; Kelley, V.; Jorgensen, M.; Steindler, D.A.; Vieweg, J.; et al. Expression of pluripotent stem cell reprogramming factors by prostate tumor initiating cells. J. Urol. 2010, 183, 2045–2053. [Google Scholar] [CrossRef] [Green Version]
- Tagscherer, K.E.; Fassl, A.; Campos, B.; Farhadi, M.; Kraemer, A.; Böck, B.C.; Macher-Goeppinger, S.; Radlwimmer, B.; Wiestler, O.D.; Herold-Mende, C.; et al. Apoptosis-based treatment of glioblastomas with ABT-737, a novel small molecule inhibitor of Bcl-2 family proteins. Oncogene 2008, 27, 6646–6656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, A.T.; Berry, P.A.; Hyde, C.; Stower, M.J.; Maitland, N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005, 65, 10946–10951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, S.K.; Islam, S.M.R.; Kwak, K.S.; Rahman, M.S.; Cho, S.G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: A multiomics analysis. Cancer Gene Ther. 2020, 27, 147–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzmán-Ramírez, N.; Völler, M.; Wetterwald, A.; Germann, M.; Cross, N.A.; Rentsch, C.A.; Schalken, J.; Thalmann, G.N.; Cecchini, M.G. In vitro propagation and characterization of neoplastic stem/progenitor-like cells from human prostate cancer tissue. Prostate 2009, 69, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Oktem, G.; Bilir, A.; Uslu, R.; Inan, S.V.; Demiray, S.B.; Atmaca, H.; Ayla, S.; Sercan, O.; Uysal, A. Expression profiling of stem Cell Signaling alters with spheroid formation in CD133(high)/CD44(high) prostate cancer stem cells. Oncol. Lett. 2014, 7, 2103–2109. [Google Scholar] [CrossRef] [Green Version]
- Yun, E.-J.; Lo, U.G.; Hsieh, J.-T. The evolving landscape of prostate cancer stem cell: Therapeutic implications and future challenges. Asian J. Urol. 2016, 3, 203–210. [Google Scholar] [CrossRef] [Green Version]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Santagata, S.; Demichelis, F.; Riva, A.; Varambally, S.; Hofer, M.D.; Kutok, J.L.; Kim, R.; Tang, J.; Montie, J.E.; Chinnaiyan, A.M.; et al. JAGGED1 Expression Is Associated with Prostate Cancer Metastasis and Recurrence. Cancer Res. 2004, 64, 6854–6857. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Zhou, X.; Redfield, S.; Lewin, J.; Miele, L. Elevated Jagged-1 and Notch-1 expression in high grade and metastatic prostate cancers. Am. J. Transl. Res. 2013, 5, 368. [Google Scholar] [CrossRef]
- Delury, C.; Hart, C.; Brown, M.; Clarke, N.; Parkin, E. Stroma-induced Jagged1 expression drives PC3 prostate cancer cell migration; disparate effects of RIP-generated proteolytic fragments on cell behaviour and Notch signaling. Biochem. Biophys. Res. Commun. 2016, 472, 255–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, Q.; Zhang, B.; Zhang, L.; Dang, T.; Rowley, D.; Ittmann, M.; Xin, L. Jagged1 upregulation in prostate epithelial cells promotes formation of reactive stroma in the Pten null mouse model for prostate cancer. Oncogene 2017, 36, 618–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arianna Calcinotto, C.S.; Spataro, C.; Zagato, E.; Di Mitri, D.; Gil, V.; Crespo, M.; De Bernardis, G.; Losa, M.; Mirenda, M.; Pasquini, E.; et al. IL-23 secreted by myeloid cells drives castration-resistant prostate cancer. Nature 2018, 559, 363–389. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Y.; Guan, W.; Huang, T.; Kang, J.; Sheng, X.; Qi, J. Androgen receptor promotes the oncogenic function of overexpressed Jagged1 in prostate cancer by enhancing cyclin B1 expression via Akt phosphorylation. Mol. Cancer Res. 2014, 12, 830–842. [Google Scholar] [CrossRef] [Green Version]
- LaVoie, M.J.; Selkoe, D.J. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J. Biol. Chem. 2003, 278, 34427–34437. [Google Scholar] [CrossRef] [Green Version]
- Kornau, H.-C.; Schenker, L.T.; Kennedy, M.B.; Seeburg, P.H. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 1995, 269, 1737–1740. [Google Scholar] [CrossRef] [PubMed]
- Niethammer, M.; Kim, E.; Sheng, M. Interaction between the C terminus of NMDA receptor subunits and multiple members of the PSD-95 family of membrane-associated guanylate kinases. J. Neurosci. 1996, 16, 2157–2163. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.R. Chromosome identity of human prostate cancer cell lines, PC-3 and PPC-1. Cytogenet. Cell Genet. 1993, 62, 183–184. [Google Scholar] [CrossRef]
- Wu, H.-C.; Hsieh, J.-T.; Gleave, M.E.; Brown, N.M.; Pathak, S.; Chung, L.W.K. Derivation of androgen-independent human LNCaP prostatic cancer cell sublines: Role of bone stromal cells. Int. J. Cancer 1994, 57, 406–412. [Google Scholar] [CrossRef]
- Thalmann, G.N.; Anezinis, P.E.; Chang, S.-M.; Zhau, H.E.; Kim, E.E.; Hopwood, V.L.; Pathak, S.; von Eschenbach, A.C.; Chung, L.W. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994, 54, 2577–2581. [Google Scholar]
- Tran, T.T.; Lee, K. TR3 Enhances AR Variant Production and Transactivation, Promoting Androgen Independence of Prostate Cancer Cells. Cancers 2022, 14, 1911. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.Y.; Park, J.H.; Ahn, R.S.; Im, S.Y.; Choi, H.S.; Soh, J.; Mellon, S.H.; Lee, K. Molecular mechanism of suppression of testicular steroidogenesis by proinflammatory cytokine tumor necrosis factor alpha. Mol. Cell Biol. 2004, 24, 2593–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.H.; Yang, S.H.; Park, E.; Cho, S.H.; Gong, E.Y.; Khadka, D.B.; Cho, W.J.; Lee, K. Structure-based virtual screening and identification of a novel androgen receptor antagonist. J. Biol. Chem. 2012, 287, 30769–30780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moilanen, A.M.; Poukka, H.; Karvonen, U.; Hakli, M.; Janne, O.A.; Palvimo, J.J. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell Biol. 1998, 18, 5128–5139. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T.; Song, C.H.; Kim, K.J.; Lee, K. A new compound targets the AF-1 of androgen receptor and decreases its activity and protein levels in prostate cancer cells. Am. J. Cancer Res. 2020, 10, 4607–4623. [Google Scholar]
- Nehring, L.C.; Miyamoto, A.; Hein, P.W.; Weinmaster, G.; Shipley, J.M. The Extracellular Matrix Protein MAGP-2 Interacts with Jagged1 and Induces Its Shedding from the Cell Surface. J. Biol. Chem. 2005, 280, 20349–20355. [Google Scholar] [CrossRef] [Green Version]
- Rao, X.; Huang, X.; Zhou, Z.; Lin, X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Jinyong Luo, Z.D.; Luo, X.; Tang, N.; Song, W.; Chen, J.; Sharff, K.A.; Luu, H.H.; Haydon, R.C.; Kinzler, K.W.; Vogelstein, B.; et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2007, 2, 1236–1247. [Google Scholar] [CrossRef] [Green Version]
- Nadiminty, N.; Tummala, R.; Liu, C.; Lou, W.; Evans, C.P.; Gao, A.C. NF-κB2/p52:c-Myc:hnRNPA1 Pathway Regulates Expression of Androgen Receptor Splice Variants and Enzalutamide Sensitivity in Prostate Cancer. Mol. Cancer Ther. 2015, 14, 1884–1895. [Google Scholar] [CrossRef] [Green Version]
- Basil, P.; Robertson, M.J.; Bingman, W.E.; Dash, A.K.; Krause, W.C.; Shafi, A.A.; Piyarathna, B.; Coarfa, C.; Weigel, N.L. Cistrome and transcriptome analysis identifies unique androgen receptor (AR) and AR-V7 splice variant chromatin binding and transcriptional activities. Sci. Rep. 2022, 12, 5351. [Google Scholar] [CrossRef]
- Sugiura, M.; Sato, H.; Okabe, A.; Fukuyo, M.; Mano, Y.; Shinohara, K.I.; Rahmutulla, B.; Higuchi, K.; Maimaiti, M.; Kanesaka, M.; et al. Identification of AR-V7 downstream genes commonly targeted by AR/AR-V7 and specifically targeted by AR-V7 in castration resistant prostate cancer. Transl. Oncol. 2021, 14, 100915. [Google Scholar] [CrossRef] [PubMed]
- Dehm, S.M.; Tindall, D.J. Ligand-independent Androgen Receptor Activity Is Activation Function-2-independent and Resistant to Antiandrogens in Androgen Refractory Prostate Cancer Cells. J. Biol. Chem. 2006, 281, 27882–27893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rybak, A.P.; Bristow, R.G.; Kapoor, A. Prostate cancer stem cells: Deciphering the origins and pathways involved in prostate tumorigenesis and aggression. Oncotarget 2015, 6, 1900–1919. [Google Scholar] [CrossRef] [PubMed]
- Ascano, J.M.; Beverly, L.J.; Capobianco, A.J. The C-terminal PDZ-ligand of JAGGED1 is essential for cellular transformation. J. Biol. Chem. 2003, 278, 8771–8779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Griend, D.J.; Karthaus, W.L.; Dalrymple, S.; Meeker, A.; DeMarzo, A.M.; Isaacs, J.T. The Role of CD133 in Normal Human Prostate Stem Cells and Malignant Cancer-Initiating Cells. Cancer Res. 2008, 68, 9703–9711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011, 396076. [Google Scholar] [CrossRef]
- Shen, Y.A.; Wang, C.Y.; Hsieh, Y.T.; Chen, Y.J.; Wei, Y.H. Metabolic reprogramming orchestrates cancer stem cell properties in nasopharyngeal carcinoma. Cell Cycle 2015, 14, 86–98. [Google Scholar] [CrossRef] [Green Version]
- Skvortsov, S.; Debbage, P.; Lukas, P.; Skvortsova, I. Crosstalk between DNA repair and cancer stem cell (CSC) associated intracellular pathways. Semin. Cancer Biol. 2015, 31, 36–42. [Google Scholar] [CrossRef]
- Vinogradov, S.; Wei, X. Cancer stem cells and drug resistance: The potential of nanomedicine. Nanomedicine 2012, 7, 597–615. [Google Scholar] [CrossRef] [Green Version]
- Nathansen, J.; Meyer, F.; Müller, L.; Schmitz, M.; Borgmann, K.; Dubrovska, A. Beyond the Double-Strand Breaks: The Role of DNA Repair Proteins in Cancer Stem-Cell Regulation. Cancers 2021, 13, 4818. [Google Scholar] [CrossRef]
- Fielden, J.; Wiseman, K.; Torrecilla, I.; Li, S.; Hume, S.; Chiang, S.C.; Ruggiano, A.; Narayan Singh, A.; Freire, R.; Hassanieh, S.; et al. TEX264 coordinates p97- and SPRTN-mediated resolution of topoisomerase 1-DNA adducts. Nat. Commun. 2020, 11, 1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Raychaudhuri, P.; Costa, R.H. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol. Cell Biol. 2007, 27, 1007–1016. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Gao, F.; Li, S.; Liu, L. FoxM1 Promotes Cell Proliferation, Invasion, and Stem Cell Properties in Nasopharyngeal Carcinoma. Front. Oncol. 2018, 8, 483. [Google Scholar] [CrossRef] [PubMed]
- Sircar, K.; Huang, H.; Hu, L.; Cogdell, D.; Dhillon, J.; Tzelepi, V.; Efstathiou, E.; Koumakpayi, I.H.; Saad, F.; Luo, D.; et al. Integrative molecular profiling reveals asparagine synthetase is a target in castration-resistant prostate cancer. Am. J. Pathol. 2012, 180, 895–903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, M.; Brooks, M.; Wicha, M.S. Asparagine and Glutamine: Co-conspirators Fueling Metastasis. Cell Metab. 2018, 27, 947–949. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Zhang, J.; Yarden, Y.; Fu, L. The key roles of cancer stem cell-derived extracellular vesicles. Signal Transduct. Target. Ther. 2021, 6, 109. [Google Scholar] [CrossRef]
- Soerohardjo, I.; Widodo, I.; Heriyanto, D.S.; Zulfiqqar, A.; Anwar, S.L. Down-regulation of RB1 and TP53 as potential predicting biomarkers for castration-resistant prostate cancer (CRPC): Indonesian retrospective cohort study. Ann. Med. Surg. 2020, 60, 549–554. [Google Scholar] [CrossRef]
- Triana-Martínez, F.; Loza, M.I.; Domínguez, E. Beyond Tumor Suppression: Senescence in Cancer Stemness and Tumor Dormancy. Cells 2020, 9, 346. [Google Scholar] [CrossRef] [Green Version]
- Tothova, Z.; Gilliland, D.G. FoxO Transcription Factors and Stem Cell Homeostasis: Insights from the Hematopoietic System. Cell Stem Cell 2007, 1, 140–152. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.X.; Chang, Y.L.; Gao, W.Q. MicroRNAs targeting prostate cancer stem cells. Exp. Biol. Med. 2015, 240, 1071–1078. [Google Scholar] [CrossRef] [Green Version]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Däbritz, J.H.M.; Zhao, Z.; Yu, Y.; Dörr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, F.; Atyah, M.M.; Horstmann, N.; Gul, S.; Vago, R.; Bruns, C.J.; Zhao, Y.; Dong, Q.-Z.; Ren, N. Characteristics of the cancer stem cell niche and therapeutic strategies. Stem Cell Res. Ther. 2022, 13, 233. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.B.; Johnson, M.; Sato, M.; Koh, S.B.S.; Mulholland, D.J.; Stout, D.; Chatziioannou, A.F.; Phelps, M.E.; Wu, H.; Wu, L. Adenovirus-mediated gene expression imaging to directly detect sentinel lymph node metastasis of prostate cancer. Nat. Med. 2008, 14, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Metrich, M.; Bezdek Pomey, A.; Berthonneche, C.; Sarre, A.; Nemir, M.; Pedrazzini, T. Jagged1 intracellular domain-mediated inhibition of Notch1 signalling regulates cardiac homeostasis in the postnatal heart. Cardiovasc. Res. 2015, 108, 74–86. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.Y.; Jung, J.; Mo, J.S.; Ann, E.J.; Ahn, J.S.; Yoon, J.H.; Park, H.S. The intracellular domain of Jagged-1 interacts with Notch1 intracellular domain and promotes its degradation through Fbw7 E3 ligase. Exp. Cell Res. 2011, 317, 2438–2446. [Google Scholar] [CrossRef]
- Kumar, S.; Park, H.-S.; Lee, K. Jagged1 intracellular domain modulates steroidogenesis in testicular Leydig cells. PLoS ONE 2021, 15, e0244553. [Google Scholar] [CrossRef]
- Liebler, S.S.; Feldner, A.; Adam, M.G.; Korff, T.; Augustin, H.G.; Fischer, A. No Evidence for a Functional Role of Bi-Directional Notch Signaling during Angiogenesis. PLoS ONE 2012, 7, e53074. [Google Scholar] [CrossRef]
- Yu, S.; Wang, X.; Ng, C.F.; Chen, S.; Chan, F.L. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res. 2007, 67, 4904–4914. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Liao, Z.; Cui, Y.; Yang, C. The relationship between homeobox B7 expression and the clinical characteristics of patient with prostate cancer. J. Cell. Biochem. 2019, 120, 6395–6401. [Google Scholar] [CrossRef]
- Waltregny, D.; Alami, Y.; Clausse, N.; de Leval, J.; Castronovo, V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate 2002, 50, 162–169. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, D.; Thomas-Ahner, J.M.; Lu, C.; Zhao, P.; Zhang, Q.; Geraghty, C.; Yan, P.S.; Hankey, W.; Sunkel, B.; et al. Diverse AR-V7 cistromes in castration-resistant prostate cancer are governed by HoxB13. Proc. Natl. Acad. Sci. USA 2018, 115, 6810–6815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, X.; Schober, M.; Tassone, E.; Khodadadi-Jamayran, A.; Sastre-Perona, A.; Zhou, H.; Tsirigos, A.; Shen, S.; Chang, M.; Melamed, J.; et al. KLF4, A Gene Regulating Prostate Stem Cell Homeostasis, Is a Barrier to Malignant Progression and Predictor of Good Prognosis in Prostate Cancer. Cell Rep. 2018, 25, 3006–3020.e3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Laterra, J. Cancer Stem Cells: Distinct Entities or Dynamically Regulated Phenotypes? Cancer Res. 2012, 72, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Eber, M.R.; Shiozawa, Y. Models of Prostate Cancer Bone Metastasis. Methods Mol. Biol. 2019, 1914, 295–308. [Google Scholar] [CrossRef]
- Palumbo-Zerr, K.; Zerr, P.; Distler, A.; Fliehr, J.; Mancuso, R.; Huang, J.; Mielenz, D.; Tomcik, M.; Furnrohr, B.G.; Scholtysek, C.; et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta 121 signaling and fibrosis. Nat. Med. 2015, 21, 150–158. [Google Scholar] [CrossRef]
- Malik, R.; Khan, A.P.; Asangani, I.A.; Cieslik, M.; Prensner, J.R.; Wang, X.; Iyer, M.K.; Jiang, X.; Borkin, D.; Escara-Wilke, J.; et al. Targeting the MLL complex in castration-resistant prostate cancer. Nat. Med. 2015, 21, 344–352. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, T.T.; Lee, K. JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells. Cancers 2022, 14, 5714. https://doi.org/10.3390/cancers14225714
Tran TT, Lee K. JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells. Cancers. 2022; 14(22):5714. https://doi.org/10.3390/cancers14225714
Chicago/Turabian StyleTran, Tuyen Thanh, and Keesook Lee. 2022. "JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells" Cancers 14, no. 22: 5714. https://doi.org/10.3390/cancers14225714
APA StyleTran, T. T., & Lee, K. (2022). JAG1 Intracellular Domain Enhances AR Expression and Signaling and Promotes Stem-like Properties in Prostate Cancer Cells. Cancers, 14(22), 5714. https://doi.org/10.3390/cancers14225714






