Insights into Biochemical Sources and Diffuse Reflectance Spectral Features for Colorectal Cancer Detection and Localization
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
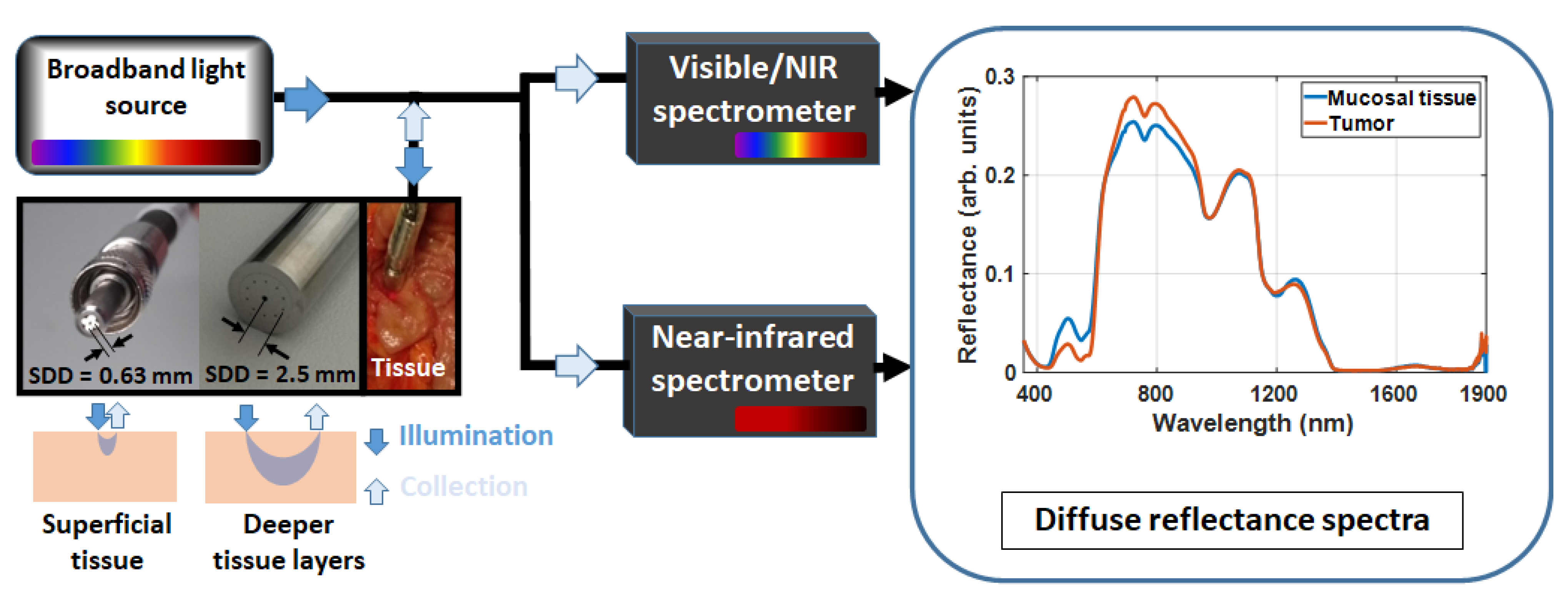
2.1. Diffuse Reflectance Spectroscopy (DRS) Instrumentation
2.2. Probing Superficial or Deeper Tissue Layers
2.3. Optical Data Collection
2.4. Clinical Protocol and Research Ethics
2.5. Data Preprocessing and Feature Selection
2.6. Extraction of Spectral Features
3. Results
3.1. Tissue Classification Features Based on PLSC Amplitudes
3.2. Wavelength Selection and Tissue Classification
3.3. Relationship between Tissue Classification Features and Tissue Biochemistry/Microstructure
4. Discussion
4.1. Impact of Depth-Resolved Determination of Wavelength Ranges and Biomarkers for Tissue Classification
4.2. Spectral Features for Colorectal Cancer (CRC) Detection
4.3. Considerations on Biomolecular Concentrations and Probed Depth for CRC Detection
4.4. Strength of the Cross-Validation of Our Model
4.5. Limitations of Our Study
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2018; Available online: https://gco.iarc.fr/ (accessed on 8 October 2022).
- Shaukat, A.; Levin, T.R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Ichkhanian, Y.; Zuchelli, T.; Watson, A.; Piraka, C. Evolving management of colorectal polyps. Ther. Adv. Gastrointest. Endosc. 2021, 14, 26317745211047010. [Google Scholar] [CrossRef] [PubMed]
- Pogue, B.W. Optics in the molecular imaging race. Opt. Photonics News 2015, 26, 24–31. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Raju, M.; Gunther, J.; Maryam, S.; Amissah, M.; Lu, H.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Accurate colorectal cancer detection and delineation by probing superficial and deeper tissue biochemistry and microstructure using diffuse reflectance spectroscopy. In Proceedings of the Molecular-Guided Surgery: Molecules, Devices, and Applications VIII, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11943. [Google Scholar]
- Nogueira, M.S.; Raju, M.; Komolibus, K.; Grygoryev, K.; Andersson-Engels, S. Assessment of tissue biochemical and optical scattering changes due to hypothermic organ preservation: A preliminary study in mouse organs. J. Phys. D Appl. Phys. 2021, 54, 37. [Google Scholar]
- Nogueira, M.S.; Raju, M.; Gunther, J.; Maryam, S.; Amissah, M.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Optical determination of superficial and deeper tissue biochemistry and microstructure for delineation and early detection of colorectal cancer. In Proceedings of the European Conference on Biomedical Optics, Munich, Germany, 20–24 June 2021; p. ETu3A-1. [Google Scholar]
- Nogueira, M.S.; Maryam, S.; Amissah, M.; Lynch, N.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Benefit of extending near-infrared wavelength range of diffuse reflectance spectroscopy for colorectal cancer detection using machine learning. In Proceedings of the European Conference on Biomedical Optics, Munich, Germany, 20–24 June 2021; p. EW4A-16. [Google Scholar]
- Nogueira, M.S.; Raju, M.; Gunther, J.; Grygoryev, K.; Komolibus, K.; Lu, H.; Andersson-Engels, S. Diffuse reflectance spectroscopy for determination of optical properties and chromophore concentrations of mice internal organs in the range of 350 nm to 1860 nm. In Proceedings of the Biophotonics: Photonic Solutions for Better Health Care VI, Strasbourg, France, 22–26 April 2018; Volume 10685. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Maryam, S.; Amissah, M.; Lu, H.; Lynch, N.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Evaluation of wavelength ranges and tissue depth probed by diffuse reflectance spectroscopy for colorectal cancer detection. Sci. Rep. 2021, 11, 798. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Raju, M.; Gunther, J.; Maryam, S.; Amissah, M.; Lu, H.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Tissue biomolecular and microstructure profiles in optical colorectal cancer delineation. J. Phys. D Appl. Phys. 2021, 54, 454002. [Google Scholar] [CrossRef]
- Mousavi, M.; Moriyama, L.T.T.; Grecco, C.; Nogueira, M.S.; Svanberg, K.; Kurachi, C.; Andersson-Engels, S. Photodynamic therapy dosimetry using multiexcitation multiemission wavelength: Toward real-time prediction of treatment outcome. J. Biomed. Opt. 2020, 25, 63812. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Maryam, S.; Amissah, M.; Lynch, N.; Killeen, S.; Lu, H.; O’Riordain, M.; Andersson-Engels, S. Improving colorectal cancer detection by extending the near-infrared wavelength range and tissue probed depth of diffuse reflectance spectroscopy: A support vector machine approach. In Proceedings of the Optical Biopsy XX: Toward Real-Time Spectroscopic Imaging and Diagnosis, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11954. [Google Scholar]
- Nogueira, M.S.; Amissah, M.; Maryam, S.; Lynch, N.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Optimization of tissue classification for colorectal cancer detection using support vector machines and diffuse reflectance spectroscopy. In Proceedings of the European Conference on Biomedical Optics, Munich, Germany, 20–24 June 2021; p. EW4A-17. [Google Scholar]
- Nogueira, M.S.; Jayet, B.; Matias, J.S.; Gunther, J.E.; Tyndall, C.; Andersson-Engels, S. Biophotonics web application for computer simulations in diffuse optics: Fostering multidisciplinary education and research. In Proceedings of the Optical Interactions with Tissue and Cells XXXIII; and Advanced Photonics in Urology, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11958. [Google Scholar]
- Saito Nogueira, M.; Gunther, J.E.; Jayet, B.; Souza Matias, J.; Tyndall, C.; Andersson-Engels, S. Biophotonics computer app: Fostering multidisciplinary distance self-paced learning with a user-friendly interface. In Proceedings of the OSA Technical Digest, San Francisco, CA, USA, 1–6 February 2021; pp. 1–2. [Google Scholar]
- Nogueira, M.S.; Gunther, J.E.; Komolibus, K.; Andersson-Engels, S. User-friendly graphical user interface for simulating tissue optical properties and fluence rates: Improving students learning in tissue optics. In Proceedings of the Optical Interactions with Tissue and Cells XXXI, San Francisco, CA, USA, 1–6 February 2020; Volume 11238. [Google Scholar]
- Mourant, J.R.; Freyer, J.P.; Hielscher, A.H.; Eick, A.A.; Shen, D.; Johnson, T.M. Mechanisms of light scattering from biological cells relevant to noninvasive optical-tissue diagnostics. Appl. Opt. 1998, 37, 3586–3593. [Google Scholar] [CrossRef]
- Mourant, J.R.; Johnson, T.M.; Carpenter, S.; Guerra, A.; Aida, T.; Freyer, J.P. Polarized angular-dependent spectroscopy of epithelial cells and epithelial cell nuclei to determine the size scale of scattering structures. J. Biomed. Opt. 2002, 7, 378–387. [Google Scholar] [CrossRef]
- Jacques, S.L. Optical properties of biological tissues: A review. Phys. Med. Biol. 2013, 58, R37. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, M.S. Fluorescence Lifetime Spectroscopy for Diagnosis of Clinically Similar Skin Lesions; Universidade de São Paulo: São Paulo, Brazil, 2016. [Google Scholar]
- Nogueira, M.S.; Kurachi, C. Assessing the photoaging process at sun exposed and non-exposed skin using fluorescence lifetime spectroscopy. In Proceedings of the Optical Biopsy XIV: Toward Real-Time Spectroscopic Imaging and Diagnosis, San Francisco, CA, USA, 13–18 February 2016; Volume 9703. [Google Scholar]
- Cosci, A.; Nogueira, M.S.; Pratavieira, S.; Takahama, A.; de Souza Azevedo, R.; Kurachi, C.; Kurachi, C. Time-resolved fluorescence spectroscopy for clinical diagnosis of actinic cheilitis: Erratum. Biomed. Opt. Express 2016, 7, 4210–4219. [Google Scholar] [CrossRef] [PubMed]
- Lacerenza, M.; Pacheco, A.; Sekar, S.K.V.; Nogueira, M.S.; Buttafava, M.; Tosi, A.; Pifferi, A.; Contini, D.; Andersson-Engels, S. Functional monitoring of lung tissue using a hybrid hyperspectral Time-Resolved GASMAS system: A systematic study on ex vivo sample. In Proceedings of the Optical Tomography and Spectroscopy, Washington, DC, USA, 20–23 April 2020; p. SW1D-2. [Google Scholar]
- Pires, L.; Nogueira, M.S.; Pratavieira, S.; Moriyama, L.T.; Kurachi, C. Time-resolved fluorescence lifetime for cutaneous melanoma detection. Biomed. Opt. Express 2014, 5, 3080. [Google Scholar] [CrossRef]
- Kurachi, C.; Pires, L.; Nogueira, M.S.; Pratavieira, S. Lifetime fluorescence for the detection of skin lesions. In Proceedings of the Biomedical Optics 2014, Miami, FL, USA, 26–30 April 2014. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Cosci, A.; Pratavieira, S.; Takahama, A., Jr.; Azevedo, R.S.; Kurachi, C. Evaluation of actinic cheilitis using fluorescence lifetime spectroscopy. In Proceedings of the Optical Biopsy XIV: Toward Real-Time Spectroscopic Imaging and Diagnosis, San Francisco, CA, USA, 13–18 February 2016; Volume 9703. [Google Scholar]
- Salvio, A.G.; Ramirez, D.P.; Inada, N.M.; Stringasci, M.D.; Nogueira, M.S.; Bagnato, V.S. Fractionated Illumination in a Single Visit Photodynamic Therapy for Basal Cell Carcinoma; Book of Abstracts. 2017. Available online: https://www.internationalphotodynamic.com/ (accessed on 8 October 2022).
- Salvio, A.G.; Ramirez, D.P.; Nogueira, M.S.; Stringasci, M.D.; Oliveira, E.R.; Inada, N.M.; Bagnato, V.S. Evaluation of Pain and Treatment Effect during Large Area Photodynamic Therapy in 140 Patients with Widespread Actinic Keratosis of Upper Limbs; Book of Abstracts. 2017. Available online: https://www.internationalphotodynamic.com/ (accessed on 8 October 2022).
- Nogueira, M.S.; Cosci, A.; Rosa, R.G.T.; Salvio, A.G.; Pratavieira, S.; Kurachi, C. Portable fluorescence lifetime spectroscopy system for in-situ interrogation of biological tissues. J. Biomed. Opt. 2017, 22, 121608. [Google Scholar]
- de Paula Campos, C.; de Paula D’Almeida, C.; Nogueira, M.S.; Moriyama, L.T.; Pratavieira, S.; Kurachi, C. Fluorescence spectroscopy in the visible range for the assessment of UVB radiation effects in hairless mice skin. Photodiagnosis Photodyn. Ther. 2017, 20, 21–27. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Guimarães, F.E.G. Photophysical processes on biological tissues and photodynamic therapy using steady-state and time-resolved fluorescence techniques: Diagnosis applications, dosimetry and photodegradation kinetics. In Livro de Resumos; SIBiUSP-Integrated USP Libraries: São Paulo, Brazil, 2016. [Google Scholar]
- Ono, B.A.; Nogueira, M.; Pires, L.; Pratavieira, S.; Kurachi, C. Subcellular localization and photodynamic activity of Photodithazine (glucosamine salt of chlorin e6) in murine melanoma B16-F10: An in vitro and in vivo study. In Proceedings of the Optical Methods for Tumor Treatment and Detection: Mechanisms and Techniques in Photodynamic Therapy XXVII, San Francisco, CA, USA, 27 January–1 February 2018; Volume 1047616, p. 44. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Bagnato, V.S.; Panhoca, V.H. Effectiveness of whitening treatments employing violet illumination alone or combined with bleaching agents. In Proceedings of the Lasers in Dentistry XXVIII, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11942. [Google Scholar]
- Panhóca, V.H.; Nogueira, M.S.; Bagnato, V.S. Treatment of facial nerve palsies with laser and endermotherapy: A report of two cases. Laser Phys. Lett. 2020, 18, 15601. [Google Scholar] [CrossRef]
- Panhóca, V.H.; Nogueira, M.S.; Bagnato, V.S. Laser and vacuum therapy for treatment of facial nerve palsies. In Proceedings of the Imaging, Therapeutics, and Advanced Technology in Head and Neck Surgery and Otolaryngology 2022, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11935, pp. 39–48. [Google Scholar]
- Bomfin, L.S.; Kitakawa, D.; Nogueira, M.S.; de Silva, L.F. Low Level Laser Therapy as adjuvant treatment for lower lip lesion. In Proceedings of the Latin America Optics and Photonics Conference, Recife, Brazil, 7–11 August 2022; p. M2B-6. [Google Scholar]
- Nogueira, M.S.; Lacerenza, M.; Sekar, S.K.V.; Buttafava, M.; Pifferi, A.; Tosi, A.; Contini, D.; Andersson-Engels, S. Broadband extraction of tissue optical properties using a portable hybrid time-resolved continuous wave instrumentation: Characterization of ex vivo organs. In Proceedings of the Clinical and Translational Biophotonics, Washington, DC, USA, 20–23 April 2020; p. TM2B-3. [Google Scholar]
- Nogueira, M.S.; Bagnato, V.S.; Panhoca, V.H. Characterization of teeth fluorescence properties due to coffee pigmentation: Towards optimization of quantitative light-induced fluorescence for tooth color assessment. In Proceedings of the Optical Interactions with Tissue and Cells XXXI, San Francisco, CA, USA, 1–6 February 2020; Volume 11238. [Google Scholar]
- Nogueira, M.S.; Pinto Junior, F.F.; Caface, R.A.; de Oliveira, K.T.; Guimarães, F.E.G. Optimization of curcumin formulations for fluorescence-based applications. In Proceedings of the Optical Interactions with Tissue and Cells XXXI, San Francisco, CA, USA, 1–6 February 2020; Volume 11238. [Google Scholar]
- Nogueira, M.S.; Junior, F.F.P.; Caface, R.A.; de Oliveira, K.T.; Bagnato, V.S.; Guimarães, F.E.G. Characterization of photophysical properties of curcumin for theranostics of neurodegenerative diseases. In Proceedings of the Optical Interactions with Tissue and Cells XXX, San Francisco, CA, USA, 2–7 February 2019; Volume 10876. [Google Scholar]
- Nogueira, M.S.; Komolibus, K.; Grygoryev, K.; Gunther, J.E.; Andersson-Engels, S. Fluorescence spectroscopy of mouse organs using ultraviolet excitation: Towards assessment of organ viability for transplantation. In Proceedings of the Optical Interactions with Tissue and Cells XXX, San Francisco, CA, USA, 2–7 February 2019; Volume 10876. [Google Scholar]
- Nogueira, M.S.; Panhóca, V.H.; Bagnato, V.S. Fluorescence spectroscopy analysis of light-induced tooth whitening. In Proceedings of the Optical Interactions with Tissue and Cells XXX, San Francisco, CA, USA, 2–7 February 2019; Volume 10876. [Google Scholar]
- De Andrade, C.T.; Nogueira, M.S.; Kanick, S.C.; Marra, K.; Gunn, J.; Andreozzi, J.; Samkoe, K.S.; Kurachi, C.; Pogue, B.W. Optical spectroscopy of radiotherapy and photodynamic therapy responses in normal rat skin shows vascular breakdown products. In Proceedings of the Optical Methods for Tumor Treatment and Detection: Mechanisms and Techniques in Photodynamic Therapy XXV, San Francisco, CA, USA, 13–18 February 2016; Volume 9694. [Google Scholar]
- Saito Nogueira, M.; Ribeiro, V.; Pires, M.; Peralta, F.; Carvalho, L.F.D.C.E.S.D. Biochemical Profiles of In Vivo Oral Mucosa by Using a Portable Raman Spectroscopy System. Optics 2021, 2, 134–147. [Google Scholar] [CrossRef]
- Carvalho, L.F.C.S.; Nogueira, M.S.; Bhattacharjee, T.; Neto, L.P.M.; Daun, L.; Mendes, T.O.; Rajasekaran, R.; Chagas, M.; Martin, A.A.; Soares, L.E.S. In vivo Raman spectroscopic characteristics of different sites of the oral mucosa in healthy volunteers. Clin. Oral Investig. 2019, 23, 3021–3031. [Google Scholar] [CrossRef]
- Leal, L.B.; Nogueira, M.S.; Mageski, J.G.A.; Martini, T.P.; Barauna, V.G.; dos Santos, L.; Carvalho, L.F.D.C.E.S.D. Diagnosis of Systemic Diseases Using Infrared Spectroscopy: Detection of Iron Overload in Plasma—Preliminary Study. Biol. Trace Element Res. 2021, 199, 3737–3751. [Google Scholar] [CrossRef]
- Carvalho, L.F.C.S.; Nogueira, M.S. New insights of Raman spectroscopy for oral clinical applications. Analyst 2018, 143, 6037–6048. [Google Scholar] [CrossRef]
- Carvalho, L.F.C.S.; Nogueira, M.S.; Neto, L.P.M.; Bhattacharjee, T.T.; Martin, A.A. Raman spectral post-processing for oral tissue discrimination—A step for an automatized diagnostic system. Biomed. Opt. Express 2017, 8, 5218, Erratum in Biomed. Opt. Express 2018, 9, 649. [Google Scholar] [CrossRef] [PubMed]
- Nogueira, M.S. Biophotonic telemedicine for disease diagnosis and monitoring during pandemics: Overcoming COVID-19 and shaping the future of healthcare. Photodiagnosis Photodyn. Ther. 2020, 31, 101836. [Google Scholar] [CrossRef] [PubMed]
- Maryam, S.; Nogueira, M.S.; Krishnamoorthy, S.; Sekar, S.K.V.; Lu, H.; Gautam, R.; Burke, R.; Andersson-Engels, S.; Riordain, R.N.; Sheahan, P. Multi-configuration Raman spectrometer for early stage diagnosis of oral cancer. In Proceedings of the Biomedical Vibrational Spectroscopy 2022: Advances in Research and Industry, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11957, pp. 20–26. [Google Scholar]
- Nogueira, M.S.; Barreto, A.L.; Furukawa, M.; Rovai, E.S.; Bastos, A.; Bertoncello, G.; e Silva de Carvalho, L.F.d.C. FTIR spectroscopy as a point of care diagnostic tool for diabetes and periodontitis: A saliva analysis approach. Photodiagn. Photodyn. Ther. 2022, 40, 103036. [Google Scholar] [CrossRef] [PubMed]
- Carnevalli, A.C.; Leal, L.; Scherma, A.; Morais, C.; Martin, F.; Bonnier, F.; Baker, M.; Byrne, H.J.; Chagas e Silva Carvalho, L.F.; Nogueira, M.S. Identification of diabetic patients via urine analysis by FTIR: Preliminary study (Conference Presentation). In Proceedings of the Photonic Diagnosis and Treatment of Infections and Inflammatory Diseases II, San Francisco, CA, USA, 2–7 February 2019; Volume 10863. [Google Scholar]
- Ferreira, M.C.C.; Monteiro, G.R.; Peralta, F.; Castro, P.A.A.; Zezell, D.; Nogueira, M.S.; Carvalho, L.F.C.S. Assessment of bound water of saliva samples by using FT-IR spectroscopy. In Proceedings of the Latin America Optics and Photonics Conference, Recife, Brazil, 7–11 August 2022; p. M4B-1. [Google Scholar]
- Nogueira, M.S.; Leal, L.B.; Marcarini, W.D.; Pimentel, R.L.; Muller, M.; Vassallo, P.F.; Campos, L.C.G.; Dos Santos, L.; Luiz, W.B.; Mill, J.G.; et al. Rapid diagnosis of COVID-19 using FT-IR ATR spectroscopy and machine learning. Sci. Rep. 2021, 11, 15409. [Google Scholar] [CrossRef]
- Chow, K.K.; Short, M.; Lam, S.; McWilliams, A.; Zeng, H. A Raman cell based on hollow core photonic crystal fiber for human breath analysis. Med. Phys. 2014, 41, 92701. [Google Scholar] [CrossRef]
- Bydlon, T.M.; Nachabé, R.; Ramanujam, N.; Sterenborg, H.J.C.M.; Hendriks, B.H.W. Chromophore based analyses of steady-state diffuse reflectance spectroscopy: Current status and perspectives for clinical adoption. J. Biophotonics 2015, 8, 9–24. [Google Scholar] [CrossRef]
- Nachabé, R.; Hendriks, B.H.W.; van der Voort, M.; Desjardins, A.E.; Sterenborg, H.J.C.M. Estimation of biological chromophores using diffuse optical spectroscopy: Benefit of extending the UV-VIS wavelength range to include 1000 to 1600 nm. Biomed. Opt. Express 2010, 1, 1432–1442. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Brugnera, J.A.; Bagnato, V.S.; Panhóca, V.H. Evaluation of the whitening effectiveness of violet illumination alone or combined with hydrogen peroxide gel. Photobiomodulation Photomed. Laser Surg. 2021, 39, 395–402. [Google Scholar] [CrossRef]
- Nachabé, R.; Evers, D.J.; Hendriks, B.H.W.; Lucassen, G.W.; van der Voort, M.; Wesseling, J.; Ruers, T.J.M. Effect of bile absorption coefficients on the estimation of liver tissue optical properties and related implications in discriminating healthy and tumorous samples. Biomed. Opt. Express 2011, 2, 600–614. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Matthews, R.; Killeen, S.; O’Riordain, M.; Andersson-Engels, S. Colorectal cancer detection based on the extraction of scattering properties and biochemical concentrations from fluorescence spectroscopy measurements. In Proceedings of the Clinical and Translational Biophotonics, Fort Lauderdale, FL, USA, 24–27 April 2022; 24–27 April 2022; p. TS2B-5. [Google Scholar] [CrossRef]
- Swartling, J.; Pifferi, A.; Enejder, A.M.K.; Andersson-Engels, S. Accelerated Monte Carlo models to simulate fluorescence spectra from layered tissues. JOSA A 2003, 20, 714–727. [Google Scholar] [CrossRef]
- Muller, M.; Hendriks, B.H.W. Recovering intrinsic fluorescence by Monte Carlo modeling. J. Biomed. Opt. 2013, 18, 27009. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chang, S.K.; Mar\’\in, N.; Follen, M.; Richards-Kortum, R.R. Model-based analysis of clinical fluorescence spectroscopy for in vivo detection of cervical intraepithelial dysplasia. J. Biomed. Opt. 2006, 11, 24008. [Google Scholar] [CrossRef] [PubMed]
- Pfefer, T.J.; Wang, Q.; Drezek, R.A. Monte Carlo modeling of time-resolved fluorescence for depth-selective interrogation of layered tissue. Comput. Methods Programs Biomed. 2011, 104, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Marcu, L.; French, P.M.W.; Elson, D.S. Fluorescence Lifetime Spectroscopy and Imaging: Principles and Applications in Biomedical Diagnostics; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Pogue, B.W.; Poplack, S.P.; McBride, T.O.; Wells, W.A.; Osterman, K.S.; Osterberg, U.L.; Paulsen, K.D. Quantitative hemoglobin tomography with diffuse near-infrared spectroscopy: Pilot results in the breast. Radiology 2001, 218, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Heffer, E.L.; Fantini, S. Quantitative oximetry of breast tumors: A near-infrared method that identifies two optimal wavelengths for each tumor. Appl. Opt. 2002, 41, 3827–3839. [Google Scholar] [CrossRef] [PubMed]
- Cerussi, A.E.; Berger, A.J.; Bevilacqua, F.; Shah, N.; Jakubowski, D.; Butler, J.; Holcombe, R.F.; Tromberg, B.J. Sources of absorption and scattering contrast for near-infrared optical mammography. Acad. Radiol. 2001, 8, 211–218. [Google Scholar] [CrossRef]
- Svensson, T.; Swartling, J.; Taroni, P.; Torricelli, A.; Lindblom, P.; Ingvar, C.; Andersson-Engels, S. Characterization of normal breast tissue heterogeneity using time-resolved near-infrared spectroscopy. Phys. Med. Biol. 2005, 50, 2559. [Google Scholar] [CrossRef]
- Chance, B.; Nioka, S.; Zhang, J.; Conant, E.F.; Hwang, E.; Briest, S.; Orel, S.G.; Schnall, M.D.; Czerniecki, B.J. Breast cancer detection based on incremental biochemical and physiological properties of breast cancers: A six-year, two-site study1. Acad. Radiol. 2005, 12, 925–933. [Google Scholar] [CrossRef]
- Nilsson, J.H.; Reistad, N.; Brange, H.; Öberg, C.-F.; Sturesson, C. Diffuse reflectance spectroscopy for surface measurement of liver pathology. Eur. Surg. Res. 2017, 58, 40–50. [Google Scholar] [CrossRef]
- Conover, D.L.; Fenton, B.M.; Foster, T.H.; Hull, E.L. An evaluation of near infrared spectroscopy and cryospectrophotometry estimates of haemoglobin oxygen saturation in a rodent mammary tumour model. Phys. Med. Biol. 2000, 45, 2685. [Google Scholar] [CrossRef]
- Steen, R.G.; Kitagishi, K.; Morgan, K. In vivo measurement of tumor blood oxygenation by near-infrared spectroscopy: Immediate effects of pentobarbital overdose or carmustine treatment. J. Neurooncol. 1994, 22, 209–220. [Google Scholar] [CrossRef]
- Kragh, M.; Quistorff, B.; Horsman, M.R.; Kristjansen, P.E.G. Acute effects of vascular modifying agents in solid tumors assessed by noninvasive laser Doppler flowmetry and near infrared spectroscopy. Neoplasia 2002, 4, 263–267. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kragh, M.; Quistorff, B.; Lund, E.L.; Kristjansen, P.E.G. Quantitative estimates of vascularity in solid tumors by non-invasive near-infrared spectroscopy. Neoplasia 2001, 3, 324–330. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gu, Y.; Chen, W.R.; Xia, M.; Jeong, S.W.; Liu, H. Effect of Photothermal Therapy on Breast Tumor Vascular Contents: Noninvasive Monitoring by Near-infrared Spectroscopy. Photochem. Photobiol. 2007, 81, 1002–1009. [Google Scholar] [CrossRef]
- Howe, F.A.; Connelly, J.P.; Robinson, S.P.; Springett, R.; Griffiths, J.R. The effects of tumour blood flow and oxygenation modifiers on subcutaneous tumours as determined by NIRS. In Oxygen Transport to Tissue XXVI; Springer: Boston, MA, USA, 2005; pp. 75–81. [Google Scholar]
- Hirosawa, N.; Sakamoto, Y.; Katayama, H.; Tonooka, S.; Yano, K. In vivo investigation of progressive alterations in rat mammary gland tumors by near-infrared spectroscopy. Anal. Biochem. 2002, 305, 156–165. [Google Scholar] [CrossRef]
- Xia, M.; Kodibagkar, V.; Liu, H.; Mason, R.P. Tumour oxygen dynamics measured simultaneously by near-infrared spectroscopy and 19F magnetic resonance imaging in rats. Phys. Med. Biol. 2005, 51, 45. [Google Scholar] [CrossRef]
- Yu, G.; Durduran, T.; Zhou, C.; Wang, H.-W.; Putt, M.E.; Saunders, H.M.; Sehgal, C.M.; Glatstein, E.; Yodh, A.G.; Busch, T.M. Noninvasive monitoring of murine tumor blood flow during and after photodynamic therapy provides early assessment of therapeutic efficacy. Clin. Cancer Res. 2005, 11, 3543–3552. [Google Scholar] [CrossRef]
- Roy, H.K.; Gomes, A.; Turzhitsky, V.; Goldberg, M.J.; Rogers, J.; Ruderman, S.; Young, K.L.; Kromine, A.; Brand, R.E.; Jameel, M.; et al. Spectroscopic microvascular blood detection from the endoscopically normal colonic mucosa: Biomarker for neoplasia risk. Gastroenterology 2008, 135, 1069–1078. [Google Scholar] [CrossRef][Green Version]
- Wang, H.-W.; Jiang, J.-K.; Lin, C.-H.; Lin, J.-K.; Huang, G.-J.; Yu, J.-S. Diffuse reflectance spectroscopy detects increased hemoglobin concentration and decreased oxygenation during colon carcinogenesis from normal to malignant tumors. Opt. Express 2009, 17, 2805–2817. [Google Scholar] [CrossRef]
- Zonios, G.; Perelman, L.T.; Backman, V.; Manoharan, R.; Fitzmaurice, M.; Van Dam, J.; Feld, M.S. Diffuse reflectance spectroscopy of human adenomatous colon polyps in vivo. Appl. Opt. 1999, 38, 6628–6637. [Google Scholar] [CrossRef]
- Dhar, A.; Johnson, K.S.; Novelli, M.R.; Bown, S.G.; Bigio, I.J.; Lovat, L.B.; Bloom, S.L. Elastic scattering spectroscopy for the diagnosis of colonic lesions: Initial results of a novel optical biopsy technique. Gastrointest. Endosc. 2006, 63, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Mourant, J.R.; Bigio, I.J.; Boyer, J.D.; Johnson, T.M.; Lacey, J.; Bohorfoush, A.G.; Mellow, M.H. Elastic scattering spectroscopy as a diagnostic tool for differentiating pathologies in the gastrointestinal tract: Preliminary testing. J. Biomed. Opt. 1996, 1, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, E.J.M.; Brouwer De Koning, S.G.; Hendriks, B.H.W.; Jóźwiak, K.; Sterenborg, H.J.C.M.; Ruers, T.J.M. Comparing in vivo and ex vivo fiberoptic diffuse reflectance spectroscopy in colorectal cancer. Transl. Biophotonics 2019, 1, e201900008. [Google Scholar] [CrossRef]
- De Koning, S.G.B.; Baltussen, E.J.M.; Karakullukcu, M.B.; Dashtbozorg, B.; Smit, L.A.; Dirven, R.; Hendriks, B.H.W.; Sterenborg, H.J.C.M.; Ruers, T.J.M. Toward complete oral cavity cancer resection using a handheld diffuse reflectance spectroscopy probe. J. Biomed. Opt. 2018, 23, 121611. [Google Scholar]
- De Boer, L.L.; Molenkamp, B.G.; Bydlon, T.M.; Hendriks, B.H.W.; Wesseling, J.; Sterenborg, H.; Ruers, T.J.M. Fat/water ratios measured with diffuse reflectance spectroscopy to detect breast tumor boundaries. Breast Cancer Res. Treat. 2015, 152, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.S.; Barman, I.; Dingari, N.C.; Volynskaya, Z.; Liu, W.; Klein, N.; Plecha, D.; Dasari, R.R.; Fitzmaurice, M. Diagnostic power of diffuse reflectance spectroscopy for targeted detection of breast lesions with microcalcifications. Proc. Natl. Acad. Sci. USA 2013, 110, 471–476. [Google Scholar] [CrossRef]
- Spliethoff, J.W.; Evers, D.J.; Klomp, H.M.; van Sandick, J.W.; Wouters, M.W.; Nachabe, R.; Lucassen, G.W.; Hendriks, B.H.W.; Wesseling, J.; Ruers, T.J.M. Improved identification of peripheral lung tumors by using diffuse reflectance and fluorescence spectroscopy. Lung Cancer 2013, 80, 165–171. [Google Scholar] [CrossRef]
- Evers, D.J.; Nachabe, R.; Hompes, D.; Van Coevorden, F.; Lucassen, G.W.; Hendriks, B.H.W.; van Velthuysen, M.-L.; Wesseling, J.; Ruers, T.J.M. Optical sensing for tumor detection in the liver. Eur. J. Surg. Oncol. 2013, 39, 68–75. [Google Scholar] [CrossRef]
- Tanis, E.; Evers, D.J.; Spliethoff, J.W.; Pully, V.V.; Kuhlmann, K.; van Coevorden, F.; Hendriks, B.H.W.; Sanders, J.; Prevoo, W.; Ruers, T.J.M. In vivo tumor identification of colorectal liver metastases with diffuse reflectance and fluorescence spectroscopy. Lasers Surg. Med. 2016, 48, 820–827. [Google Scholar] [CrossRef]
- Baltussen, E.J.M.; Snæbjörnsson, P.; De Koning, S.G.B.; Sterenborg, H.J.C.M.; Aalbers, A.G.J.; Kok, N.; Beets, G.L.; Hendriks, B.H.W.; Kuhlmann, K.F.D.; Ruers, T.J.M. Diffuse reflectance spectroscopy as a tool for real-time tissue assessment during colorectal cancer surgery. J. Biomed. Opt. 2017, 22, 106014. [Google Scholar] [CrossRef]
- Baltussen, E.J.M.; Kok, E.N.D.; de Koning, S.G.B.; Sanders, J.; Aalbers, A.G.J.; Kok, N.F.M.; Beets, G.L.; Flohil, C.C.; Bruin, S.C.; Kuhlmann, K.F.D.; et al. Hyperspectral imaging for tissue classification, a way toward smart laparoscopic colorectal surgery. J. Biomed. Opt. 2019, 24, 16002. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, E.J.M.; de Koning, S.G.; Sanders, J.; Aalbers, A.G.J.; Kok, N.F.M.; Beets, G.L.; Hendriks, B.H.W.; Sterenborg, H.J.C.M.; Kuhlmann, K.F.D.; Ruers, T.J.M. Using Diffuse Reflectance Spectroscopy to Distinguish Tumor Tissue from Fibrosis in Rectal Cancer Patients as a Guide to Surgery. Lasers Surg. Med. 2019, 52, 604–611. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, E.J.M.; De Koning, S.G.B.; Sanders, J.; Aalbers, A.G.J.; Kok, N.F.M.; Beets, G.L.; Hendriks, B.H.W.; Sterenborg, H.J.C.M.; Kuhlmann, K.F.D.; Ruers, T.J.M. Tissue diagnosis during colorectal cancer surgery using optical sensing: An in vivo study. J. Transl. Med. 2019, 17, 333. [Google Scholar] [CrossRef] [PubMed]
- Baltussen, E.J.M.; Sterenborg, H.J.C.M.; Ruers, T.J.M.; Dashtbozorg, B. Optimizing algorithm development for tissue classification in colorectal cancer based on diffuse reflectance spectra. Biomed. Opt. Express 2019, 10, 6096–6113. [Google Scholar] [CrossRef] [PubMed]
- Langhout, G.C.; Spliethoff, J.W.; Schmitz, S.J.; Aalbers, A.G.J.; van Velthuysen, M.-L.; Hendriks, B.H.W.; Ruers, T.J.M.; Kuhlmann, K.F.D. Differentiation of healthy and malignant tissue in colon cancer patients using optical spectroscopy: A tool for image-guided surgery. Lasers Surg. Med. 2015, 47, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Langhout, G.C.; Spliethoff, J.W.; Aalbers, A.G.J.; Verwaal, V.J.; Hendriks, B.H.W.; Ruers, T.J.M.; Kuhlmann, K.F.D. Colorectal Cancer Identified Using Optical Spectroscopy. Ann. Oncol. 2014, 25, iv206. [Google Scholar] [CrossRef][Green Version]
- Kumashiro, R.; Konishi, K.; Chiba, T.; Akahoshi, T.; Nakamura, S.; Murata, M.; Tomikawa, M.; Matsumoto, T.; Maehara, Y.; Hashizume, M. Integrated endoscopic system based on optical imaging and hyperspectral data analysis for colorectal cancer detection. Anticancer Res. 2016, 36, 3925–3932. [Google Scholar]
- Han, Z.; Zhang, A.; Wang, X.; Sun, Z.; Wang, M.D.; Xie, T. In vivo use of hyperspectral imaging to develop a noncontact endoscopic diagnosis support system for malignant colorectal tumors. J. Biomed. Opt. 2016, 21, 16001. [Google Scholar] [CrossRef]
- Yuan, X.; Zhang, D.; Wang, C.; Dai, B.; Zhao, M.; Li, B. Hyperspectral Imaging and SPA--LDA Quantitative Analysis for Detection of Colon Cancer Tissue. J. Appl. Spectrosc. 2018, 85, 307–312. [Google Scholar] [CrossRef]
- Ge, Z.; Schomacker, K.T.; Nishioka, N.S. Identification of colonic dysplasia and neoplasia by diffuse reflectance spectroscopy and pattern recognition techniques. Appl. Spectrosc. 1998, 52, 833–839. [Google Scholar] [CrossRef]
- Rodriguez-Diaz, E.; Huang, Q.; Cerda, S.R.; O’Brien, M.J.; Bigio, I.J.; Singh, S.K. Endoscopic histological assessment of colonic polyps by using elastic scattering spectroscopy. Gastrointest. Endosc. 2015, 81, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lin, Z.; Wu, H.; Wang, L.; Wu, T.; Tan, C. Diagnosis of colorectal cancer by near-infrared optical fiber spectroscopy and random forest. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 135, 185–191. [Google Scholar] [CrossRef]
- Chen, H.; Lin, Z.; Mo, L.; Tan, C. Identification of colorectal cancer using near-infrared spectroscopy and adaboost with decision stump. Anal. Lett. 2017, 50, 2608–2618. [Google Scholar] [CrossRef]
- Chen, H.; Tan, C.; Wu, H.; Lin, Z.; Wu, T. Feasibility of rapid diagnosis of colorectal cancer by near-infrared spectroscopy and support vector machine. Anal. Lett. 2014, 47, 2580–2593. [Google Scholar] [CrossRef]
- Ehlen, L.; Zabarylo, U.J.; Speichinger, F.; Bogomolov, A.; Belikova, V.; Bibikova, O.; Artyushenko, V.; Minet, O.; Beyer, K.; Kreis, M.E.; et al. Synergy of Fluorescence and Near-Infrared Spectroscopy in Detection of Colorectal Cancer. J. Surg. Res. 2019, 242, 349–356. [Google Scholar] [CrossRef]
- Claridge, E.; Hidović-Rowe, D. Model based inversion for deriving maps of histological parameters characteristic of cancer from ex-vivo multispectral images of the colon. IEEE Trans. Med. Imaging 2013, 33, 822–835. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef] [PubMed]
- Brereton, R.G.; Lloyd, G.R. Partial least squares discriminant analysis: Taking the magic away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Gromski, P.S.; Muhamadali, H.; Ellis, D.I.; Xu, Y.; Correa, E.; Turner, M.L.; Goodacre, R. A tutorial review: Metabolomics and partial least squares-discriminant analysis--a marriage of convenience or a shotgun wedding. Anal. Chim. Acta 2015, 879, 10–23. [Google Scholar] [CrossRef]
- ElMasry, G.; Nakauchi, S. Prediction of meat spectral patterns based on optical properties and concentrations of the major constituents. Food Sci. Nutr. 2016, 4, 269–283. [Google Scholar] [CrossRef]
- Maruo, K.; Yamada, Y. Near-infrared noninvasive blood glucose prediction without using multivariate analyses: Introduction of imaginary spectra due to scattering change in the skin. J. Biomed. Opt. 2015, 20, 47003. [Google Scholar] [CrossRef] [PubMed]
- Tromberg, B.J.; Shah, N.; Lanning, R.; Cerussi, A.; Espinoza, J.; Pham, T.; Svaasand, L.; Butler, J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia 2000, 2, 26–40. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-Y.; Chen, C.-T.; Chiang, C.-P.; Young, S.-T.; Chow, S.-N.; Chiang, H.K. Partial least-squares discriminant analysis on autofluorescence spectra of oral carcinogenesis. Appl. Spectrosc. 1998, 52, 1190–1196. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Cosci, A.; Kurachi, C. Assessment of oxidative stress and metabolic rates in liver grafts using time-resolved fluorescence spectroscopy. In Proceedings of the Biophotonics: Photonic Solutions for Better Health Care VI, Strasbourg, France, 22–26 April 2018; Volume 10685. [Google Scholar] [CrossRef]
- Nogueira, M.S.; Rosa, R.G.T.; Pratavieira, S.; D’almeida, C.D.P.; Kurachi, C. Assembly and characterization of a fluorescence lifetime spectroscopy system for skin lesions diagnostic. Biophotonics S. Am. 2015, 9531, 95313D. [Google Scholar] [CrossRef]
- Almeida, C.D.P.D.; Campos, C.; Nogueira, M.S.; Kurachi, C. Time-resolved and steady-state fluorescence spectroscopy for the assessment of skin photoaging process. In Proceedings of the Biophotonics South America, Rio de Janeiro, Brazil, 23–25 May 2015; Volume 9531. [Google Scholar] [CrossRef]
- Baker, M.J.; Byrne, H.J.; Chalmers, J.; Gardner, P.; Goodacre, R.; Henderson, A.; Kazarian, S.G.; Martin, F.L.; Moger, J.; Stone, N.; et al. Clinical applications of infrared and Raman spectroscopy: State of play and future challenges. Analyst 2018, 143, 1735–1757. [Google Scholar] [CrossRef] [PubMed]
- Coelho, M.C.D.M.S.; Leal, L.B.; Nogueira, M.S.; Castro, P.A.; Peralta, F.; Zezell, D.M. Biochemical characterization of saliva of smoking and non-smoking patients by using Fourier-transform infrared spectroscopy. In Proceedings of the Biomedical Vibrational Spectroscopy 2022: Advances in Research and Industry, San Francisco, CA, USA, 22 January–28 February 2022; Volume 11957, p. 119570E. [Google Scholar] [CrossRef]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef]
- Bravo, J.J.; Paulsen, K.D.; Roberts, D.W.; Kanick, S.C. Sub-diffuse optical biomarkers characterize localized microstructure and function of cortex and malignant tumor. Opt. Lett. 2016, 41, 781–784. [Google Scholar] [CrossRef]
- Kirkpatrick, N.D.; Brewer, M.A.; Utzinger, U. Endogenous optical biomarkers of ovarian cancer evaluated with multiphoton microscopy. Cancer Epidemiol. Prev. Biomark. 2007, 16, 2048–2057. [Google Scholar] [CrossRef]
- Petrik, V.; Loosemore, A.; Howe, F.A.; Bell, B.A.; Papadopoulos, M.C. OMICS and brain tumour biomarkers. Br. J. Neurosurg. 2006, 20, 275–280. [Google Scholar] [CrossRef]
- Georgakoudi, I.; Quinn, K.P. Optical imaging using endogenous contrast to assess metabolic state. Annu. Rev. Biomed. Eng. 2012, 14, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O. Non-coding RNAs as biomarkers for colorectal cancer screening and early detection. In Non-Coding RNAs in Colorectal Cancer; Springer: Boston, MA USA, 2016; pp. 153–170. [Google Scholar]
- Custodio, A.; Barriuso, J.; De Castro, J.; Martínez-Marín, V.; Moreno, V.; Rodríguez-Salas, N.; Feliu, J. Molecular markers to predict outcome to antiangiogenic therapies in colorectal cancer: Current evidence and future perspectives. Cancer Treat. Rev. 2013, 39, 908–924. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Wang, L.; Liu, H.-L.; Wu, X.-B.; Wang, H.-S.; Liu, Z.-H.; Li, Y.; Diao, D.-C.; Chen, H.-L.; Peng, J.-S. Tissue metabolomic fingerprinting reveals metabolic disorders associated with human gastric cancer morbidity. Oncol. Rep. 2011, 26, 431–438. [Google Scholar] [PubMed]
- Gadducci, A.; Guerrieri, M.E.; Greco, C. Tissue biomarkers as prognostic variables of cervical cancer. Crit. Rev. Oncol. Hematol. 2013, 86, 104–129. [Google Scholar] [CrossRef] [PubMed]
- Beger, R.D. A review of applications of metabolomics in cancer. Metabolites 2013, 3, 552–574. [Google Scholar] [CrossRef] [PubMed]
- Langhout, G.C.; Bydlon, T.M.; van der Voort, M.; Müller, M.; Kortsmit, J.; Lucassen, G.; Balthasar, A.J.R.; van Geffen, G.-J.; Steinfeldt, T.; Sterenborg, H.J.C.M.; et al. Nerve detection using optical spectroscopy, an evaluation in four different models: In human and swine, in-vivo, and post mortem. Lasers Surg. Med. 2018, 50, 253–261. [Google Scholar] [CrossRef]
- Salomatina, E.; Yaroslavsky, A.N. Evaluation of the in vivo and ex vivo optical properties in a mouse ear model. Phys. Med. Biol. 2008, 53, 2797. [Google Scholar] [CrossRef]




| Patient and Cancer Characteristics | Number of Patients/Tumors | |
|---|---|---|
| Total | 47 | |
| Gender | Male | 32 |
| Female | 15 | |
| Age (years) | Median | 69 |
| Minimum | 40 | |
| Maximum | 89 | |
| Interquartile range | 13.5 | |
| Cancer types | Adenocarcinoma | 47 |
| T (tumor) stage | pT1 | 5 |
| pT2 | 7 | |
| pT3 | 26 | |
| pT4 | 9 | |
| N (lymph node) stage | N0 | 19 |
| N1a | 9 | |
| N1b | 12 | |
| N1c | 1 | |
| N2 | 1 | |
| N2a | 4 | |
| N2b | 1 |
| Wavelengths | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|
| 350–540 nm, 540–590 nm | (78.2 ± 0.9)% | (75.8 ± 1.6)% | (77.1 ± 1.0)% | (0.854 ± 0.007) |
| 350–590 nm | (78.4 ± 1.1)% | (74.9 ± 1.4)% | (76.7 ± 0.8)% | (0.85 ± 0.005) |
| 600–1230 nm | (79.8 ± 0.9)% | (84.4 ± 1.4)% | (81.9 ± 0.8)% | (0.894 ± 0.005) |
| 350–590 nm, 600–1230 nm | (78.6 ± 0.7)% | (75.4 ± 1.0)% | (77.1 ± 0.7)% | (0.854 ± 0.006) |
| 1530–1700 nm | (70.9 ± 1.1)% | (67.0 ± 1.6)% | (69.1 ± 1.0)% | (0.771 ± 0.009) |
| 1730–1850 nm | (69.1 ± 1.0)% | (69.5 ± 1.3)% | (69.3 ± 0.9)% | (0.765 ± 0.007) |
| 1530–1700 nm, 1730–1850 nm | (76.4 ± 0.9)% | (77.7 ± 1.1)% | (77.0 ± 0.7)% | (0.845 ± 0.006) |
| 350–590 nm, 600–1230 nm, 1530–1700 nm, 1730–1850 nm | (85.5 ± 0.8)% | (84.0 ± 1.0)% | (84.8 ± 0.7)% | (0.919 ± 0.004) |
| 350–1920 nm | (85.6 ± 0.9)% | (80.4 ± 1.1)% | (83.2 ± 0.8)% | (0.905 ± 0.005) |
| Wavelengths | Sensitivity | Specificity | Accuracy | AUC |
|---|---|---|---|---|
| 380–400 nm | (86.0 ± 0.9)% | (85.0 ± 0.9)% | (85.6 ± 0.7)% | (0.925 ± 0.004) |
| 420–610 nm | (85.6 ± 0.5)% | (87.2 ± 0.6)% | (86.3 ± 0.3)% | (0.93 ± 0.004) |
| 650–950 nm | (89.6 ± 0.6)% | (89.7 ± 1.0)% | (89.7 ± 0.6)% | (0.96 ± 0.004) |
| 380–400 nm, 420–610 nm, 650–950 nm | (87.0 ± 0.8)% | (85.5 ± 0.8)% | (86.3 ± 0.7)% | (0.931 ± 0.003) |
| 1200–1220 nm | (67.8 ± 1.2)% | (63.3 ± 1.9)% | (65.7 ± 1.1)% | (0.707 ± 0.013) |
| 1250–1380 nm | (77.1 ± 1.0)% | (80.7 ± 1.0)% | (78.8 ± 0.7)% | (0.87 ± 0.006) |
| 1600–1690 nm | (62.4 ± 1.1)% | (58.3 ± 1.6)% | (60.4 ± 0.8)% | (0.654 ± 0.008) |
| 1200–1220 nm, 1250–1380 nm, 1600–1690 nm | (77.6 ± 1.0)% | (84.7 ± 1.1)% | (81.0 ± 0.8)% | (0.883 ± 0.006) |
| 380–400 nm, 420–610 nm, 650–950 nm, 1200–1220 nm, 1250–1380 nm, 1600–1690 nm | (89.1 ± 0.7)% | (90.2 ± 0.7)% | (89.6 ± 0.5)% | (0.957 ± 0.004) |
| 350–1920 nm | (89.3 ± 0.6)% | (90.2 ± 0.7)% | (89.7 ± 0.5)% | (0.959 ± 0.003) |
| Scattering and Absorption Features | ||||||||
|---|---|---|---|---|---|---|---|---|
| PLS (Short-SDD probe) | VIS Scat | NIR Scat | Hb | HbO2 | MetHb | Water | Lipid | |
| PLSC1 | X | X | X | |||||
| PLSC2 | X | X | X | X | X | |||
| PLSC3 | X | X | X | X | ||||
| PLSC4 | X | X | X | X | ||||
| PLS (Long-SDD probe) | PLSC1 | X | X | X | X | |||
| PLSC2 | X | X | X | X | X | X | ||
| PLSC3 | X | X | X | X | ||||
| PLSC4 | X | X | X | X | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saito Nogueira, M.; Maryam, S.; Amissah, M.; McGuire, A.; Spillane, C.; Killeen, S.; Andersson-Engels, S.; O’Riordain, M. Insights into Biochemical Sources and Diffuse Reflectance Spectral Features for Colorectal Cancer Detection and Localization. Cancers 2022, 14, 5715. https://doi.org/10.3390/cancers14225715
Saito Nogueira M, Maryam S, Amissah M, McGuire A, Spillane C, Killeen S, Andersson-Engels S, O’Riordain M. Insights into Biochemical Sources and Diffuse Reflectance Spectral Features for Colorectal Cancer Detection and Localization. Cancers. 2022; 14(22):5715. https://doi.org/10.3390/cancers14225715
Chicago/Turabian StyleSaito Nogueira, Marcelo, Siddra Maryam, Michael Amissah, Andrew McGuire, Chloe Spillane, Shane Killeen, Stefan Andersson-Engels, and Micheal O’Riordain. 2022. "Insights into Biochemical Sources and Diffuse Reflectance Spectral Features for Colorectal Cancer Detection and Localization" Cancers 14, no. 22: 5715. https://doi.org/10.3390/cancers14225715
APA StyleSaito Nogueira, M., Maryam, S., Amissah, M., McGuire, A., Spillane, C., Killeen, S., Andersson-Engels, S., & O’Riordain, M. (2022). Insights into Biochemical Sources and Diffuse Reflectance Spectral Features for Colorectal Cancer Detection and Localization. Cancers, 14(22), 5715. https://doi.org/10.3390/cancers14225715







