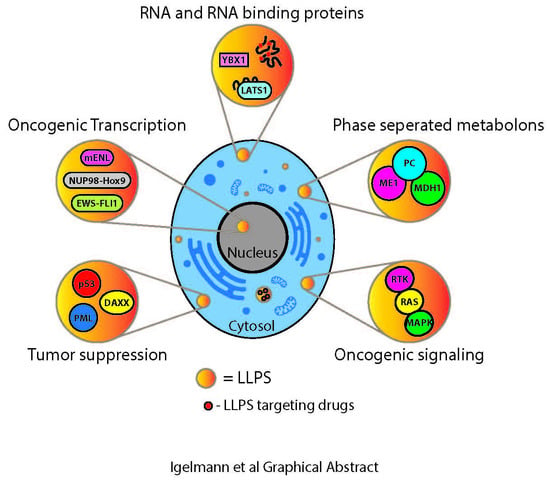
Liquid–Liquid Phase Separation in Cancer Signaling, Metabolism and Anticancer Therapy
Abstract
:Simple Summary
Abstract
1. Introduction
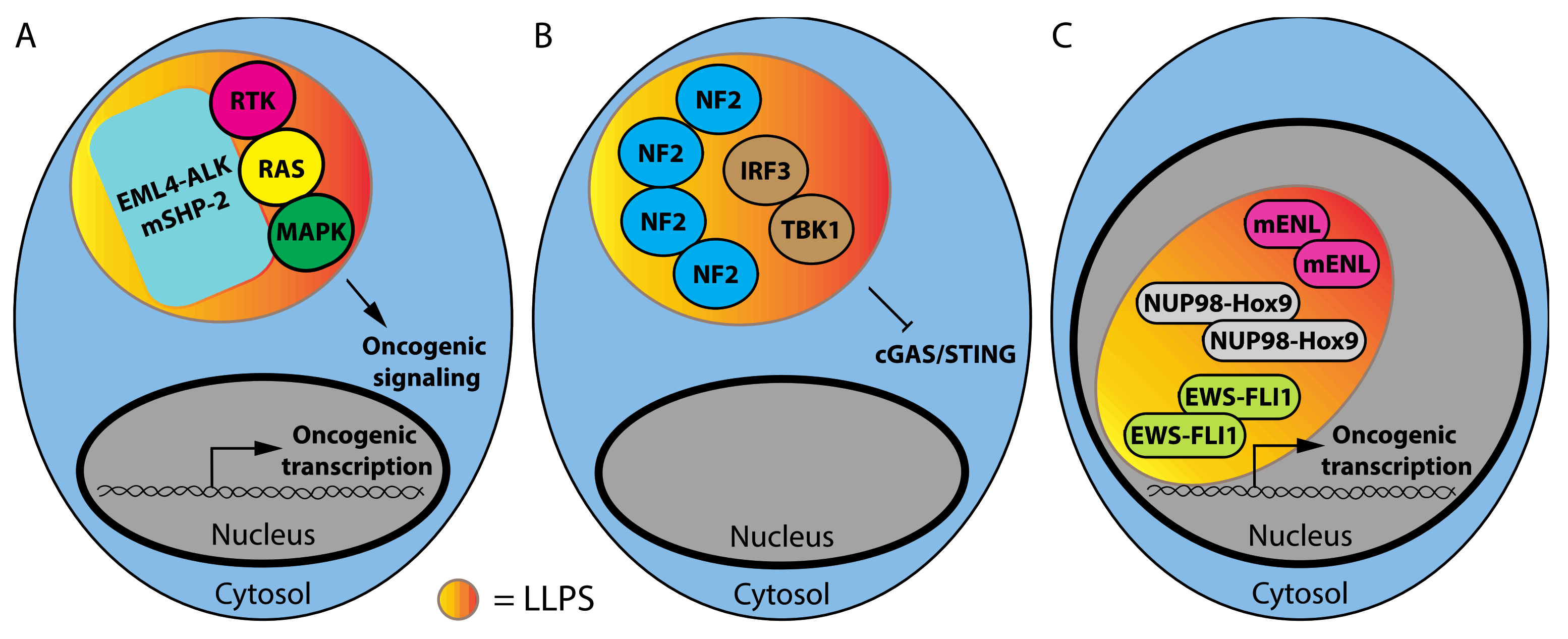
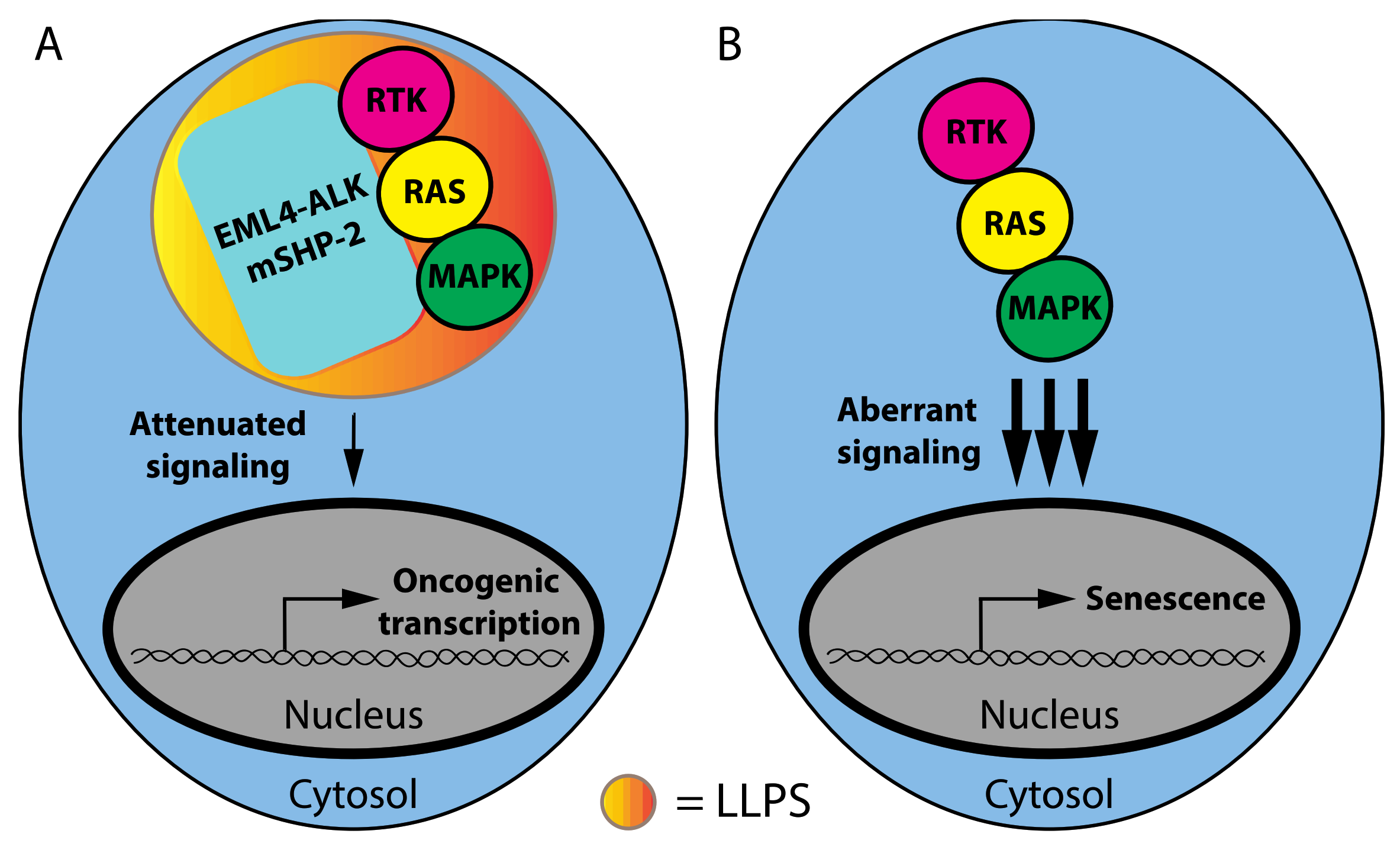
1.1. Phase Separation and Mutant Oncoproteins
1.2. Phase Separation and Tumor Suppressors
1.3. RNA-Binding Proteins and Non-Coding RNAs That Affect LLPS in Cancer
1.4. Phase Separation and Cancer Metabolism
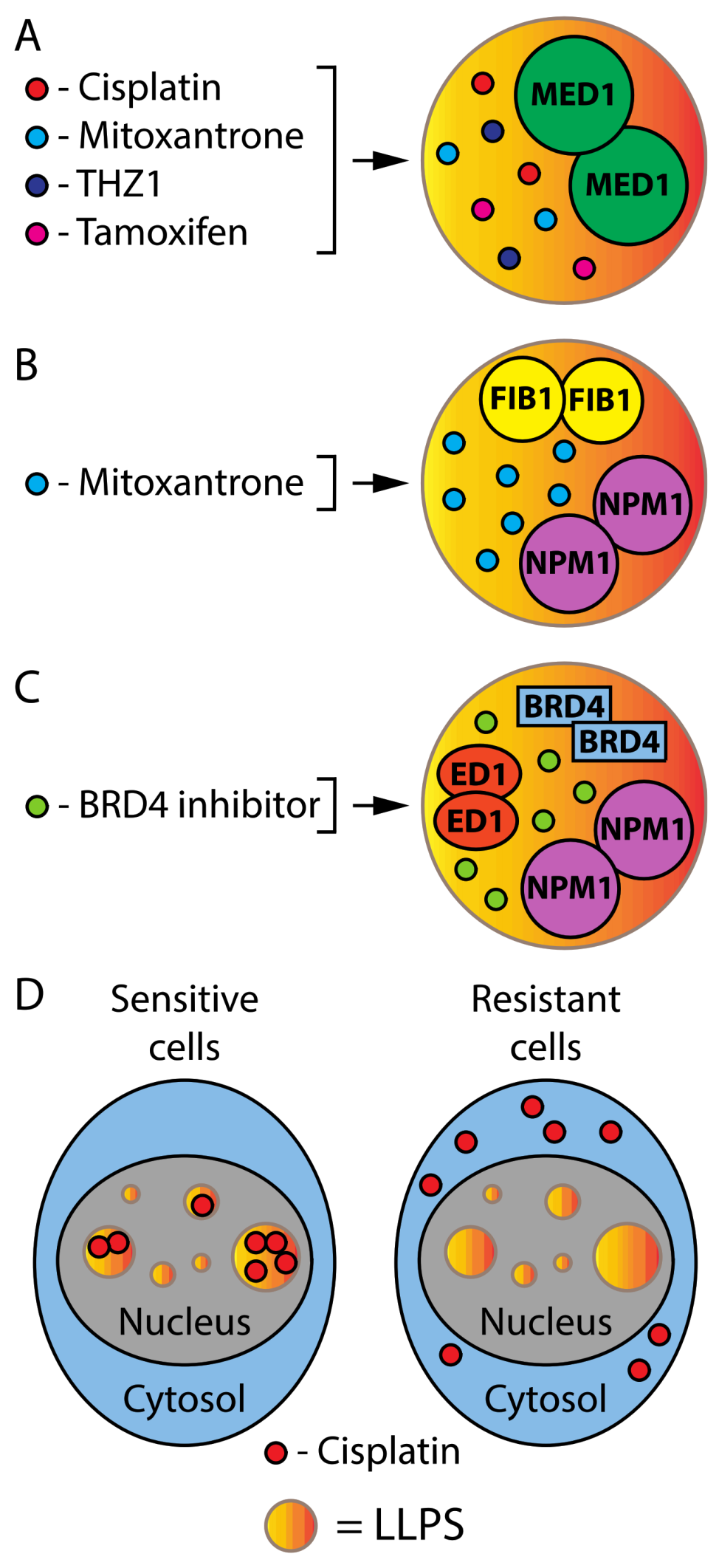
1.5. Phase Separation in Cancer Therapy
2. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lau, Y.; Oamen, H.P.; Caudron, F. Protein Phase Separation during Stress Adaptation and Cellular Memory. Cells 2020, 9, 1302. [Google Scholar] [CrossRef] [PubMed]
- Javed, K.; Jullien, J.; Agarwal, G.; Lawrence, N.; Butler, R.; Ioannou, P.S.; Nazir, F.; Gurdon, J.B. DNA-induced spatial entrapment of general transcription machinery can stabilize gene expression in a nondividing cell. Proc. Natl. Acad. Sci. USA 2022, 119, e2116091119. [Google Scholar] [CrossRef] [PubMed]
- Dine, E.; Gil, A.A.; Uribe, G.; Brangwynne, C.P.; Toettcher, J.E. Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Syst. 2018, 6, 655–663.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyon, A.S.; Peeples, W.B.; Rosen, M.K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 2021, 22, 215–235. [Google Scholar] [CrossRef]
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Boeynaems, S.; Bogaert, E.; Kovacs, D.; Konijnenberg, A.; Timmerman, E.; Volkov, A.; Guharoy, M.; De Decker, M.; Jaspers, T.; Ryan, V.H.; et al. Phase Separation of C9orf72 Dipeptide Repeats Perturbs Stress Granule Dynamics. Mol. Cell 2017, 65, 1044–1055.e5. [Google Scholar] [CrossRef] [Green Version]
- Rhine, K.; Makurath, M.A.; Liu, J.; Skanchy, S.; Lopez, C.; Catalan, K.F.; Ma, Y.; Fare, C.M.; Shorter, J.; Ha, T.; et al. ALS/FTLD-Linked Mutations in FUS Glycine Residues Cause Accelerated Gelation and Reduced Interactions with Wild-Type FUS. Mol. Cell 2020, 80, 666–681.e8. [Google Scholar] [CrossRef]
- McSwiggen, D.T.; Mir, M.; Darzacq, X.; Tjian, R. Evaluating phase separation in live cells: Diagnosis, caveats, and functional consequences. Genes Dev. 2019, 33, 1619–1634. [Google Scholar] [CrossRef]
- Brady, J.P.; Farber, P.J.; Sekhar, A.; Lin, Y.H.; Huang, R.; Bah, A.; Nott, T.J.; Chan, H.S.; Baldwin, A.J.; Forman-Kay, J.D.; et al. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. USA 2017, 114, E8194–E8203. [Google Scholar] [CrossRef] [Green Version]
- Murthy, A.C.; Dignon, G.L.; Kan, Y.; Zerze, G.H.; Parekh, S.H.; Mittal, J.; Fawzi, N.L. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 2019, 26, 637–648. [Google Scholar] [CrossRef]
- Murakami, K.; Kajimoto, S.; Shibata, D.; Kuroi, K.; Fujii, F.; Nakabayashi, T. Observation of liquid-liquid phase separation of ataxin-3 and quantitative evaluation of its concentration in a single droplet using Raman microscopy. Chem. Sci. 2021, 12, 7411–7418. [Google Scholar] [CrossRef]
- Ellis, R.J. Macromolecular crowding: An important but neglected aspect of the intracellular environment. Curr. Opin. Struct. Biol. 2001, 11, 114–119. [Google Scholar] [CrossRef]
- Freeman Rosenzweig, E.S.; Xu, B.; Kuhn Cuellar, L.; Martinez-Sanchez, A.; Schaffer, M.; Strauss, M.; Cartwright, H.N.; Ronceray, P.; Plitzko, J.M.; Forster, F.; et al. The Eukaryotic CO2-Concentrating Organelle Is Liquid-like and Exhibits Dynamic Reorganization. Cell 2017, 171, 148–162.e19. [Google Scholar] [CrossRef] [Green Version]
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Q.; McAtee, C.K.; Su, X. Phase separation in immune signalling. Nat. Rev. Immunol. 2021, 22, 188–199. [Google Scholar] [CrossRef]
- Su, X.; Ditlev, J.A.; Hui, E.; Xing, W.; Banjade, S.; Okrut, J.; King, D.S.; Taunton, J.; Rosen, M.K.; Vale, R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352, 595–599. [Google Scholar] [CrossRef] [Green Version]
- Deviri, D.; Safran, S.A. Physical theory of biological noise buffering by multicomponent phase separation. Proc. Natl. Acad. Sci. USA 2021, 118, e2100099118. [Google Scholar] [CrossRef]
- Ouyang, M.; Li, X.; Zhang, J.; Feng, P.; Pu, H.; Kong, L.; Bai, Z.; Rong, L.; Xu, X.; Chi, W.; et al. Liquid-Liquid Phase Transition Drives Intra-chloroplast Cargo Sorting. Cell 2020, 180, 1144–1159.e20. [Google Scholar] [CrossRef]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol. 2020, 219, e201907157. [Google Scholar] [CrossRef]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Benard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef]
- Lafontaine, D.L.J.; Riback, J.A.; Bascetin, R.; Brangwynne, C.P. The nucleolus as a multiphase liquid condensate. Nat. Rev. Mol. Cell Biol. 2021, 22, 165–182. [Google Scholar] [CrossRef] [PubMed]
- Deniz, A.A. Networking and Dynamic Switches in Biological Condensates. Cell 2020, 181, 228–230. [Google Scholar] [CrossRef] [PubMed]
- Guillen-Boixet, J.; Kopach, A.; Holehouse, A.S.; Wittmann, S.; Jahnel, M.; Schlussler, R.; Kim, K.; Trussina, I.; Wang, J.; Mateju, D.; et al. RNA-Induced Conformational Switching and Clustering of G3BP Drive Stress Granule Assembly by Condensation. Cell 2020, 181, 346–361.e17. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.W.; Kedersha, N.; Lee, D.S.W.; Strom, A.R.; Drake, V.; Riback, J.A.; Bracha, D.; Eeftens, J.M.; Iwanicki, A.; Wang, A.; et al. Competing Protein-RNA Interaction Networks Control Multiphase Intracellular Organization. Cell 2020, 181, 306–324.e28. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Mathieu, C.; Kolaitis, R.M.; Zhang, P.; Messing, J.; Yurtsever, U.; Yang, Z.; Wu, J.; Li, Y.; Pan, Q.; et al. G3BP1 Is a Tunable Switch that Triggers Phase Separation to Assemble Stress Granules. Cell 2020, 181, 325–345.e28. [Google Scholar] [CrossRef]
- Fox, A.H.; Nakagawa, S.; Hirose, T.; Bond, C.S. Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem. Sci. 2018, 43, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Gibson, B.A.; Doolittle, L.K.; Schneider, M.W.G.; Jensen, L.E.; Gamarra, N.; Henry, L.; Gerlich, D.W.; Redding, S.; Rosen, M.K. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e21. [Google Scholar] [CrossRef]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [Green Version]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361, eaar3958. [Google Scholar] [CrossRef] [Green Version]
- Lukas, C.; Savic, V.; Bekker-Jensen, S.; Doil, C.; Neumann, B.; Pedersen, R.S.; Grofte, M.; Chan, K.L.; Hickson, I.D.; Bartek, J.; et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011, 13, 243–253. [Google Scholar] [CrossRef]
- Ochs, F.; Karemore, G.; Miron, E.; Brown, J.; Sedlackova, H.; Rask, M.B.; Lampe, M.; Buckle, V.; Schermelleh, L.; Lukas, J.; et al. Stabilization of chromatin topology safeguards genome integrity. Nature 2019, 574, 571–574. [Google Scholar] [CrossRef]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194. [Google Scholar] [CrossRef] [Green Version]
- Cai, D.; Liu, Z.; Lippincott-Schwartz, J. Biomolecular Condensates and Their Links to Cancer Progression. Trends Biochem. Sci. 2021, 46, 535–549. [Google Scholar] [CrossRef]
- Zhu, G.; Xie, J.; Kong, W.; Xie, J.; Li, Y.; Du, L.; Zheng, Q.; Sun, L.; Guan, M.; Li, H.; et al. Phase Separation of Disease-Associated SHP2 Mutants Underlies MAPK Hyperactivation. Cell 2020, 183, 490–502.e18. [Google Scholar] [CrossRef]
- Tulpule, A.; Guan, J.; Neel, D.S.; Allegakoen, H.R.; Lin, Y.P.; Brown, D.; Chou, Y.T.; Heslin, A.; Chatterjee, N.; Perati, S.; et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 2021, 184, 2649–2664.e18. [Google Scholar] [CrossRef]
- Witzel, F.; Maddison, L.; Bluthgen, N. How scaffolds shape MAPK signaling: What we know and opportunities for systems approaches. Front. Physiol. 2012, 3, 475. [Google Scholar] [CrossRef] [Green Version]
- Deschenes-Simard, X.; Gaumont-Leclerc, M.F.; Bourdeau, V.; Lessard, F.; Moiseeva, O.; Forest, V.; Igelmann, S.; Mallette, F.A.; Saba-El-Leil, M.K.; Meloche, S.; et al. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Genes Dev. 2013, 27, 900–915. [Google Scholar] [CrossRef] [Green Version]
- Deschenes-Simard, X.; Kottakis, F.; Meloche, S.; Ferbeyre, G. ERKs in cancer: Friends or foes? Cancer Res. 2014, 74, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Malleshaiah, M.K.; Shahrezaei, V.; Swain, P.S.; Michnick, S.W. The scaffold protein Ste5 directly controls a switch-like mating decision in yeast. Nature 2010, 465, 101–105. [Google Scholar] [CrossRef]
- Koshland, D.E., Jr.; Goldbeter, A.; Stock, J.B. Amplification and adaptation in regulatory and sensory systems. Science 1982, 217, 220–225. [Google Scholar] [CrossRef]
- Meng, F.; Yu, Z.; Zhang, D.; Chen, S.; Guan, H.; Zhou, R.; Wu, Q.; Zhang, Q.; Liu, S.; Venkat Ramani, M.K.; et al. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol. Cell 2021, 81, 4147–4164.e7. [Google Scholar] [CrossRef]
- Dou, Z.; Ghosh, K.; Vizioli, M.G.; Zhu, J.; Sen, P.; Wangensteen, K.J.; Simithy, J.; Lan, Y.; Lin, Y.; Zhou, Z.; et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550, 402–406. [Google Scholar] [CrossRef] [Green Version]
- Gluck, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef]
- An, X.; Zhu, Y.; Zheng, T.; Wang, G.; Zhang, M.; Li, J.; Ji, H.; Li, S.; Yang, S.; Xu, D.; et al. An Analysis of the Expression and Association with Immune Cell Infiltration of the cGAS/STING Pathway in Pan-Cancer. Mol. Ther. Nucleic Acids 2019, 14, 80–89. [Google Scholar] [CrossRef] [Green Version]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Chong, S.; Xuan, F.; Liang, A.; Cui, X.; Gates, L.; Carroll, T.S.; Li, Y.; Feng, L.; Chen, G.; et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 2020, 577, 121–126. [Google Scholar] [CrossRef]
- Ahn, J.H.; Davis, E.S.; Daugird, T.A.; Zhao, S.; Quiroga, I.Y.; Uryu, H.; Li, J.; Storey, A.J.; Tsai, Y.H.; Keeley, D.P.; et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 2021, 595, 591–595. [Google Scholar] [CrossRef]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e19. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Woo, A.J.; Chu, J.; Snow, J.W.; Fujiwara, Y.; Kim, C.G.; Cantor, A.B.; Orkin, S.H. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 2010, 143, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Wong, D.J.; Liu, H.; Ridky, T.W.; Cassarino, D.; Segal, E.; Chang, H.Y. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Eaton, E.N.; Li, S.H.; Chaffer, C.L.; Reinhardt, F.; Kah, K.J.; Bell, G.; Guo, W.; Rubin, J.; Richardson, A.L.; et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011, 145, 926–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petronilho, E.C.; Pedrote, M.M.; Marques, M.A.; Passos, Y.M.; Mota, M.F.; Jakobus, B.; de Sousa, G.D.S.; da Costa, F.P.; Felix, A.L.; Ferretti, G.D.S.; et al. Phase separation of p53 precedes aggregation and is affected by oncogenic mutations and ligands. Chem. Sci. 2021, 12, 7334–7349. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, J.J.; Otero, J.H.; Scott, D.C.; Szulc, E.; Martin, E.W.; Sabri, N.; Granata, D.; Marzahn, M.R.; Lindorff-Larsen, K.; Salvatella, X.; et al. Cancer Mutations of the Tumor Suppressor SPOP Disrupt the Formation of Active, Phase-Separated Compartments. Mol. Cell 2018, 72, 19–36.e8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.Z.; Lu, T.W.; Stolerman, L.M.; Tenner, B.; Yang, J.R.; Zhang, J.F.; Falcke, M.; Rangamani, P.; Taylor, S.S.; Mehta, S.; et al. Phase Separation of a PKA Regulatory Subunit Controls cAMP Compartmentation and Oncogenic Signaling. Cell 2020, 182, 1531–1544.e15. [Google Scholar] [CrossRef]
- Bourdeau, V.; Baudry, D.; Ferbeyre, G. PML links aberrant cytokine signaling and oncogenic stress to cellular senescence. Front. Biosci. 2009, 14, 475–485. [Google Scholar] [CrossRef] [Green Version]
- Jin, J. Interplay between ubiquitylation and SUMOylation: Empowered by phase separation. J. Biol. Chem. 2019, 294, 15235–15236. [Google Scholar] [CrossRef] [Green Version]
- McManus, F.P.; Bourdeau, V.; Acevedo, M.; Lopes-Paciencia, S.; Mignacca, L.; Lamoliatte, F.; Rojas Pino, J.W.; Ferbeyre, G.; Thibault, P. Quantitative SUMO proteomics reveals the modulation of several PML nuclear body associated proteins and an anti-senescence function of UBC9. Sci. Rep. 2018, 8, 7754. [Google Scholar] [CrossRef]
- Vernier, M.; Bourdeau, V.; Gaumont-Leclerc, M.F.; Moiseeva, O.; Begin, V.; Saad, F.; Mes-Masson, A.M.; Ferbeyre, G. Regulation of E2Fs and senescence by PML nuclear bodies. Genes Dev. 2011, 25, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Talluri, S.; Dick, F.A. The retinoblastoma protein and PML collaborate to organize heterochromatin and silence E2F-responsive genes during senescence. Cell Cycle 2013, 13, 641–651. [Google Scholar] [CrossRef] [Green Version]
- de The, H.; Pandolfi, P.P.; Chen, Z. Acute Promyelocytic Leukemia: A Paradigm for Oncoprotein-Targeted Cure. Cancer Cell 2017, 32, 552–560. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Li, W.; Song, Y.; Wang, Z.; Ju, R.; Ulman, A.; Hu, J.; Palomba, F.; Zhao, Y.; Le, J.P.; et al. UTX condensation underlies its tumour-suppressive activity. Nature 2021, 597, 726–731. [Google Scholar] [CrossRef]
- Yao, R.W.; Xu, G.; Wang, Y.; Shan, L.; Luan, P.F.; Wang, Y.; Wu, M.; Yang, L.Z.; Xing, Y.H.; Yang, L.; et al. Nascent Pre-rRNA Sorting via Phase Separation Drives the Assembly of Dense Fibrillar Components in the Human Nucleolus. Mol. Cell 2019, 76, 767–783.e11. [Google Scholar] [CrossRef]
- Wiedner, H.J.; Giudice, J. It’s not just a phase: Function and characteristics of RNA-binding proteins in phase separation. Nat. Struct. Mol. Biol. 2021, 28, 465–473. [Google Scholar] [CrossRef]
- Li, W.; Hu, J.; Shi, B.; Palomba, F.; Digman, M.A.; Gratton, E.; Jiang, H. Biophysical properties of AKAP95 protein condensates regulate splicing and tumorigenesis. Nat. Cell Biol. 2020, 22, 960–972. [Google Scholar] [CrossRef]
- Hu, X.; Harvey, S.E.; Zheng, R.; Lyu, J.; Grzeskowiak, C.L.; Powell, E.; Piwnica-Worms, H.; Scott, K.L.; Cheng, C. The RNA-binding protein AKAP8 suppresses tumor metastasis by antagonizing EMT-associated alternative splicing. Nat. Commun. 2020, 11, 486. [Google Scholar] [CrossRef]
- Liu, X.M.; Ma, L.; Schekman, R. Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. eLife 2021, 10, e71982. [Google Scholar] [CrossRef]
- Jayavelu, A.K.; Schnoder, T.M.; Perner, F.; Herzog, C.; Meiler, A.; Krishnamoorthy, G.; Huber, N.; Mohr, J.; Edelmann-Stephan, B.; Austin, R.; et al. Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 2020, 588, 157–163. [Google Scholar] [CrossRef]
- Perner, F.; Schnoeder, T.M.; Xiong, Y.; Jayavelu, A.K.; Mashamba, N.; Santamaria, N.T.; Huber, N.; Todorova, K.; Hatton, C.; Perner, B.; et al. YBX1 mediates translation of oncogenic transcripts to control cell competition in AML. Leukemia 2022, 36, 426–437. [Google Scholar] [CrossRef]
- Somasekharan, S.P.; El-Naggar, A.; Leprivier, G.; Cheng, H.; Hajee, S.; Grunewald, T.G.; Zhang, F.; Ng, T.; Delattre, O.; Evdokimova, V.; et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J. Cell Biol. 2015, 208, 913–929. [Google Scholar] [CrossRef]
- Li, R.H.; Tian, T.; Ge, Q.W.; He, X.Y.; Shi, C.Y.; Li, J.H.; Zhang, Z.; Liu, F.Z.; Sang, L.J.; Yang, Z.Z.; et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021, 31, 1088–1105. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, W.; Pickering, B.F.; Chu, K.L.; Savino, A.M.; Yang, X.; Luo, H.; Nguyen, D.T.; Mo, S.; Barin, E.; et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell 2021, 39, 958–972.e8. [Google Scholar] [CrossRef]
- Srere, P.A. Complexes of sequential metabolic enzymes. Annu. Rev. Biochem. 1987, 56, 89–124. [Google Scholar] [CrossRef]
- Wheeldon, I.; Minteer, S.D.; Banta, S.; Barton, S.C.; Atanassov, P.; Sigman, M. Substrate channelling as an approach to cascade reactions. Nat. Chem. 2016, 8, 299–309. [Google Scholar] [CrossRef]
- Kerfeld, C.A.; Sawaya, M.R.; Tanaka, S.; Nguyen, C.V.; Phillips, M.; Beeby, M.; Yeates, T.O. Protein structures forming the shell of primitive bacterial organelles. Science 2005, 309, 936–938. [Google Scholar] [CrossRef]
- Zang, K.; Wang, H.; Hartl, F.U.; Hayer-Hartl, M. Scaffolding protein CcmM directs multiprotein phase separation in beta-carboxysome biogenesis. Nat. Struct. Mol. Biol. 2021, 28, 909–922. [Google Scholar] [CrossRef]
- Wang, H.; Yan, X.; Aigner, H.; Bracher, A.; Nguyen, N.D.; Hee, W.Y.; Long, B.M.; Price, G.D.; Hartl, F.U.; Hayer-Hartl, M. Rubisco condensate formation by CcmM in beta-carboxysome biogenesis. Nature 2019, 566, 131–135. [Google Scholar] [CrossRef] [Green Version]
- Narayanaswamy, R.; Levy, M.; Tsechansky, M.; Stovall, G.M.; O’Connell, J.D.; Mirrielees, J.; Ellington, A.D.; Marcotte, E.M. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 2009, 106, 10147–10152. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.Y.; Zhao, H.; Pugh, R.J.; Pedley, A.M.; French, J.; Jones, S.A.; Zhuang, X.; Jinnah, H.; Huang, T.J.; Benkovic, S.J. Purinosome formation as a function of the cell cycle. Proc. Natl. Acad. Sci. USA 2015, 112, 1368–1373. [Google Scholar] [CrossRef] [Green Version]
- Doigneaux, C.; Pedley, A.M.; Mistry, I.N.; Papayova, M.; Benkovic, S.J.; Tavassoli, A. Hypoxia drives the assembly of the multienzyme purinosome complex. J. Biol. Chem. 2020, 295, 9551–9566. [Google Scholar] [CrossRef]
- Jin, M.; Fuller, G.G.; Han, T.; Yao, Y.; Alessi, A.F.; Freeberg, M.A.; Roach, N.P.; Moresco, J.J.; Karnovsky, A.; Baba, M.; et al. Glycolytic Enzymes Coalesce in G Bodies under Hypoxic Stress. Cell Rep. 2017, 20, 895–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuller, G.G.; Han, T.; Freeberg, M.A.; Moresco, J.J.; Ghanbari Niaki, A.; Roach, N.P.; Yates, J.R., 3rd; Myong, S.; Kim, J.K. RNA promotes phase separation of glycolysis enzymes into yeast G bodies in hypoxia. eLife 2020, 9, e48480. [Google Scholar] [CrossRef] [PubMed]
- Kohnhorst, C.L.; Kyoung, M.; Jeon, M.; Schmitt, D.L.; Kennedy, E.L.; Ramirez, J.; Bracey, S.M.; Luu, B.T.; Russell, S.J.; An, S. Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem. 2017, 292, 9191–9203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igelmann, S.; Lessard, F.; Uchenunu, O.; Bouchard, J.; Fernandez-Ruiz, A.; Rowell, M.C.; Lopes-Paciencia, S.; Papadopoli, D.; Fouillen, A.; Ponce, K.J.; et al. A hydride transfer complex reprograms NAD metabolism and bypasses senescence. Mol. Cell 2021, 81, 3848–3865.e19. [Google Scholar] [CrossRef]
- Uriarte, M.; Sen Nkwe, N.; Tremblay, R.; Ahmed, O.; Messmer, C.; Mashtalir, N.; Barbour, H.; Masclef, L.; Voide, M.; Viallard, C.; et al. Starvation-induced proteasome assemblies in the nucleus link amino acid supply to apoptosis. Nat. Commun. 2021, 12, 6984. [Google Scholar] [CrossRef]
- Liu, Q.; Li, J.; Zhang, W.; Xiao, C.; Zhang, S.; Nian, C.; Li, J.; Su, D.; Chen, L.; Zhao, Q.; et al. Glycogen accumulation and phase separation drives liver tumor initiation. Cell 2021, 184, 5559–5576.e19. [Google Scholar] [CrossRef]
- Wolfe, K.; Kofuji, S.; Yoshino, H.; Sasaki, M.; Okumura, K.; Sasaki, A.T. Dynamic compartmentalization of purine nucleotide metabolic enzymes at leading edge in highly motile renal cell carcinoma. Biochem. Biophys. Res. Commun. 2019, 516, 50–56. [Google Scholar] [CrossRef]
- Heller, G.T.; Sormanni, P.; Vendruscolo, M. Targeting disordered proteins with small molecules using entropy. Trends Biochem. Sci. 2015, 40, 491–496. [Google Scholar] [CrossRef] [Green Version]
- Klein, I.A.; Boija, A.; Afeyan, L.K.; Hawken, S.W.; Fan, M.; Dall’Agnese, A.; Oksuz, O.; Henninger, J.E.; Shrinivas, K.; Sabari, B.R.; et al. Partitioning of cancer therapeutics in nuclear condensates. Science 2020, 368, 1386–1392. [Google Scholar] [CrossRef]
- Rovira-Clave, X.; Jiang, S.; Bai, Y.; Zhu, B.; Barlow, G.; Bhate, S.; Coskun, A.F.; Han, G.; Ho, C.K.; Hitzman, C.; et al. Subcellular localization of biomolecules and drug distribution by high-definition ion beam imaging. Nat. Commun. 2021, 12, 4628. [Google Scholar] [CrossRef]
- Ali, A.; Abouleila, Y.; Shimizu, Y.; Hiyama, E.; Watanabe, T.M.; Yanagida, T.; Germond, A. Single-Cell Screening of Tamoxifen Abundance and Effect Using Mass Spectrometry and Raman-Spectroscopy. Anal. Chem. 2019, 91, 2710–2718. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Igelmann, S.; Lessard, F.; Ferbeyre, G. Liquid–Liquid Phase Separation in Cancer Signaling, Metabolism and Anticancer Therapy. Cancers 2022, 14, 1830. https://doi.org/10.3390/cancers14071830
Igelmann S, Lessard F, Ferbeyre G. Liquid–Liquid Phase Separation in Cancer Signaling, Metabolism and Anticancer Therapy. Cancers. 2022; 14(7):1830. https://doi.org/10.3390/cancers14071830
Chicago/Turabian StyleIgelmann, Sebastian, Frédéric Lessard, and Gerardo Ferbeyre. 2022. "Liquid–Liquid Phase Separation in Cancer Signaling, Metabolism and Anticancer Therapy" Cancers 14, no. 7: 1830. https://doi.org/10.3390/cancers14071830
APA StyleIgelmann, S., Lessard, F., & Ferbeyre, G. (2022). Liquid–Liquid Phase Separation in Cancer Signaling, Metabolism and Anticancer Therapy. Cancers, 14(7), 1830. https://doi.org/10.3390/cancers14071830