Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information
Abstract
:Simple Summary
Abstract
1. Introduction

2. Materials and Methods
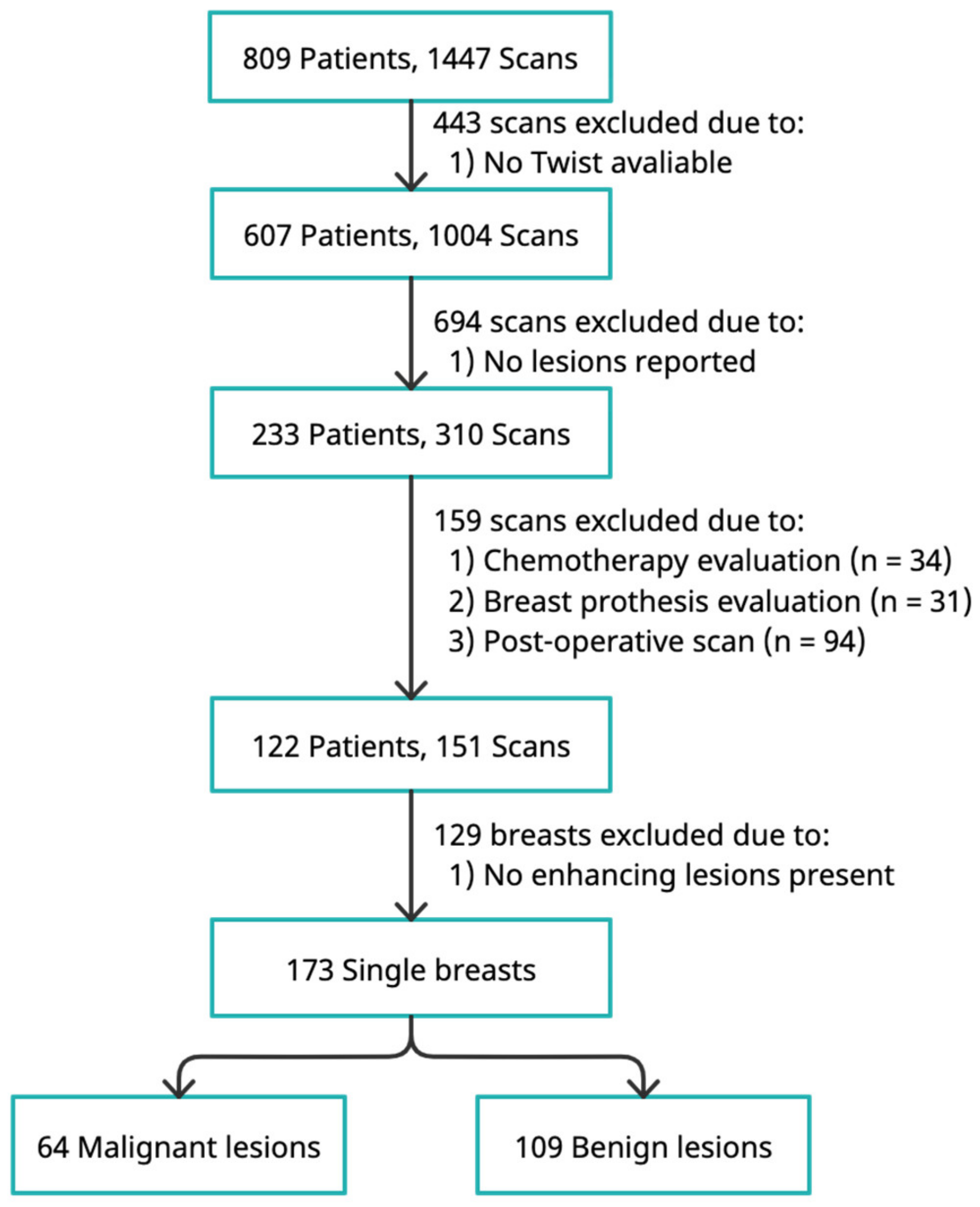
2.1. Study Population
2.2. MRI Protocol
2.3. Development of the Deep-Learning-Based Classification System
3. Results
3.1. Patient and Lesion Characteristics
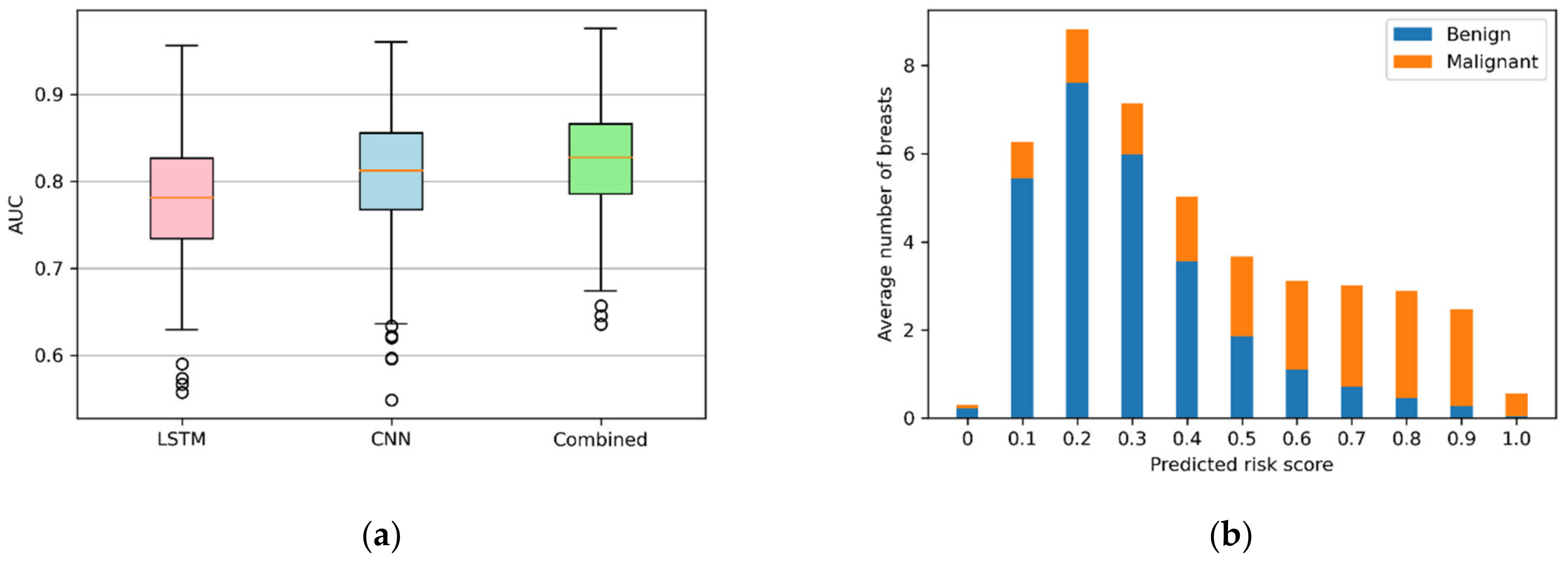
3.2. Performance of Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vandermeer, F.Q.; Bluemke, D.A. Breast MRI: State of the art. Cancer Investig. 2007, 25, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Bakker, M.F.; de Lange, S.V.; Pijnappel, R.M.; Mann, R.M.; Peeters, P.H.M.; Monninkhof, E.M.; Emaus, M.J.; Loo, C.E.; Bisschops, R.H.C.; Lobbes, M.B.I.; et al. Supplemental MRI Screening for Women with Extremely Dense Breast Tissue. N. Engl. J. Med. 2019, 381, 2091–2102. [Google Scholar] [CrossRef] [PubMed]
- Saadatmand, S.; Geuzinge, H.A.; Rutgers, E.J.T.; Mann, R.M.; de Roy van Zuidewijn, D.B.W.; Zonderland, H.M.; Tollenaar, R.A.E.M.; Lobbes, M.B.I.; Ausems, M.G.E.M.; van ’t Riet, M.; et al. MRI versus mammography for breast cancer screening in women with familial risk (FaMRIsc): A multicentre, randomised, controlled trial. Lancet Oncol. 2019, 2045, 1136–1147. [Google Scholar] [CrossRef] [Green Version]
- Kuhl, C.K. A Call for Improved Breast Cancer Screening Strategies, Not Only for Women with Dense Breasts. JAMA Netw. Open 2021, 4, e2121492. [Google Scholar] [CrossRef]
- Mann, R.M.; Kuhl, C.K.; Moy, L. Contrast-enhanced MRI for breast cancer screening. J. Magn. Reson. Imaging 2019, 50, 377–390. [Google Scholar] [CrossRef]
- Chhor, C.M.; Mercado, C.L. Abbreviated MRI protocols: Wave of the future for breast cancer screening. Am. J. Roentgenol. 2017, 208, 284–289. [Google Scholar] [CrossRef]
- Kuhl, C.K.; Schrading, S.; Strobel, K.; Schild, H.H.; Hilgers, R.D.; Bieling, H.B. Abbreviated breast Magnetic Resonance Imaging (MRI): First postcontrast subtracted images and maximum-intensity projection—A novel approach to breast cancer screening with MRI. J. Clin. Oncol. 2014, 32, 2304–2310. [Google Scholar] [CrossRef]
- Pinker, K.; Moy, L.; Sutton, E.J.; Mann, R.M.; Weber, M.; Thakur, S.B.; Jochelson, M.S.; Bago-Horvath, Z.; Morris, E.A.; Baltzer, P.A.; et al. Diffusion-Weighted Imaging with Apparent Diffusion Coefficient Mapping for Breast Cancer Detection as a Stand-Alone Parameter: Comparison with Dynamic Contrast-Enhanced and Multiparametric Magnetic Resonance Imaging. Investig. Radiol. 2018, 53, 587–595. [Google Scholar] [CrossRef]
- Van Zelst, J.C.M.; Vreemann, S.; Witt, H.J.; Gubern-Merida, A.; Dorrius, M.D.; Duvivier, K.; Lardenoije-Broker, S.; Lobbes, M.B.I.; Loo, C.; Veldhuis, W.; et al. Multireader Study on the Diagnostic Accuracy of Ultrafast Breast Magnetic Resonance Imaging for Breast Cancer Screening. Investig. Radiol. 2018, 53, 579–586. [Google Scholar] [CrossRef]
- Partovi, S.; Sin, D.; Lu, Z.; Sieck, L.; Marshall, H.; Pham, R.; Plecha, D. Fast MRI breast cancer screening—Ready for prime time. Clin. Imaging 2020, 60, 160–168. [Google Scholar] [CrossRef]
- Kim, E.S.; Cho, N.; Kim, S.Y.; Kwon, B.R.; Yi, A.; Ha, S.M.; Lee, S.H.; Chang, J.M.; Moon, W.K. Comparison of abbreviated mri and full diagnostic mri in distinguishing between benign and malignant lesions detected by breast mri: A multireader study. Korean J. Radiol. 2021, 22, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Grimm, L.J.; Soo, M.S.; Yoon, S.; Kim, C.; Ghate, S.V.; Johnson, K.S. Abbreviated Screening Protocol for Breast MRI. A Feasibility Study. Acad. Radiol. 2015, 22, 1157–1162. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C.; Di Carlo, P.A.; Lee, B.; Obadina, E.; Sippo, D.; Mullen, L. An Abbreviated Protocol for High-Risk Screening Breast MRI Saves Time and Resources. J. Am. Coll. Radiol. 2016, 13, R74–R80. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, K.H.; Baltzer, P.A.; Dietzel, M.; Krumbein, I.; Geppert, C.; Kaiser, W.A.; Reichenbach, J.R. Resolving arterial phase and temporal enhancement characteristics in DCE MRM at high spatial resolution with TWIST acquisition. J. Magn. Reson. Imaging 2011, 34, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Le, Y.; Kipfer, H.; Majidi, S.; Holz, S.; Dale, B.; Geppert, C.; Kroeker, R.; Lin, C. Application of time-resolved angiography with stochastic trajectories (twist)-dixon in dynamic contrast-enhanced (dce) breast mri. J. Magn. Reson. Imaging 2013, 38, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Mori, N.; Tsuchiya, K.; Schacht, D.V.; Pineda, F.D.; Jiang, Y.; Karczmar, G.S. Kinetic analysis of benign and malignant breast lesions with ultrafast dynamic contrast-enhanced MRI: Comparison with standard kinetic assessment. Am. J. Roentgenol. 2016, 207, 1159–1166. [Google Scholar] [CrossRef]
- Goto, M.; Sakai, K.; Yokota, H.; Kiba, M.; Yoshida, M.; Imai, H.; Weiland, E.; Yokota, I.; Yamada, K. Diagnostic performance of initial enhancement analysis using ultra-fast dynamic contrast-enhanced MRI for breast lesions. Eur. Radiol. 2019, 29, 1164–1174. [Google Scholar] [CrossRef]
- Onishi, N.; Sadinski, M.; Gibbs, P.; Gallagher, K.M.; Hughes, M.C.; Ko, E.S.; Dashevsky, B.Z.; Shanbhag, D.D.; Fung, M.M.; Hunt, T.M.; et al. Differentiation between subcentimeter carcinomas and benign lesions using kinetic parameters derived from ultrafast dynamic contrast-enhanced breast MRI. Eur. Radiol. 2020, 30, 756–766. [Google Scholar] [CrossRef]
- Mus, R.D.; Borelli, C.; Bult, P.; Weiland, E.; Karssemeijer, N.; Barentsz, J.O.; Gubern-Mérida, A.; Platel, B.; Mann, R.M. Time to enhancement derived from ultrafast breast MRI as a novel parameter to discriminate benign from malignant breast lesions. Eur. J. Radiol. 2017, 89, 90–96. [Google Scholar] [CrossRef]
- Codari, M.; Schiaffino, S.; Sardanelli, F.; Trimboli, R.M. Artificial intelligence for breast MRI in 2008-2018: A systematic mapping review. Am. J. Roentgenol. 2019, 212, 280–292. [Google Scholar] [CrossRef]
- Sheth, D.; Giger, M.L. Artificial intelligence in the interpretation of breast cancer on MRI. J. Magn. Reson. Imaging 2020, 51, 1310–1324. [Google Scholar] [CrossRef] [PubMed]
- Pötsch, N.; Dietzel, M.; Kapetas, P.; Clauser, P.; Pinker, K.; Ellmann, S.; Uder, M.; Helbich, T.; Baltzer, P.A.T.; An, A.I. classifier derived from 4D radiomics of dynamic contrast-enhanced breast MRI data: Potential to avoid unnecessary breast biopsies. Eur. Radiol. 2021, 31, 5866–5876. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Wu, G.; Cheng, H.; Zhang, J.; Shao, G.; Li, L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Radiol. 2017, 94, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Dalmış, M.U.; Gubern-Mérida1, A.; Vreemann, S.; Bult, P.; Karssemeijer, N.; Mann, R.; Teuwen, J.; Dalmiş, M.U.; Gubern-Mérida, A.; Vreemann, S.; et al. Artificial Intelligence Based Classification of Breast Lesions Imaged with a Multi-Parametric Breast MRI Protocol With ultrafast DCE-MRI, T2 and DWI. Investig. Radiol. 2019, 54, 325–332. [Google Scholar] [CrossRef]
- Greff, K.; Srivastava, R.K.; Koutnik, J.; Steunebrink, B.R.; Schmidhuber, J. LSTM: A Search Space Odyssey. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 2222–2232. [Google Scholar] [CrossRef] [Green Version]
- Benou, A.; Veksler, R.; Friedman, A.; Riklin Raviv, T. Ensemble of expert deep neural networks for spatio-temporal denoising of contrast-enhanced MRI sequences. Med. Image Anal. 2017, 42, 145–159. [Google Scholar] [CrossRef]
- Huynh, B.; Giger, M.L. Breast lesion classification based on dynamic contrast-enhanced magnetic resonance images sequences with long short-term memory networks. J. Med. Imaging 2018, 6, 1. [Google Scholar] [CrossRef]
- Feng, C.; Elazab, A.; Yang, P.; Wang, T.; Zhou, F.; Hu, H.; Xiao, X.; Lei, B. Deep Learning Framework for Alzheimer’s Disease Diagnosis via 3D-CNN and FSBi-LSTM. IEEE Access 2019, 7, 63605–63618. [Google Scholar] [CrossRef]
- Zou, J.; Balter, J.M.; Cao, Y. Estimation of pharmacokinetic parameters from DCE-MRI by extracting long and short time-dependent features using an LSTM network. Med. Phys. 2020, 47, 3447–3457. [Google Scholar] [CrossRef]
- Zhang, J.; Saha, A.; Zhu, Z.; Mazurowski, M.A. Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI with Application to Radiogenomics. IEEE Trans. Med. Imaging 2018, 38, 435–447. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; IEEE: Manhattan, NY, USA, 2016; pp. 770–778. [Google Scholar]
- Platel, B.; Mus, R.; Welte, T.; Karssemeijer, N.; Mann, R. Automated characterization of breast lesions imaged with an ultrafast DCE-MR protocol. IEEE Trans. Med. Imaging 2014, 33, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Boca, I.; Ciurea, A.I.; Ciortea, C.A.; Ștefan, P.A.; Lisencu, L.A.; Dudea, S.M. Differentiating breast tumors from background parenchymal enhancement at contrast-enhanced mammography: The role of radiomics—A pilot reader study. Diagnostics 2021, 11, 1248. [Google Scholar] [CrossRef] [PubMed]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef] [Green Version]


| Parameter | TWIST | T1-Weighted | ||
|---|---|---|---|---|
| 1.5 T | 3.0 T | 1.5 T | 3.0 T | |
| TR/TE (ms) | 2.50/0.90 | 4.12/2.08 | 5.27/2.39 | 4.50/1.60 |
| Flip angle (°) | 20 | 20 | 10 | 10 |
| Phase oversampling (%) | 26 | 20 | N/A | N/A |
| Slice oversampling (%) | 20 | 0 | N/A | N/A |
| Voxel size (mm3) | 0.68 × 0.68 × 3.0 | 0.91 × 0.91 × 3.0 | 0.84 × 0.84 × 1.2 | 0.89 × 0.89 × 1.2 |
| Temporal resolution (s) | 5.2 | 4.3 | 120 | 120 |
| Field of view (mm) | 350 | 350 | 350 | 370 |
| Fat suppression | None | None | SPAIR | SPAIR |
| Characteristics | Value (Proportion) |
|---|---|
| Benign lesions | 109 (0.63) |
| Adenosis | 24 (0.14) |
| Fibroadenoma | 19 (0.11) |
| Hyperplasia | 6 (0.03) |
| Glandular tissue | 4 (0.02) |
| Cyst | 3 (0.02) |
| Inflammation | 1 (0.01) |
| Other 1 | 51 (0.29) |
| Malignant lesions | 64 (0.37) |
| Invasive ductal carcinoma | 51 (0.29) |
| Invasive lobular carcinoma | 4 (0.02) |
| Ductal carcinoma in situ | 4 (0.02) |
| Micropapillary carcinoma | 2 (0.01) |
| Apocrine carcinoma | 1 (0.01) |
| Mucinous carcinoma | 2 (0.01) |
| Lesion size (mm) 2 | |
| Overall | 19.9 ± 18.4 |
| Malignant | 28.6 ± 20.8 |
| Benign | 13.9 ± 13.6 |
| Threshold | 2D CNN | LSTM | Combined | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| 0.1 | 0.92 (0.91, 0.93) | 0.25 (0.23, 0.27) | 0.92 (0.91, 0.93) | 0.22 (0.20, 0.24) | 0.96 (0.95, 0.96) | 0.14 (0.13, 0.16) |
| 0.2 | 0.82 (0.81, 0.84) | 0.56 (0.54, 0.58) | 0.81 (0.80, 0.83) | 0.48 (0.46, 0.50) | 0.87 (0.86, 0.88) | 0.48 (0.46, 0.50) |
| 0.3 | 0.73 (0.72, 0.74) | 0.76 (0.75, 0.77) | 0.73 (0.72, 0.75) | 0.65 (0.63, 0.67) | 0.78 (0.77, 0.79) | 0.72 (0.71, 0.74) |
| 0.4 | 0.64 (0.63, 0.66) | 0.88 (0.87, 0.88) | 0.66 (0.64, 0.68) | 0.78 (0.77, 0.79) | 0.68 (0.67, 0.70) | 0.86 (0.85, 0.86) |
| 0.5 | 0.56 (0.55, 0.57) | 0.93 (0.92, 0.93) | 0.59 (0.57, 0.61) | 0.88 (0.87, 0.88) | 0.57 (0.55, 0.58) | 0.92 (0.91, 0.93) |
| 0.6 | 0.44 (0.43, 0.46) | 0.95 (0.94, 0.95) | 0.46 (0.44, 0.48) | 0.92 (0.91, 0.93) | 0.44 (0.43, 0.46) | 0.96 (0.95, 0.96) |
| 0.7 | 0.34 (0.33, 0.35) | 0.96 (0.96, 0.97) | 0.34 (0.33, 0.36) | 0.94 (0.94, 0.95) | 0.32 (0.31, 0.33) | 0.98 (0.97, 0.98) |
| 0.8 | 0.25 (0.23, 0.26) | 0.98 (0.97, 0.98) | 0.24 (0.22, 0.25) | 0.97 (0.96, 0.97) | 0.18 (0.17, 0.19) | 0.99 (0.99, 0.99) |
| 0.9 | 0.15 (0.14, 0.16) | 0.99 (0.99, 0.99) | 0.12 (0.10, 0.13) | 0.99 (0.98, 0.99) | 0.07 (0.06, 0.08) | 1.0 N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jing, X.; Dorrius, M.D.; Wielema, M.; Sijens, P.E.; Oudkerk, M.; van Ooijen, P. Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information. Cancers 2022, 14, 2042. https://doi.org/10.3390/cancers14082042
Jing X, Dorrius MD, Wielema M, Sijens PE, Oudkerk M, van Ooijen P. Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information. Cancers. 2022; 14(8):2042. https://doi.org/10.3390/cancers14082042
Chicago/Turabian StyleJing, Xueping, Monique D. Dorrius, Mirjam Wielema, Paul E. Sijens, Matthijs Oudkerk, and Peter van Ooijen. 2022. "Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information" Cancers 14, no. 8: 2042. https://doi.org/10.3390/cancers14082042
APA StyleJing, X., Dorrius, M. D., Wielema, M., Sijens, P. E., Oudkerk, M., & van Ooijen, P. (2022). Breast Tumor Identification in Ultrafast MRI Using Temporal and Spatial Information. Cancers, 14(8), 2042. https://doi.org/10.3390/cancers14082042







