Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance
Abstract
Simple Summary
Abstract
1. Introduction
2. Biogenesis and Localization of LncRNAs
3. Classifications of LncRNAs
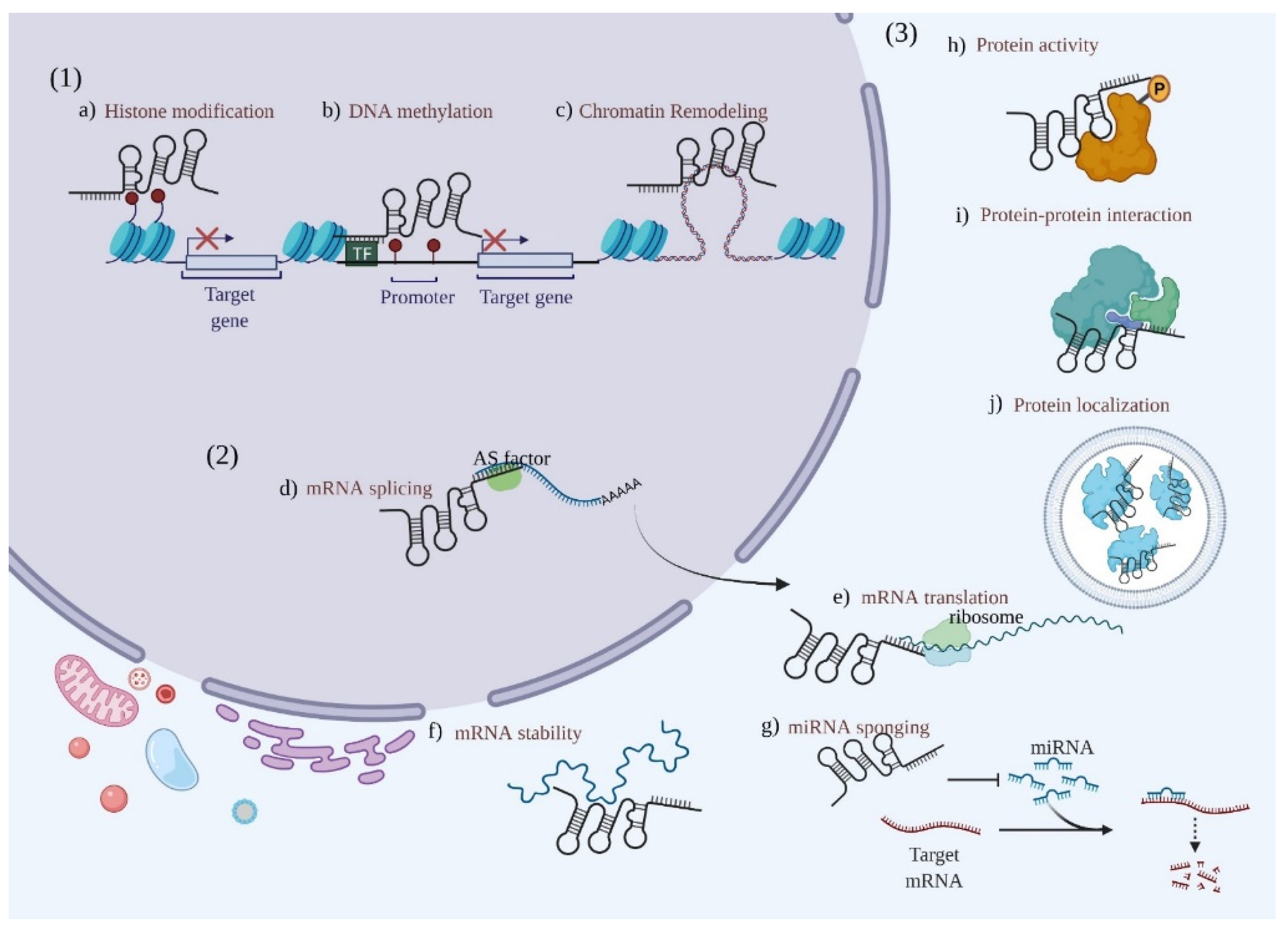
4. Roles of LncRNAs in PC
4.1. LncRNAs Act as Histone Modulators
4.2. LncRNAs Function as ceRNAs
4.3. LncRNAs Bind to Proteins
4.4. LncRNAs Regulate the EMT Pathway in PC
4.5. LncRNAs Modulate CSC Properties
5. Clinical Significance of LncRNAs in PC
5.1. Diagnostic Biomarkers for PC
5.2. Prognostic Biomarkers for PC
6. Future Expectations
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Oettle, H.; Neuhaus, P.; Hochhaus, A.; Hartmann, J.T.; Gellert, K.; Ridwelski, K.; Niedergethmann, M.; Zulke, C.; Fahlke, J.; Arning, M.B.; et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: The CONKO-001 randomized trial. JAMA 2013, 310, 1473–1481. [Google Scholar] [CrossRef]
- Neoptolemos, J.P.; Stocken, D.D.; Friess, H.; Bassi, C.; Dunn, J.A.; Hickey, H.; Beger, H.; Fernandez-Cruz, L.; Dervenis, C.; Lacaine, F.; et al. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N. Engl. J. Med. 2004, 350, 1200–1210. [Google Scholar] [CrossRef]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef]
- Yang, Y.; Wen, L.; Zhu, H. Unveiling the hidden function of long non-coding RNA by identifying its major partner-protein. Cell Biosci. 2015, 5, 59. [Google Scholar] [CrossRef]
- Gutschner, T.; Diederichs, S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012, 9, 703–719. [Google Scholar] [CrossRef]
- Brunner, A.L.; Beck, A.H.; Edris, B.; Sweeney, R.T.; Zhu, S.X.; Li, R.; Montgomery, K.; Varma, S.; Gilks, T.; Guo, X.; et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol. 2012, 13, R75. [Google Scholar] [CrossRef]
- Wang, K.; Liu, C.Y.; Zhou, L.Y.; Wang, J.X.; Wang, M.; Zhao, B.; Zhao, W.K.; Xu, S.J.; Fan, L.H.; Zhang, X.J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Guo, C.J.; Ma, X.K.; Xing, Y.H.; Zheng, C.C.; Xu, Y.F.; Shan, L.; Zhang, J.; Wang, S.; Wang, Y.; Carmichael, G.G.; et al. Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 2020, 181, 621–636.e622. [Google Scholar] [CrossRef]
- Schlackow, M.; Nojima, T.; Gomes, T.; Dhir, A.; Carmo-Fonseca, M.; Proudfoot, N.J. Distinctive Patterns of Transcription and RNA Processing for Human lincRNAs. Mol. Cell 2017, 65, 25–38. [Google Scholar] [CrossRef]
- Yin, Y.; Lu, J.Y.; Zhang, X.; Shao, W.; Xu, Y.; Li, P.; Hong, Y.; Cui, L.; Shan, G.; Tian, B.; et al. U1 snRNP regulates chromatin retention of noncoding RNAs. Nature 2020, 580, 147–150. [Google Scholar] [CrossRef]
- Melé, M.; Mattioli, K.; Mallard, W.; Shechner, D.M.; Gerhardinger, C.; Rinn, J.L. Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017, 27, 27–37. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ulitsky, I. Predictive models of subcellular localization of long RNAs. RNA 2019, 25, 557–572. [Google Scholar] [CrossRef]
- Rosenberg, A.B.; Patwardhan, R.P.; Shendure, J.; Seelig, G. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell 2015, 163, 698–711. [Google Scholar] [CrossRef]
- Azam, S.; Hou, S.; Zhu, B.; Wang, W.; Hao, T.; Bu, X.; Khan, M.; Lei, H. Nuclear retention element recruits U1 snRNP components to restrain spliced lncRNAs in the nucleus. RNA Biol. 2019, 16, 1001–1009. [Google Scholar] [CrossRef]
- Shukla, C.J.; McCorkindale, A.L.; Gerhardinger, C.; Korthauer, K.D.; Cabili, M.N.; Shechner, D.M.; Irizarry, R.A.; Maass, P.G.; Rinn, J.L. High-throughput identification of RNA nuclear enrichment sequences. EMBO J. 2018, 37, e98452. [Google Scholar] [CrossRef]
- Lubelsky, Y.; Ulitsky, I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 2018, 555, 107–111. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ron, M.; Mikl, M.; Segal, E.; Ulitsky, I. Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol. Cell 2020, 79, 251–267.e256. [Google Scholar] [CrossRef]
- Carlevaro-Fita, J.; Rahim, A.; Guigó, R.; Vardy, L.A.; Johnson, R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 2016, 22, 867–882. [Google Scholar] [CrossRef]
- Zeng, C.; Hamada, M. Identifying sequence features that drive ribosomal association for lncRNA. BMC Genom. 2018, 19, 906. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef]
- Rackham, O.; Shearwood, A.M.; Mercer, T.R.; Davies, S.M.; Mattick, J.S.; Filipovska, A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 2011, 17, 2085–2093. [Google Scholar] [CrossRef]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef]
- Gudenas, B.L.; Wang, L. Prediction of LncRNA Subcellular Localization with Deep Learning from Sequence Features. Sci. Rep. 2018, 8, 16385. [Google Scholar] [CrossRef]
- Statello, L.; Maugeri, M.; Garre, E.; Nawaz, M.; Wahlgren, J.; Papadimitriou, A.; Lundqvist, C.; Lindfors, L.; Collén, A.; Sunnerhagen, P.; et al. Identification of RNA-binding proteins in exosomes capable of interacting with different types of RNA: RBP-facilitated transport of RNAs into exosomes. PLoS ONE 2018, 13, e0195969. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef]
- Alessio, E.; Bonadio, R.S.; Buson, L.; Chemello, F.; Cagnin, S. A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 302. [Google Scholar] [CrossRef]
- Mondal, T.; Juvvuna, P.K.; Kirkeby, A.; Mitra, S.; Kosalai, S.T.; Traxler, L.; Hertwig, F.; Wernig-Zorc, S.; Miranda, C.; Deland, L.; et al. Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 Determines Neuroblastoma Susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell 2018, 33, 417–434.e417. [Google Scholar] [CrossRef]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Ning, S.; Zhang, J.; Wang, P.; Zhi, H.; Wang, J.; Liu, Y.; Gao, Y.; Guo, M.; Yue, M.; Wang, L.; et al. Lnc2Cancer: A manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic Acids Res. 2016, 44, D980–D985. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Bayoumi, A.S.; Sayed, A.; Broskova, Z.; Teoh, J.P.; Wilson, J.; Su, H.; Tang, Y.L.; Kim, I.M. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int. J. Mol. Sci. 2016, 17, 356. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Wang, Y.; Xie, Y.; Li, L.; He, Y.; Zheng, D.; Yu, P.; Yu, L.; Tang, L.; Wang, Y.; Wang, Z. EZH2 RIP-seq Identifies Tissue-specific Long Non-coding RNAs. Curr. Gene Ther. 2018, 18, 275–285. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Sun, X.; Du, P.; Yuan, W.; Du, Z.; Yu, M.; Yu, X.; Hu, T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015, 6, e1907. [Google Scholar] [CrossRef]
- Abbosh, P.H.; Montgomery, J.S.; Starkey, J.A.; Novotny, M.; Zuhowski, E.G.; Egorin, M.J.; Moseman, A.P.; Golas, A.; Brannon, K.M.; Balch, C.; et al. Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res. 2006, 66, 5582–5591. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Yang, H.; Hu, L.B.; Lei, Y.H.; Qin, Y.; Li, J.; Bi, C.W.; Wang, J.S.; Huo, Q. Effect of siRNA targeting EZH2 on cell viability and apoptosis of bladder cancer T24 cells. Genet. Mol. Res. 2014, 13, 9939–9950. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Li, X.; Zhou, Z.; Wu, C.L.; Dai, M.; Bai, X. EZH2 promotes tumor progression via regulating VEGF-A/AKT signaling in non-small cell lung cancer. Cancer Lett. 2015, 359, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef]
- Kim, K.; Jutooru, I.; Chadalapaka, G.; Johnson, G.; Frank, J.; Burghardt, R.; Kim, S.; Safe, S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene 2013, 32, 1616–1625. [Google Scholar] [CrossRef]
- Rogers, M.A.; Kalter, V.; Strowitzki, M.; Schneider, M.; Lichter, P. IGF2 knockdown in two colorectal cancer cell lines decreases survival, adhesion and modulates survival-associated genes. Tumour Biol. 2016, 37, 12485–12495. [Google Scholar] [CrossRef]
- Dong, Y.; Li, J.; Han, F.; Chen, H.; Zhao, X.; Qin, Q.; Shi, R.; Liu, J. High IGF2 expression is associated with poor clinical outcome in human ovarian cancer. Oncol. Rep. 2015, 34, 936–942. [Google Scholar] [CrossRef][Green Version]
- Cai, H.; An, Y.; Chen, X.; Sun, D.; Chen, T.; Peng, Y.; Zhu, F.; Jiang, Y.; He, X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget 2016, 7, 86857–86870. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Xiao, Z.; Tong, J.H.; To, K.F.; Fang, X.; Cheng, A.S.; Chen, Y. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J. Cancer 2017, 140, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Yang, L.; Qi, X.; Wang, T.; Li, M.; Xu, K. Inhibition of long non-coding RNA HOTAIR enhances radiosensitivity via regulating autophagy in pancreatic cancer. Cancer Manag. Res. 2018, 10, 5261–5271. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Hu, M.; Zhou, L.; Ling, S.; Li, Y.; Kong, B.; Huang, P. Long non-coding RNA HOTAIR promotes cancer cell energy metabolism in pancreatic adenocarcinoma by upregulating hexokinase-2. Oncol. Lett. 2019, 18, 2212–2219. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Quagliata, L.; Matter, M.S.; Piscuoglio, S.; Arabi, L.; Ruiz, C.; Procino, A.; Kovac, M.; Moretti, F.; Makowska, Z.; Boldanova, T.; et al. Long noncoding RNA HOTTIP/HOXA13 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Hepatology 2014, 59, 911–923. [Google Scholar] [CrossRef]
- Hu, P.; Qiao, O.; Wang, J.; Li, J.; Jin, H.; Li, Z.; Jin, Y. rs1859168 A > C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J. Surg. Oncol. 2017, 15, 155. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, X.; Zhou, Y.; Liu, Y.; Zhou, Q.; Ye, H.; Wang, Y.; Zeng, J.; Song, Y.; Gao, W.; et al. The long non-coding RNA HOTTIP promotes progression and gemcitabine resistance by regulating HOXA13 in pancreatic cancer. J. Transl. Med. 2015, 13, 84. [Google Scholar] [CrossRef]
- Cheng, Y.; Jutooru, I.; Chadalapaka, G.; Corton, J.C.; Safe, S. The long non-coding RNA HOTTIP enhances pancreatic cancer cell proliferation, survival and migration. Oncotarget 2015, 6, 10840–10852. [Google Scholar] [CrossRef]
- Wong, C.H.; Li, C.H.; He, Q.; Chan, S.L.; Tong, J.H.; To, K.F.; Lin, L.Z.; Chen, Y. Ectopic HOTTIP expression induces noncanonical transactivation pathways to promote growth and invasiveness in pancreatic ductal adenocarcinoma. Cancer Lett. 2020, 477, 1–9. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, C.; Zhou, Q.; Wang, Y.; Zhao, Y.; Zhao, X.; Li, W.; Zheng, S.; Ye, H.; Wang, L.; et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017, 410, 68–81. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Hämmerle, M.; Diederichs, S. MALAT1—a paradigm for long noncoding RNA function in cancer. J. Mol. Med. 2013, 91, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hamblin, M.H.; Yin, K.J. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol. 2017, 14, 1705–1714. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, H.; Wang, L.; Liu, T.; Du, L.; Yang, Y.; Wang, C. MALAT1 Is Associated with Poor Response to Oxaliplatin-Based Chemotherapy in Colorectal Cancer Patients and Promotes Chemoresistance through EZH2. Mol. Cancer Ther. 2017, 16, 739–751. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Chen, G.; Dang, Y.W.; Li, C.J.; Luo, D.Z. Expression and prognostic significance of lncRNA MALAT1 in pancreatic cancer tissues. Asian Pac. J. Cancer Prev. 2014, 15, 2971–2977. [Google Scholar] [CrossRef]
- Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jiang, W.; Jin, Z.; Guo, Z.; Wang, L. Elevated expression level of long noncoding RNA MALAT-1 facilitates cell growth, migration and invasion in pancreatic cancer. Oncol. Rep. 2014, 32, 2485–2492. [Google Scholar] [CrossRef]
- Jiao, F.; Hu, H.; Han, T.; Yuan, C.; Wang, L.; Jin, Z.; Guo, Z.; Wang, L. Long noncoding RNA MALAT-1 enhances stem cell-like phenotypes in pancreatic cancer cells. Int. J. Mol. Sci. 2015, 16, 6677–6693. [Google Scholar] [CrossRef]
- Han, T.; Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jin, Z.L.; Song, W.F.; Wang, L.W. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget 2016, 7, 11194–11207. [Google Scholar] [CrossRef]
- Cheng, Y.; Imanirad, P.; Jutooru, I.; Hedrick, E.; Jin, U.H.; Rodrigues Hoffman, A.; Leal de Araujo, J.; Morpurgo, B.; Golovko, A.; Safe, S. Role of metastasis-associated lung adenocarcinoma transcript-1 (MALAT-1) in pancreatic cancer. PLoS ONE 2018, 13, e0192264. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Shan, T.; Ding, W.; Hua, Z.; Shen, Y.; Lu, Z.; Chen, B.; Dai, T. Study on mechanism about long noncoding RNA MALAT1 affecting pancreatic cancer by regulating Hippo-YAP signaling. J. Cell. Physiol. 2018, 233, 5805–5814. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lu, Z.; Cai, J.; Huang, K.; Chen, B.; Li, G.; Dong, P.; Zheng, J. MALAT1 functions as a competing endogenous RNA to mediate Rac1 expression by sequestering miR-101b in liver fibrosis. Cell Cycle 2015, 14, 3885–3896. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; He, H.; Xiao, W.; Liu, Q.; Deng, Z.; Lu, Y.; Wang, Q.; Zheng, Q.; Li, Y. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget 2016, 7, 54733–54743. [Google Scholar] [CrossRef]
- Liu, P.; Yang, H.; Zhang, J.; Peng, X.; Lu, Z.; Tong, W.; Chen, J. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci. Rep. 2017, 7, 5186. [Google Scholar] [CrossRef]
- Zhuo, M.; Yuan, C.; Han, T.; Cui, J.; Jiao, F.; Wang, L. A novel feedback loop between high MALAT-1 and low miR-200c-3p promotes cell migration and invasion in pancreatic ductal adenocarcinoma and is predictive of poor prognosis. BMC Cancer 2018, 18, 1032. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, X.; Shi, M.; Wen, C.; Shen, B. MiR-216a decreases MALAT1 expression, induces G2/M arrest and apoptosis in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2017, 483, 816–822. [Google Scholar] [CrossRef]
- Hao, Y.; Crenshaw, T.; Moulton, T.; Newcomb, E.; Tycko, B. Tumour-suppressor activity of H19 RNA. Nature 1993, 365, 764–767. [Google Scholar] [CrossRef]
- Chung, W.Y.; Yuan, L.; Feng, L.; Hensle, T.; Tycko, B. Chromosome 11p15.5 regional imprinting: Comparative analysis of KIP2 and H19 in human tissues and Wilms’ tumors. Hum. Mol. Genet. 1996, 5, 1101–1108. [Google Scholar] [CrossRef]
- Lynch, C.A.; Tycko, B.; Bestor, T.H.; Walsh, C.P. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histone hyperacetylation. Mol. Cancer 2002, 1, 2. [Google Scholar] [CrossRef]
- Yoshimizu, T.; Miroglio, A.; Ripoche, M.A.; Gabory, A.; Vernucci, M.; Riccio, A.; Colnot, S.; Godard, C.; Terris, B.; Jammes, H.; et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. USA 2008, 105, 12417–12422. [Google Scholar] [CrossRef] [PubMed]
- Ariel, I.; Sughayer, M.; Fellig, Y.; Pizov, G.; Ayesh, S.; Podeh, D.; Libdeh, B.A.; Levy, C.; Birman, T.; Tykocinski, M.L.; et al. The imprinted H19 gene is a marker of early recurrence in human bladder carcinoma. Mol. Pathol. 2000, 53, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yu, B.; Li, J.; Su, L.; Yan, M.; Zhu, Z.; Liu, B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014, 5, 2318–2329. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef]
- Ma, L.; Tian, X.; Wang, F.; Zhang, Z.; Du, C.; Xie, X.; Kornmann, M.; Yang, Y. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol. Ther. 2016, 17, 1051–1061. [Google Scholar] [CrossRef]
- Ma, C.; Nong, K.; Zhu, H.; Wang, W.; Huang, X.; Yuan, Z.; Ai, K. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014, 35, 9163–9169. [Google Scholar] [CrossRef]
- Sun, Y.; Zhu, Q.; Yang, W.; Shan, Y.; Yu, Z.; Zhang, Q.; Wu, H. LncRNA H19/miR-194/PFTK1 axis modulates the cell proliferation and migration of pancreatic cancer. J. Cell. Biochem. 2019, 120, 3874–3886. [Google Scholar] [CrossRef]
- Ma, L.; Tian, X.; Guo, H.; Zhang, Z.; Du, C.; Wang, F.; Xie, X.; Gao, H.; Zhuang, Y.; Kornmann, M.; et al. Long noncoding RNA H19 derived miR-675 regulates cell proliferation by down-regulating E2F-1 in human pancreatic ductal adenocarcinoma. J. Cancer 2018, 9, 389–399. [Google Scholar] [CrossRef]
- Wang, F.; Rong, L.; Zhang, Z.; Li, M.; Ma, L.; Ma, Y.; Xie, X.; Tian, X.; Yang, Y. LncRNA H19-Derived miR-675-3p Promotes Epithelial-Mesenchymal Transition and Stemness in Human Pancreatic Cancer Cells by targeting the STAT3 Pathway. J. Cancer 2020, 11, 4771–4782. [Google Scholar] [CrossRef]
- Wang, F.; Li, X.; Xie, X.; Zhao, L.; Chen, W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008, 582, 1919–1927. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, W.; Yang, C.; Wu, W.; Wu, S.; Qin, X.; Li, X. Long non-coding RNA UCA1a(CUDR) promotes proliferation and tumorigenesis of bladder cancer. Int. J. Oncol. 2012, 41, 276–284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huang, J.; Zhou, N.; Watabe, K.; Lu, Z.; Wu, F.; Xu, M.; Mo, Y.Y. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis. 2014, 5, e1008. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yang, Y.N.; Yuan, H.H.; Zhang, T.T.; Sui, H.; Wei, X.L.; Liu, L.; Huang, P.; Zhang, W.J.; Bai, Y.X. UCA1, a long non-coding RNA up-regulated in colorectal cancer influences cell proliferation, apoptosis and cell cycle distribution. Pathology 2014, 46, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wu, F.; Dai, W.Y.; Zheng, D.C.; Zheng, C.; Ye, H.; Zhou, B.; Chen, J.J.; Chen, P. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin. Transl. Oncol. 2015, 17, 640–646. [Google Scholar] [CrossRef]
- Yao, K.; Wang, Q.; Jia, J.; Zhao, H. A competing endogenous RNA network identifies novel mRNA, miRNA and lncRNA markers for the prognosis of diabetic pancreatic cancer. Tumour Biol. 2017, 39, 1010428317707882. [Google Scholar] [CrossRef]
- Fu, X.L.; Liu, D.J.; Yan, T.T.; Yang, J.Y.; Yang, M.W.; Li, J.; Huo, Y.M.; Liu, W.; Zhang, J.F.; Hong, J.; et al. Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci. Rep. 2016, 6, 33535. [Google Scholar] [CrossRef]
- Chen, P.; Wan, D.; Zheng, D.; Zheng, Q.; Wu, F.; Zhi, Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed. Pharmacother. 2016, 83, 1220–1226. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, D.; Li, G.; Ming, X.; Tu, Y.; Tian, J.; Lu, H.; Yu, B. Long non coding RNA-UCA1 contributes to cardiomyocyte apoptosis by suppression of p27 expression. Cell. Physiol. Biochem. 2015, 35, 1986–1998. [Google Scholar] [CrossRef]
- Gong, J.; Lu, X.; Xu, J.; Xiong, W.; Zhang, H.; Yu, X. Coexpression of UCA1 and ITGA2 in pancreatic cancer cells target the expression of miR-107 through focal adhesion pathway. J. Cell. Physiol. 2019, 234, 12884–12896. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, Y.; Ding, W.; Hua, Z.; Wang, L.; Zhu, Y.; Qian, H.; Dai, T. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life 2018, 70, 276–290. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, W.; Gu, S.; Wang, H.; Zhang, Y.; Chen, W.; Xu, W.; Lin, C.; Gong, A.; Xu, M. The UCA1/KRAS axis promotes human pancreatic ductal adenocarcinoma stem cell properties and tumor growth. Am. J. Cancer Res. 2019, 9, 496–510. [Google Scholar] [PubMed]
- Guo, S.; Fesler, A.; Wang, H.; Ju, J. microRNA based prognostic biomarkers in pancreatic Cancer. Biomark. Res. 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, H.; Sun, W.; Kong, J.; Zhang, L.; Tang, J.; Wang, J.; Xu, E.; Lai, M.; Zhang, H. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol. Cancer 2018, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Raulefs, S.; Bruns, P.; Afonso-Grunz, F.; Plötner, A.; Thermann, R.; Jäger, C.; Schlitter, A.M.; Kong, B.; Regel, I.; et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol. Cancer 2015, 14, 94. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, Y.; Sui, W.; Zou, Y.; Lv, Z. Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma. Cancer Med. 2017, 6, 673–680. [Google Scholar] [CrossRef]
- Zhu, H.; Shan, Y.; Ge, K.; Lu, J.; Kong, W.; Jia, C. LncRNA CYTOR promotes pancreatic cancer cell proliferation and migration by sponging miR-205-5p. Pancreatology 2020, 20, 1139–1148. [Google Scholar] [CrossRef]
- Yuan, Z.J.; Yu, C.; Hu, X.F.; He, Y.; Chen, P.; Ouyang, S.X. LINC00152 promotes pancreatic cancer cell proliferation, migration and invasion via targeting miR-150. Am. J. Transl. Res. 2020, 12, 2241–2256. [Google Scholar]
- Zhou, L.; Li, J.; Shao, Q.Q.; Guo, J.C.; Liang, Z.Y.; Zhou, W.X.; Zhang, T.P.; You, L.; Zhao, Y.P. Expression and Significances of MTSS1 in Pancreatic Cancer. Pathol. Oncol. Res. 2016, 22, 7–14. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, F.; Xu, F.; Fang, K.; Fang, Z.; Shuai, X.; Cai, K.; Chen, J.; Hu, P.; Chen, D.; et al. A reciprocal feedback of Myc and lncRNA MTSS1-AS contributes to extracellular acidity-promoted metastasis of pancreatic cancer. Theranostics 2020, 10, 10120–10140. [Google Scholar] [CrossRef]
- Xia, E.; Bhandari, A.; Shen, Y.; Zhou, X.; Wang, O. lncRNA LINC00673 induces proliferation, metastasis and epithelial-mesenchymal transition in thyroid carcinoma via Kruppel-like factor 2. Int. J. Oncol. 2018, 53, 1927–1938. [Google Scholar] [CrossRef]
- Yu, J.; Liu, Y.; Gong, Z.; Zhang, S.; Guo, C.; Li, X.; Tang, Y.; Yang, L.; He, Y.; Wei, F.; et al. Overexpression long non-coding RNA LINC00673 is associated with poor prognosis and promotes invasion and metastasis in tongue squamous cell carcinoma. Oncotarget 2017, 8, 16621–16632. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Huang, X.; Tan, W.; Yu, D.; Du, Z.; Chang, J.; Wei, L.; Han, Y.; Wang, C.; Che, X.; et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat. Genet. 2016, 48, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Dai, H.S.; Shu, J.J.; Liu, W.; Bie, P.; Zhang, L.D. LNC00673 suppresses proliferation and metastasis of pancreatic cancer via target miR-504/HNF1A. J. Cancer 2020, 11, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Sboner, A.; Nair, S.S.; Giannopoulou, E.; Li, R.; Hennig, S.; Mosquera, J.M.; Pauwels, J.; Park, K.; Kossai, M.; et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 2014, 5, 5383. [Google Scholar] [CrossRef]
- Chen, X.; Kong, J.; Ma, Z.; Gao, S.; Feng, X. Up regulation of the long non-coding RNA NEAT1 promotes esophageal squamous cell carcinoma cell progression and correlates with poor prognosis. Am. J. Cancer Res. 2015, 5, 2808–2815. [Google Scholar]
- Fu, J.W.; Kong, Y.; Sun, X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J. Cancer Res. Clin. Oncol. 2016, 142, 1571–1579. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, L.; Zhao, J.; Li, C.; Nie, J.; Liu, F.; Zhuo, C.; Zheng, Y.; Li, B.; Wang, Z.; et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol. Cancer 2015, 14, 191. [Google Scholar] [CrossRef]
- Jiang, P.; Wu, X.; Wang, X.; Huang, W.; Feng, Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016, 7, 43337–43351. [Google Scholar] [CrossRef]
- Huang, B.; Liu, C.; Wu, Q.; Zhang, J.; Min, Q.; Sheng, T.; Wang, X.; Zou, Y. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem. Biophys. Res. Commun. 2017, 482, 828–834. [Google Scholar] [CrossRef]
- Luo, Z.; Yi, Z.J.; Ou, Z.L.; Han, T.; Wan, T.; Tang, Y.C.; Wang, Z.C.; Huang, F.Z. RELA/NEAT1/miR-302a-3p/RELA feedback loop modulates pancreatic ductal adenocarcinoma cell proliferation and migration. J. Cell. Physiol. 2019, 234, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Xu, M.; Xu, J.; Wu, K.; Fang, Q.; Liang, Y.; Zhou, S.; Cen, D.; Ji, L.; Han, W.; et al. ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Dis. 2018, 9, 387. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Gao, L.; Cui, G.; Cao, Y. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am. J. Cancer Res. 2020, 10, 237–248. [Google Scholar] [PubMed]
- Zhang, B.; Li, C.; Sun, Z. Long non-coding RNA LINC00346, LINC00578, LINC00673, LINC00671, LINC00261, and SNHG9 are novel prognostic markers for pancreatic cancer. Am. J. Transl. Res. 2018, 10, 2648–2658. [Google Scholar] [PubMed]
- Shi, W.; Zhang, C.; Ning, Z.; Hua, Y.; Li, Y.; Chen, L.; Liu, L.; Chen, Z.; Meng, Z. Long non-coding RNA LINC00346 promotes pancreatic cancer growth and gemcitabine resistance by sponging miR-188-3p to derepress BRD4 expression. J. Exp. Clin. Cancer Res. 2019, 38, 60. [Google Scholar] [CrossRef]
- Peng, W.X.; He, R.Z.; Zhang, Z.; Yang, L.; Mo, Y.Y. LINC00346 promotes pancreatic cancer progression through the CTCF-mediated Myc transcription. Oncogene 2019, 38, 6770–6780. [Google Scholar] [CrossRef]
- Mitra, A.; Mishra, L.; Li, S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget 2015, 6, 10697–10711. [Google Scholar] [CrossRef]
- Karamitopoulou, E. Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front. Oncol. 2012, 2, 209. [Google Scholar] [CrossRef]
- Zhang, Q.; Geng, P.L.; Yin, P.; Wang, X.L.; Jia, J.P.; Yao, J. Down-regulation of long non-coding RNA TUG1 inhibits osteosarcoma cell proliferation and promotes apoptosis. Asian Pac. J. Cancer Prev. 2013, 14, 2311–2315. [Google Scholar] [CrossRef]
- Han, Y.; Liu, Y.; Gui, Y.; Cai, Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J. Surg. Oncol. 2013, 107, 555–559. [Google Scholar] [CrossRef]
- Yang, F.; Li, X.; Zhang, L.; Cheng, L.; Li, X. LncRNA TUG1 promoted viability and associated with gemcitabine resistant in pancreatic ductal adenocarcinoma. J. Pharmacol. Sci. 2018, 137, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Sun, H.; Kong, H.; Chen, Z.; Chen, B.; Zhou, M. The Lncrna-TUG1/EZH2 Axis Promotes Pancreatic Cancer Cell Proliferation, Migration and EMT Phenotype Formation Through Sponging Mir-382. Cell. Physiol. Biochem. 2017, 42, 2145–2158. [Google Scholar] [CrossRef] [PubMed]
- Hui, B.; Xu, Y.; Zhao, B.; Ji, H.; Ma, Z.; Xu, S.; He, Z.; Wang, K.; Lu, J. Overexpressed long noncoding RNA TUG1 affects the cell cycle, proliferation, and apoptosis of pancreatic cancer partly through suppressing RND3 and MT2A. Onco Targets Ther. 2019, 12, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.F.; Zhao, F.L. Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2377–2384. [Google Scholar]
- Lu, Y.; Tang, L.; Zhang, Z.; Li, S.; Liang, S.; Ji, L.; Yang, B.; Liu, Y.; Wei, W. Long Noncoding RNA TUG1/miR-29c Axis Affects Cell Proliferation, Invasion, and Migration in Human Pancreatic Cancer. Dis. Markers 2018, 2018, 6857042. [Google Scholar] [CrossRef]
- Ou, Z.L.; Luo, Z.; Lu, Y.B. Long non-coding RNA HULC as a diagnostic and prognostic marker of pancreatic cancer. World J. Gastroenterol. 2019, 25, 6728–6742. [Google Scholar] [CrossRef]
- Feng, H.; Wei, B.; Zhang, Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int. J. Biol. Macromol. 2019, 126, 891–898. [Google Scholar] [CrossRef]
- Takahashi, K.; Ota, Y.; Kogure, T.; Suzuki, Y.; Iwamoto, H.; Yamakita, K.; Kitano, Y.; Fujii, S.; Haneda, M.; Patel, T.; et al. Circulating extracellular vesicle-encapsulated HULC is a potential biomarker for human pancreatic cancer. Cancer Sci. 2020, 111, 98–111. [Google Scholar] [CrossRef]
- Takahashi, K.; Koyama, K.; Ota, Y.; Iwamoto, H.; Yamakita, K.; Fujii, S.; Kitano, Y. The Interaction Between Long Non-coding RNA HULC and MicroRNA-622 via Transfer by Extracellular Vesicles Regulates Cell Invasion and Migration in Human Pancreatic Cancer. Front. Oncol. 2020, 10, 1013. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, W.; Jung, Y.S.; Chen, X. PUMA Cooperates with p21 to Regulate Mammary Epithelial Morphogenesis and Epithelial-To-Mesenchymal Transition. PLoS ONE 2013, 8, e66464. [Google Scholar] [CrossRef]
- Huang, C.; Yu, W.; Wang, Q.; Cui, H.; Wang, Y.; Zhang, L.; Han, F.; Huang, T. Increased expression of the lncRNA PVT1 is associated with poor prognosis in pancreatic cancer patients. Minerva Med. 2015, 106, 143–149. [Google Scholar] [PubMed]
- Zhou, Q.; Chen, F.; Zhao, J.; Li, B.; Liang, Y.; Pan, W.; Zhang, S.; Wang, X.; Zheng, D. Long non-coding RNA PVT1 promotes osteosarcoma development by acting as a molecular sponge to regulate miR-195. Oncotarget 2016, 7, 82620–82633. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Liu, S.; Wang, H.; Zhang, Z.; Yang, Q.; Gao, F. LncRNA PVT1 overexpression is a poor prognostic biomarker and regulates migration and invasion in small cell lung cancer. Am. J. Transl. Res. 2016, 8, 5025–5034. [Google Scholar] [PubMed]
- Xu, M.D.; Wang, Y.; Weng, W.; Wei, P.; Qi, P.; Zhang, Q.; Tan, C.; Ni, S.J.; Dong, L.; Yang, Y.; et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin. Cancer Res. 2017, 23, 2071–2080. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Sun, M.; Liu, G.J.; Wei, C.C.; Zhang, E.B.; Kong, R.; Xu, T.P.; Huang, M.D.; Wang, Z.X. Long Noncoding RNA PVT1 Promotes Non-Small Cell Lung Cancer Cell Proliferation through Epigenetically Regulating LATS2 Expression. Mol. Cancer Ther. 2016, 15, 1082–1094. [Google Scholar] [CrossRef]
- You, L.; Chang, D.; Du, H.Z.; Zhao, Y.P. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2011, 407, 1–6. [Google Scholar] [CrossRef]
- Wu, B.Q.; Jiang, Y.; Zhu, F.; Sun, D.L.; He, X.Z. Long Noncoding RNA PVT1 Promotes EMT and Cell Proliferation and Migration Through Downregulating p21 in Pancreatic Cancer Cells. Technol. Cancer Res. Treat. 2017, 16, 819–827. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, W.; Zhang, J.; Ge, L.; Zhang, Y.; Jiang, X.; Peng, W.; Wang, D.; Gong, A.; Xu, M. Long non-coding RNA PVT1 promotes epithelial-mesenchymal transition via the TGF-β/Smad pathway in pancreatic cancer cells. Oncol. Rep. 2018, 40, 1093–1102. [Google Scholar] [CrossRef]
- Zhao, L.; Kong, H.; Sun, H.; Chen, Z.; Chen, B.; Zhou, M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J. Cell. Physiol. 2018, 233, 4044–4055. [Google Scholar] [CrossRef]
- Sun, J.; Zhang, P.; Yin, T.; Zhang, F.; Wang, W. Upregulation of LncRNA PVT1 Facilitates Pancreatic Ductal Adenocarcinoma Cell Progression and Glycolysis by Regulating MiR-519d-3p and HIF-1A. J. Cancer 2020, 11, 2572–2579. [Google Scholar] [CrossRef]
- Huang, F.; Chen, W.; Peng, J.; Li, Y.; Zhuang, Y.; Zhu, Z.; Shao, C.; Yang, W.; Yao, H.; Zhang, S. LncRNA PVT1 triggers Cyto-protective autophagy and promotes pancreatic ductal adenocarcinoma development via the miR-20a-5p/ULK1 Axis. Mol. Cancer 2018, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.K.; Desai, N.S. Cancer Stem Cells: Acquisition, Characteristics, Therapeutic Implications, Targeting Strategies and Future Prospects. Stem Cell Rev. Rep. 2019, 15, 331–355. [Google Scholar] [CrossRef] [PubMed]
- Dalerba, P.; Clarke, M.F. Cancer stem cells and tumor metastasis: First steps into uncharted territory. Cell Stem Cell 2007, 1, 241–242. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.; Fojo, T.; Bates, S. Tumour stem cells and drug resistance. Nat. Rev. Cancer 2005, 5, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Gopalan, V.; Lam, A.K. Identification of Cancer Stem Cells in Esophageal Adenocarcinoma. Methods Mol. Biol. 2018, 1756, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Xiao, R.; Pan, S.; Yang, X.; Yuan, W.; Tu, Z.; Xu, M.; Zhu, Y.; Yin, Q.; Wu, Y.; et al. Uncovering the roles of long non-coding RNAs in cancer stem cells. J. Hematol. Oncol. 2017, 10, 62. [Google Scholar] [CrossRef]
- Lathia, J.; Liu, H.; Matei, D. The Clinical Impact of Cancer Stem Cells. Oncologist 2019, 25, 123–131. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef]
- Prasad, S.; Ramachandran, S.; Gupta, N.; Kaushik, I.; Srivastava, S.K. Cancer cells stemness: A doorstep to targeted therapy. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165424. [Google Scholar] [CrossRef]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010, 42, 1113–1117. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell 2013, 25, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.C.; Lin, H. Repressing the repressor: A lincRNA as a MicroRNA sponge in embryonic stem cell self-renewal. Dev. Cell 2013, 25, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.X.; Wang, Y.; Li, C.; Xu, J.W.; Zhou, B.; Zhu, J.K.; Han, H.F.; Wang, L.; Wang, Y.S.; Hu, S.Y. LincRNA-ROR promotes invasion, metastasis and tumor growth in pancreatic cancer through activating ZEB1 pathway. Cancer Lett. 2016, 374, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Wellner, U.; Schubert, J.; Burk, U.C.; Schmalhofer, O.; Zhu, F.; Sonntag, A.; Waldvogel, B.; Vannier, C.; Darling, D.; zur Hausen, A.; et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat. Cell Biol. 2009, 11, 1487–1495. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Sun, Y.; Ma, L. ZEB1: At the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle 2015, 14, 481–487. [Google Scholar] [CrossRef]
- Hill, L.; Browne, G.; Tulchinsky, E. ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. Int. J. Cancer 2013, 132, 745–754. [Google Scholar] [CrossRef]
- Chen, W.; Wang, H.; Liu, Y.; Xu, W.; Ling, C.; Li, Y.; Liu, J.; Chen, M.; Zhang, Y.; Chen, B.; et al. Linc-RoR promotes proliferation, migration, and invasion via the Hippo/YAP pathway in pancreatic cancer cells. J. Cell. Biochem. 2020, 121, 632–641. [Google Scholar] [CrossRef]
- Zhulina, Y.; Cao, Y.; Amcoff, K.; Carlson, M.; Tysk, C.; Halfvarson, J. The prognostic significance of faecal calprotectin in patients with inactive inflammatory bowel disease. Aliment. Pharmacol. Ther. 2016, 44, 495–504. [Google Scholar] [CrossRef]
- Gao, S.; Wang, P.; Hua, Y.; Xi, H.; Meng, Z.; Liu, T.; Chen, Z.; Liu, L. ROR functions as a ceRNA to regulate Nanog expression by sponging miR-145 and predicts poor prognosis in pancreatic cancer. Oncotarget 2016, 7, 1608–1618. [Google Scholar] [CrossRef]
- Fu, Z.; Li, G.; Li, Z.; Wang, Y.; Zhao, Y.; Zheng, S.; Ye, H.; Luo, Y.; Zhao, X.; Wei, L.; et al. Endogenous miRNA Sponge LincRNA-ROR promotes proliferation, invasion and stem cell-like phenotype of pancreatic cancer cells. Cell Death Discov. 2017, 3, 17004. [Google Scholar] [CrossRef]
- Bejerano, G.; Lowe, C.B.; Ahituv, N.; King, B.; Siepel, A.; Salama, S.R.; Rubin, E.M.; Kent, W.J.; Haussler, D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 2006, 441, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Katzman, S.; Kern, A.D.; Bejerano, G.; Fewell, G.; Fulton, L.; Wilson, R.K.; Salama, S.R.; Haussler, D. Human genome ultraconserved elements are ultraselected. Science 2007, 317, 915. [Google Scholar] [CrossRef] [PubMed]
- Sana, J.; Hankeova, S.; Svoboda, M.; Kiss, I.; Vyzula, R.; Slaby, O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology 2012, 82, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Ferdin, J.; Nishida, N.; Wu, X.; Nicoloso, M.S.; Shah, M.Y.; Devlin, C.; Ling, H.; Shimizu, M.; Kumar, K.; Cortez, M.A.; et al. HINCUTs in cancer: Hypoxia-induced noncoding ultraconserved transcripts. Cell Death Differ. 2013, 20, 1675–1687. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, J.; Yuan, X.; Qian, W.; Zhang, B.; Shi, M.; Xie, J.; Shen, B.; Xu, H.; Hou, Z.; et al. Long noncoding RNA uc.345 promotes tumorigenesis of pancreatic cancer by upregulation of hnRNPL expression. Oncotarget 2016, 7, 71556–71566. [Google Scholar] [CrossRef] [PubMed]
- Dery, K.J.; Gaur, S.; Gencheva, M.; Yen, Y.; Shively, J.E.; Gaur, R.K. Mechanistic control of carcinoembryonic antigen-related cell adhesion molecule-1 (CEACAM1) splice isoforms by the heterogeneous nuclear ribonuclear proteins hnRNP L, hnRNP A1, and hnRNP M. J. Biol. Chem. 2011, 286, 16039–16051. [Google Scholar] [CrossRef]
- Goehe, R.W.; Shultz, J.C.; Murudkar, C.; Usanovic, S.; Lamour, N.F.; Massey, D.H.; Zhang, L.; Camidge, D.R.; Shay, J.W.; Minna, J.D.; et al. hnRNP L regulates the tumorigenic capacity of lung cancer xenografts in mice via caspase-9 pre-mRNA processing. J. Clin. Investig. 2010, 120, 3923–3939. [Google Scholar] [CrossRef]
- Shivapurkar, N.; Weiner, L.M.; Marshall, J.L.; Madhavan, S.; Deslattes Mays, A.; Juhl, H.; Wellstein, A. Recurrence of early stage colon cancer predicted by expression pattern of circulating microRNAs. PLoS ONE 2014, 9, e84686. [Google Scholar] [CrossRef]
- Wu, X.B.; Feng, X.; Chang, Q.M.; Zhang, C.W.; Wang, Z.F.; Liu, J.; Hu, Z.Q.; Liu, J.Z.; Wu, W.D.; Zhang, Z.P.; et al. Cross-talk among AFAP1-AS1, ACVR1 and microRNA-384 regulates the stemness of pancreatic cancer cells and tumorigenicity in nude mice. J. Exp. Clin. Cancer Res. 2019, 38, 107. [Google Scholar] [CrossRef]
- Ye, Y.; Chen, J.; Zhou, Y.; Fu, Z.; Zhou, Q.; Wang, Y.; Gao, W.; Zheng, S.; Zhao, X.; Chen, T.; et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J. Transl. Med. 2015, 13, 137. [Google Scholar] [CrossRef]
- Lou, S.; Xu, J.; Wang, B.; Li, S.; Ren, J.; Hu, Z.; Xu, B.; Luo, F. Downregulation of lncRNA AFAP1-AS1 by oridonin inhibits the epithelial-to-mesenchymal transition and proliferation of pancreatic cancer cells. Acta Biochim. Biophys. Sin. 2019, 51, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, Q.; Zhou, Y.; Wang, X.; Zhang, Q.; Wang, Y.; Zhuang, H.; Jiang, X.; Xiong, W. The long coding RNA AFAP1-AS1 promotes tumor cell growth and invasion in pancreatic cancer through upregulating the IGF1R oncogene via sequestration of miR-133a. Cell Cycle 2018, 17, 1949–1966. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, M.; Chen, Y.; Xu, S.; Guo, Y.; Zhao, L. Cucurbitacin B suppresses proliferation of pancreatic cancer cells by ceRNA: Effect of miR-146b-5p and lncRNA-AFAP1-AS1. J. Cell. Physiol. 2019, 234, 4655–4667. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol. 2012, 48, R45–R53. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Xia, R.; Jin, F.; Xu, T.; Liu, Z.; De, W.; Liu, X. Downregulated long noncoding RNA MEG3 is associated with poor prognosis and promotes cell proliferation in gastric cancer. Tumour Biol. 2014, 35, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Kruer, T.L.; Dougherty, S.M.; Reynolds, L.; Long, E.; de Silva, T.; Lockwood, W.W.; Clem, B.F. Expression of the lncRNA Maternally Expressed Gene 3 (MEG3) Contributes to the Control of Lung Cancer Cell Proliferation by the Rb Pathway. PLoS ONE 2016, 11, e0166363. [Google Scholar] [CrossRef]
- Xiong, Y.; Liu, T.; Wang, S.; Chi, H.; Chen, C.; Zheng, J. Cyclophosphamide promotes the proliferation inhibition of mouse ovarian granulosa cells and premature ovarian failure by activating the lncRNA-Meg3-p53-p66Shc pathway. Gene 2017, 596, 1–8. [Google Scholar] [CrossRef]
- Chak, W.P.; Lung, R.W.; Tong, J.H.; Chan, S.Y.; Lun, S.W.; Tsao, S.W.; Lo, K.W.; To, K.F. Downregulation of long non-coding RNA MEG3 in nasopharyngeal carcinoma. Mol. Carcinog. 2017, 56, 1041–1054. [Google Scholar] [CrossRef]
- Gu, L.; Zhang, J.; Shi, M.; Zhan, Q.; Shen, B.; Peng, C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed. Pharmacother. 2017, 89, 1269–1276. [Google Scholar] [CrossRef]
- Ma, L.; Wang, F.; Du, C.; Zhang, Z.; Guo, H.; Xie, X.; Gao, H.; Zhuang, Y.; Kornmann, M.; Gao, H.; et al. Long non-coding RNA MEG3 functions as a tumour suppressor and has prognostic predictive value in human pancreatic cancer. Oncol. Rep. 2018, 39, 1132–1140. [Google Scholar] [CrossRef]
- Fantes, J.; Ragge, N.K.; Lynch, S.A.; McGill, N.I.; Collin, J.R.; Howard-Peebles, P.N.; Hayward, C.; Vivian, A.J.; Williamson, K.; van Heyningen, V.; et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 2003, 33, 461–463. [Google Scholar] [CrossRef] [PubMed]
- Shahryari, A.; Rafiee, M.R.; Fouani, Y.; Oliae, N.A.; Samaei, N.M.; Shafiee, M.; Semnani, S.; Vasei, M.; Mowla, S.J. Two novel splice variants of SOX2OT, SOX2OT-S1, and SOX2OT-S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells 2014, 32, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Askarian-Amiri, M.E.; Seyfoddin, V.; Smart, C.E.; Wang, J.; Kim, J.E.; Hansji, H.; Baguley, B.C.; Finlay, G.J.; Leung, E.Y. Emerging role of long non-coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS ONE 2014, 9, e102140. [Google Scholar] [CrossRef] [PubMed]
- Hou, Z.; Zhao, W.; Zhou, J.; Shen, L.; Zhan, P.; Xu, C.; Chang, C.; Bi, H.; Zou, J.; Yao, X.; et al. A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int. J. Biochem. Cell Biol. 2014, 53, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.M.; Teng, F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int. J. Clin. Exp. Pathol. 2015, 8, 4008–4014. [Google Scholar]
- Chen, L.; Zhang, J.; Chen, Q.; Ge, W.; Meng, L.; Huang, X.; Shen, P.; Yuan, H.; Shi, G.; Miao, Y.; et al. Long noncoding RNA SOX2OT promotes the proliferation of pancreatic cancer by binding to FUS. Int. J. Cancer 2020, 147, 175–188. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef]
- Amaral, P.P.; Dinger, M.E.; Mattick, J.S. Non-coding RNAs in homeostasis, disease and stress responses: An evolutionary perspective. Brief Funct. Genom. 2013, 12, 254–278. [Google Scholar] [CrossRef]
- Su, X.; Malouf, G.G.; Chen, Y.; Zhang, J.; Yao, H.; Valero, V.; Weinstein, J.N.; Spano, J.P.; Meric-Bernstam, F.; Khayat, D.; et al. Comprehensive analysis of long non-coding RNAs in human breast cancer clinical subtypes. Oncotarget 2014, 5, 9864–9876. [Google Scholar] [CrossRef]
- Melo, C.P.; Campos, C.B.; Rodrigues Jde, O.; Aguirre-Neto, J.C.; Atalla, Â.; Pianovski, M.A.; Carbone, E.K.; Lares, L.B.; Moraes-Souza, H.; Octacílio-Silva, S.; et al. Long non-coding RNAs: Biomarkers for acute leukaemia subtypes. Br. J. Haematol. 2016, 173, 318–320. [Google Scholar] [CrossRef]
- Xie, Z.; Chen, X.; Li, J.; Guo, Y.; Li, H.; Pan, X.; Jiang, J.; Liu, H.; Wu, B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget 2016, 7, 25408–25419. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.C.; Dang, Y.W.; Wei, D.M.; Chen, P.; Tang, R.X.; Huang, Q.; Liu, J.H.; Luo, D.Z. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: A comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases. Onco Targets Ther. 2017, 10, 3991–4005. [Google Scholar] [CrossRef] [PubMed]
- Wolpin, B.M.; Rizzato, C.; Kraft, P.; Kooperberg, C.; Petersen, G.M.; Wang, Z.; Arslan, A.A.; Beane-Freeman, L.; Bracci, P.M.; Buring, J.; et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat. Genet. 2014, 46, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, G.Q.; Chen, H.; Zhao, Z.J.; Chen, H.Z.; Liu, H.; Wang, G.; Jia, Y.H.; Pan, S.H.; Kong, R.; et al. Plasma and tumor levels of Linc-pint are diagnostic and prognostic biomarkers for pancreatic cancer. Oncotarget 2016, 7, 71773–71781. [Google Scholar] [CrossRef]
- Lu, H.; Yang, D.; Zhang, L.; Lu, S.; Ye, J.; Li, M.; Hu, W. Linc-pint inhibits early stage pancreatic ductal adenocarcinoma growth through TGF-β pathway activation. Oncol. Lett. 2019, 17, 4633–4639. [Google Scholar] [CrossRef]
- Moldovan, G.L.; Pfander, B.; Jentsch, S. PCNA, the maestro of the replication fork. Cell 2007, 129, 665–679. [Google Scholar] [CrossRef]
- Cory, S.; Adams, J.M. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2002, 2, 647–656. [Google Scholar] [CrossRef]
- Johnson, N.A.; Savage, K.J.; Ludkovski, O.; Ben-Neriah, S.; Woods, R.; Steidl, C.; Dyer, M.J.; Siebert, R.; Kuruvilla, J.; Klasa, R.; et al. Lymphomas with concurrent BCL2 and MYC translocations: The critical factors associated with survival. Blood 2009, 114, 2273–2279. [Google Scholar] [CrossRef]
- Yang, J.; Jiang, B.; Hai, J.; Duan, S.; Dong, X.; Chen, C. Long noncoding RNA opa-interacting protein 5 antisense transcript 1 promotes proliferation and invasion through elevating integrin α6 expression by sponging miR-143-3p in cervical cancer. J. Cell. Biochem. 2019, 120, 907–916. [Google Scholar] [CrossRef]
- Wang, M.; Sun, X.; Yang, Y.; Jiao, W. Long non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells and leads to poor prognosis by targeting miR-378a-3p. Thorac. Cancer 2018, 9, 939–949. [Google Scholar] [CrossRef]
- Zou, Y.; Yao, S.; Chen, X.; Liu, D.; Wang, J.; Yuan, X.; Rao, J.; Xiong, H.; Yu, S.; Yuan, X.; et al. LncRNA OIP5-AS1 regulates radioresistance by targeting DYRK1A through miR-369-3p in colorectal cancer cells. Eur. J. Cell Biol. 2018, 97, 369–378. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, F.; Yang, F.; Liu, Y. Kockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed. Pharmacother. 2018, 101, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Liu, Y.; Guo, C.; Shao, Y. LncRNA OIP5-AS1 promotes the malignancy of pancreatic ductal adenocarcinoma via regulating miR-429/FOXD1/ERK pathway. Cancer Cell Int. 2020, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Ma, J.; Wang, B.; Wu, X.; Liu, Z. Long non-coding RNA OIP5-AS1 promotes pancreatic cancer cell growth through sponging miR-342-3p via AKT/ERK signaling pathway. J. Physiol. Biochem. 2020, 76, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Ye, L.F.; Zhang, C.; Peng, T.; Zhou, X.H. Long non-coding RNA XIST exerts oncogenic functions in human nasopharyngeal carcinoma by targeting miR-34a-5p. Gene 2016, 592, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.L.; Ju, H.Q.; Lu, Y.X.; Chen, L.Z.; Zeng, Z.L.; Zhang, D.S.; Luo, H.Y.; Wang, F.; Qiu, M.Z.; Wang, D.S.; et al. Long non-coding RNA XIST regulates gastric cancer progression by acting as a molecular sponge of miR-101 to modulate EZH2 expression. J. Exp. Clin. Cancer Res. 2016, 35, 142. [Google Scholar] [CrossRef]
- Qin, Y.; Dang, X.; Li, W.; Ma, Q. miR-133a functions as a tumor suppressor and directly targets FSCN1 in pancreatic cancer. Oncol. Res. 2013, 21, 353–363. [Google Scholar] [CrossRef]
- Gong, Y.; Ren, J.; Liu, K.; Tang, L.M. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J. Gastroenterol. 2015, 21, 2949–2958. [Google Scholar] [CrossRef]
- Wei, W.; Liu, Y.; Lu, Y.; Yang, B.; Tang, L. LncRNA XIST Promotes Pancreatic Cancer Proliferation Through miR-133a/EGFR. J. Cell. Biochem. 2017, 118, 3349–3358. [Google Scholar] [CrossRef]
- Palmieri, G.; Paliogiannis, P.; Sini, M.C.; Manca, A.; Palomba, G.; Doneddu, V.; Tanda, F.; Pascale, M.R.; Cossu, A. Long non-coding RNA CASC2 in human cancer. Crit. Rev. Oncol. Hematol. 2017, 111, 31–38. [Google Scholar] [CrossRef]
- He, X.; Liu, Z.; Su, J.; Yang, J.; Yin, D.; Han, L.; De, W.; Guo, R. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and regulates cell proliferation in non-small cell lung cancer. Tumour Biol. 2016, 37, 9503–9510. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Wu, X.; Li, S.; Xu, X.; Zhu, H.; Chen, X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci. Rep. 2016, 6, 26524. [Google Scholar] [CrossRef] [PubMed]
- Baldinu, P.; Cossu, A.; Manca, A.; Satta, M.P.; Sini, M.C.; Rozzo, C.; Dessole, S.; Cherchi, P.; Gianfrancesco, F.; Pintus, A.; et al. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum. Mutat. 2004, 23, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, Y.H.; Yao, Y.L.; Li, Z.; Li, Z.Q.; Ma, J.; Xue, Y.X. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21. Cell. Signal. 2015, 27, 275–282. [Google Scholar] [CrossRef]
- Cao, Y.; Xu, R.; Xu, X.; Zhou, Y.; Cui, L.; He, X. Downregulation of lncRNA CASC2 by microRNA-21 increases the proliferation and migration of renal cell carcinoma cells. Mol. Med. Rep. 2016, 14, 1019–1025. [Google Scholar] [CrossRef]
- Yu, Y.; Liang, S.; Zhou, Y.; Li, S.; Li, Y.; Liao, W. HNF1A/CASC2 regulates pancreatic cancer cell proliferation through PTEN/Akt signaling. J. Cell. Biochem. 2019, 120, 2816–2827. [Google Scholar] [CrossRef]
- Xu, D.F.; Wang, L.S.; Zhou, J.H. Long non-coding RNA CASC2 suppresses pancreatic cancer cell growth and progression by regulating the miR-24/MUC6 axis. Int. J. Oncol. 2020, 56, 494–507. [Google Scholar] [CrossRef]
- Zhang, H.; Feng, X.; Zhang, M.; Liu, A.; Tian, L.; Bo, W.; Wang, H.; Hu, Y. Long non-coding RNA CASC2 upregulates PTEN to suppress pancreatic carcinoma cell metastasis by downregulating miR-21. Cancer Cell Int. 2019, 19, 18. [Google Scholar] [CrossRef]
- Qu, S.; Niu, K.; Wang, J.; Dai, J.; Ganguly, A.; Gao, C.; Tian, Y.; Lin, Z.; Yang, X.; Zhang, X.; et al. LINC00671 suppresses cell proliferation and metastasis in pancreatic cancer by inhibiting AKT and ERK signaling pathway. Cancer Gene Ther. 2020, 28, 221–233. [Google Scholar] [CrossRef]
- Kleger, A.; Perkhofer, L.; Seufferlein, T. Smarter drugs emerging in pancreatic cancer therapy. Ann. Oncol. 2014, 25, 1260–1270. [Google Scholar] [CrossRef]
- Uccello, M.; Moschetta, M.; Mak, G.; Alam, T.; Henriquez, C.M.; Arkenau, H.T. Towards an optimal treatment algorithm for metastatic pancreatic ductal adenocarcinoma (PDA). Curr. Oncol. 2018, 25, e90–e94. [Google Scholar] [CrossRef] [PubMed]
- Binenbaum, Y.; Na’ara, S.; Gil, Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist. Updat. 2015, 23, 55–68. [Google Scholar] [CrossRef]
- Zhang, X.W.; Bu, P.; Liu, L.; Zhang, X.Z.; Li, J. Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance. Biochem. Biophys. Res. Commun. 2015, 462, 227–232. [Google Scholar] [CrossRef]
- You, L.; Wang, H.; Yang, G.; Zhao, F.; Zhang, J.; Liu, Z.; Zhang, T.; Liang, Z.; Liu, C.; Zhao, Y. Gemcitabine exhibits a suppressive effect on pancreatic cancer cell growth by regulating processing of PVT1 to miR1207. Mol. Oncol. 2018, 12, 2147–2164. [Google Scholar] [CrossRef]
- Zhou, C.; Yi, C.; Yi, Y.; Qin, W.; Yan, Y.; Dong, X.; Zhang, X.; Huang, Y.; Zhang, R.; Wei, J.; et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/β-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol. Cancer 2020, 19, 118. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Helms, C.; Liao, W.; Zaba, L.C.; Duan, S.; Gardner, J.; Wise, C.; Miner, A.; Malloy, M.J.; Pullinger, C.R.; et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008, 4, e1000041. [Google Scholar] [CrossRef]
- Chen, J.; Zhao, D.; Meng, Q. Knockdown of HCP5 exerts tumor-suppressive functions by up-regulating tumor suppressor miR-128-3p in anaplastic thyroid cancer. Biomed. Pharmacother. 2019, 116, 108966. [Google Scholar] [CrossRef]
- Wang, L.; Luan, T.; Zhou, S.; Lin, J.; Yang, Y.; Liu, W.; Tong, X.; Jiang, W. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Med. 2019, 8, 4389–4403. [Google Scholar] [CrossRef]
- Yang, C.; Sun, J.; Liu, W.; Yang, Y.; Chu, Z.; Yang, T.; Gui, Y.; Wang, D. Long noncoding RNA HCP5 contributes to epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation and interacting with miR-139-5p. Am. J. Transl. Res. 2019, 11, 953–963. [Google Scholar] [PubMed]
- Yu, Y.; Shen, H.M.; Fang, D.M.; Meng, Q.J.; Xin, Y.H. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4812–4819. [Google Scholar] [CrossRef]
- Wang, W.; Lou, W.; Ding, B.; Yang, B.; Lu, H.; Kong, Q.; Fan, W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging 2019, 11, 2610–2627. [Google Scholar] [CrossRef] [PubMed]
- Yuan, B.; Guan, Q.; Yan, T.; Zhang, X.; Xu, W.; Li, J. LncRNA HCP5 Regulates Pancreatic Cancer Progression by miR-140-5p/CDK8 Axis. Cancer Biother. Radiopharm. 2020, 35, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, J.; Dong, L.; Xia, L.; Zhu, H.; Li, Z.; Yu, X. Long Noncoding RNA HCP5 Regulates Pancreatic Cancer Gemcitabine (GEM) Resistance By Sponging Hsa-miR-214-3p To Target HDGF. Onco Targets Ther. 2019, 12, 8207–8216. [Google Scholar] [CrossRef]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef]
- Tani, H.; Torimura, M.; Akimitsu, N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE 2013, 8, e55684. [Google Scholar] [CrossRef]
- Gao, Z.Q.; Wang, J.F.; Chen, D.H.; Ma, X.S.; Wu, Y.; Tang, Z.; Dang, X.W. Long non-coding RNA GAS5 suppresses pancreatic cancer metastasis through modulating miR-32-5p/PTEN axis. Cell Biosci. 2017, 7, 66. [Google Scholar] [CrossRef]
- Lu, X.; Fang, Y.; Wang, Z.; Xie, J.; Zhan, Q.; Deng, X.; Chen, H.; Jin, J.; Peng, C.; Li, H.; et al. Downregulation of gas5 increases pancreatic cancer cell proliferation by regulating CDK6. Cell Tissue Res. 2013, 354, 891–896. [Google Scholar] [CrossRef]
- Sharma, N.S.; Gnamlin, P.; Durden, B.; Gupta, V.K.; Kesh, K.; Garrido, V.T.; Dudeja, V.; Saluja, A.; Banerjee, S. Long non-coding RNA GAS5 acts as proliferation “brakes” in CD133+ cells responsible for tumor recurrence. Oncogenesis 2019, 8, 68. [Google Scholar] [CrossRef]
- Gao, Z.Q.; Wang, J.F.; Chen, D.H.; Ma, X.S.; Yang, W.; Zhe, T.; Dang, X.W. Long non-coding RNA GAS5 antagonizes the chemoresistance of pancreatic cancer cells through down-regulation of miR-181c-5p. Biomed. Pharmacother. 2018, 97, 809–817. [Google Scholar] [CrossRef]
- Liu, B.; Wu, S.; Ma, J.; Yan, S.; Xiao, Z.; Wan, L.; Zhang, F.; Shang, M.; Mao, A. lncRNA GAS5 Reverses EMT and Tumor Stem Cell-Mediated Gemcitabine Resistance and Metastasis by Targeting miR-221/SOCS3 in Pancreatic Cancer. Mol. Ther. Nucleic Acids 2018, 13, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zheng, S.; Hu, C.; Li, G.; Lin, H.; Xia, R.; Ye, Y.; He, R.; Li, Z.; Lin, Q.; et al. Cancer-associated fibroblast-induced lncRNA UPK1A-AS1 confers platinum resistance in pancreatic cancer via efficient double-strand break repair. Oncogene 2022, 41, 2372–2389. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.; Andersen, M.; Stagsted, L.; Ebbesen, K.; Hansen, T.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Mizrahi, A.; Czerniak, A.; Levy, T.; Amiur, S.; Gallula, J.; Matouk, I.; Abu-lail, R.; Sorin, V.; Birman, T.; de Groot, N.; et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J. Transl. Med. 2009, 7, 69. [Google Scholar] [CrossRef]
- Lavie, O.; Edelman, D.; Levy, T.; Fishman, A.; Hubert, A.; Segev, Y.; Raveh, E.; Gilon, M.; Hochberg, A. A phase 1/2a, dose-escalation, safety, pharmacokinetic, and preliminary efficacy study of intraperitoneal administration of BC-819 (H19-DTA) in subjects with recurrent ovarian/peritoneal cancer. Arch. Gynecol. Obstet. 2017, 295, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Gofrit, O.; Benjamin, S.; Halachmi, S.; Leibovitch, I.; Dotan, Z.; Lamm, D.; Ehrlich, N.; Yutkin, V.; Ben-Am, M.; Hochberg, A. DNA based therapy with diphtheria toxin-A BC-819: A phase 2b marker lesion trial in patients with intermediate risk nonmuscle invasive bladder cancer. J. Urol. 2014, 191, 1697–1702. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Zhuo, C.; Hou, W.; Hu, L.; Lin, C.; Chen, C.; Lin, X. Genomic Editing of Non-Coding RNA Genes with CRISPR/Cas9 Ushers in a Potential Novel Approach to Study and Treat Schizophrenia. Front. Mol. Neurosci. 2017, 10, 28. [Google Scholar] [CrossRef]
- Yang, J.; Meng, X.; Pan, J.; Jiang, N.; Zhou, C.; Wu, Z.; Gong, Z. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. RNA Biol. 2018, 15, 35–43. [Google Scholar] [CrossRef]
- Zhen, S.; Hua, L.; Liu, Y.; Sun, X.; Jiang, M.; Chen, W.; Zhao, L.; Li, X. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 2017, 8, 9634–9646. [Google Scholar] [CrossRef]
- Wolfbeis, O. An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, K.; Aoki, Y.; Koo, T.; McClorey, G.; Benner, L.; Coenen-Stass, A.; O’Donovan, L.; Lehto, T.; Garcia-Guerra, A.; Nordin, J.; et al. Self-Assembly into Nanoparticles Is Essential for Receptor Mediated Uptake of Therapeutic Antisense Oligonucleotides. Nano Lett. 2015, 15, 4364–4373. [Google Scholar] [CrossRef] [PubMed]
- Ruivo, C.; Adem, B.; Silva, M.; Melo, S. The Biology of Cancer Exosomes: Insights and New Perspectives. Cancer Res. 2017, 77, 6480–6488. [Google Scholar] [CrossRef] [PubMed]
- Al-Shaheri, F.; Alhamdani, M.; Bauer, A.; Giese, N.; Büchler, M.; Hackert, T.; Hoheisel, J. Blood biomarkers for differential diagnosis and early detection of pancreatic cancer. Cancer Treat. Rev. 2021, 96, 102193. [Google Scholar] [CrossRef] [PubMed]


| Criterial | Classification |
|---|---|
| genome location | intergenic lncRNAs |
| intronic lncRNAs | |
| exonic lncRNAs | |
| transcriptional orientation | sense lncRNAs |
| antisense lncRNAs | |
| subcellular localization | nuclear lncRNAs |
| cytoplasmic lncRNAs | |
| mode of action | cis-acting lncRNAs |
| trans-acting lncRNAs |
| No | Lnc | Vitro Functions | Specimen | Expression | Reference PMID | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prolife a | Cycle b | Apopt c | Migra d | Invas e | Angio f | CSC g | Drug Resistance | Other | |||||
| 1 | GAS5 | + | + | BXPC3/PANC1/ASPC1/HS766T | down | 24026436 | |||||||
| 2 | ENST0000048073 | + | ASPC1/BXPC3/CFPAC1/PANC1/SW1990 | down | 25314054 | ||||||||
| 3 | linc00673 | + | + | BXPC3/CFPAC1 | down | 27213290 | |||||||
| 4 | ENSG00000218510 | + | ASPC1/BXPC3/CAPAN2/CFPAC1/HPAC/MIA PACA2/PANC1/SW1990 | down | 27628540 | ||||||||
| 5 | F11AS1 | SW1990/BXPC3 | down | 28220683 | |||||||||
| linc00226 | |||||||||||||
| 6 | MEG3 | + | + | + | + | + | PANC1 | down | 28320094 | ||||
| 7 | CASC2 | + | SW1990/BXPC3/ASPC1/CFPAC1/PANC1 | down | 28865121 | ||||||||
| 8 | DAPK1 | + | + | PANC1/HS766T | down | 31966799 | |||||||
| 9 | GAS5 | + | GEM/5-FU h | PATU8988/SW1990/PATU89885-FU/SW1990-GEM | down | 29112934 | |||||||
| 10 | DGCR5 | + | + | + | 5-FU | HPAC/PANC1 | down | 29207609 | |||||
| 11 | GAS5 | + | + | + | + | BXPC3/PANC1 | down | 29225772 | |||||
| 12 | MEG3 | + | + | + | + | GEM | MIAPACA2/PANC1/T3M4 | down | 29328401 | ||||
| 13 | XLOC_000647 | + | + | BXPC3/MIAPACA2 | down | 29386037 | |||||||
| 14 | AB209630 | + | GEM | BXPC3-GEM/PANC1-GEM | down | 29526843 | |||||||
| 15 | BC032020 | + | + | + | + | ASPC1/PANC1 | down | 29532883 | |||||
| 16 | PCTST | + | + | BXPC3/MIAPACA2 | down | 29978472 | |||||||
| 17 | KCNK15AS1 | + | + | BXPC3/MIAPACA2 | down | 30032148 | |||||||
| 18 | linc00671/00261/SNHG9 | + | BXPC3/HPAC | down | 30210701 | ||||||||
| 19 | NONHSAT105177 | + | + | cholesterol pathway/melittin/exosome | PATU8988/SW1990 | down | 30237397 | ||||||
| 20 | GAS5 | + | + | + | + | GEM | PANC1 | down | 30388621 | ||||
| 21 | GLSAS | + | + | + | glycolysis/glutamate | BXPC3/PANC1 | down | 30563888 | |||||
| 22 | CASC2 | + | + | PANC1 | down | 30675129 | |||||||
| 23 | linc00052 | + | + | + | + | + | ASPC1/SW1990 | down | 30712321 | ||||
| 24 | TUSC7 | + | + | + | + | + | CAPAN2/PANC1 | down | 30714151 | ||||
| 25 | linc-pint | + | HPNE/PL45 | down | 30944652 | ||||||||
| 26 | linc01197 | + | ASPC1/BXPC3/HPNE/PANC1 | down | 31027497 | ||||||||
| 27 | PXNAS1 | + | + | ASPC1/PACA2 | down | 31488171 | |||||||
| 28 | GAS5 | + | + | + | GEM | oxidation reaction | CD133HI(SU86.86/KPC001/S2VP10)/CD133LO(MIAPACA2) | down | 31740660 | ||||
| 29 | linc01111 | + | + | + | + | PANC1/MIAPACA2 | down | 31767833 | |||||
| 30 | CASC2 | + | + | + | + | ASPC1/PANC1 | down | 31894271 | |||||
| 31 | linc00673 | + | + | + | CAPAN1 | down | 31949497 | ||||||
| 32 | linc00261 | + | + | PANC1/MIAPACA2 | down | 32020223 | |||||||
| 33 | linc285194 | + | + | involved in the inhibition of PANC1 by propofol | PANC1 | down | 32303144 | ||||||
| 34 | linc00261 | + | + | A549/PANC1 | down | 32414223 | |||||||
| 35 | linc00261 | + | + | + | CFPAC1/BXPC3/PANC1/ASPC1 | down | 32590069 | ||||||
| 36 | DGCR5 | + | SW1990/PANC1 | down | 32626951 | ||||||||
| 37 | linc00671 | + | + | + | PANC1/CAPAN2/CAPAN1/ASPC1/SW1990/BXPC3 | down | 32801328 | ||||||
| 38 | MTSS1-AS | + | + | ASPC1/PANC1/SW1990/BXPC3 | down | 32929338 | |||||||
| No | Lnc | Position | Location | Mechanism | Animal Model | Vivo Functions | Pheno f | Reference PMID | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miRNA c | Target | RBPs d | Pathways | Upstream | Grow | Meta e | |||||||
| 1 | GAS5 | CDK6 | suppr g | 24026436 | |||||||||
| 2 | ENST0000048073 | OS-9/HIF-1a | suppr | 25314054 | |||||||||
| 3 | linc00673 | 17q24.3 | PTPN11 | SRC/ERK; STAT1 | suppr | 27213290 | |||||||
| 4 | ENSG00000218510 | suppr | 27628540 | ||||||||||
| 5 | F11-AS1 | suppr | 28220683 | ||||||||||
| linc00226 | |||||||||||||
| 6 | MEG3 | PI3K | PI3K/AKT | suppr | 28320094 | ||||||||
| 7 | CASC2 | PTEN/AKT | HNF1A | suppr | 28865121 | ||||||||
| 8 | DAPK1 | miR-812 | ROCK-1/RhoA | suppr | 31966799 | ||||||||
| 9 | GAS5 | miR-181c-5p | Hippo | subcutaneous xenograft | + | suppr | 29112934 | ||||||
| 10 | DGCR5 | miR-320a | EMT | suppr | 29207609 | ||||||||
| 11 | GAS5 | miR-32-5p | PTEN | subcutaneous xenograft | + | suppr | 29225772 | ||||||
| 12 | MEG3 | 14q32.3 | EMT; PCNA | suppr | 29328401 | ||||||||
| 13 | XLOC_000647 | NLRP3 | EMT | subcutaneous xenograft | + | suppr | 29386037 | ||||||
| 14 | AB209630 | PI3K/AKT | suppr | 29526843 | |||||||||
| 15 | BC032020 | ZENF451/TGF-β | subcutaneous xenograft | + | suppr | 29532883 | |||||||
| 16 | PCTST | TACC-3 | EMT | subcutaneous xenograft | + | suppr | 29978472 | ||||||
| 17 | KCNK15-AS1 | Nucl a | EMT | ALKBH5 | suppr | 30032148 | |||||||
| 18 | linc00671/00261/SNHG9 | suppr | 30210701 | ||||||||||
| 19 | NONHSAT105177 | nucl | EMT | subcutaneous xenograft | + | suppr | 30237397 | ||||||
| 20 | GAS5 | miR-221 | SOCS3 | EMT | subcutaneous xenograft | + | + | suppr | 30388621 | ||||
| 21 | GLS-AS | 2 | nucl | GLS premRNA | ADAR1/Dicer | Myc | subcutaneous xenograft | + | + | suppr | 30563888 | ||
| 22 | CASC2 | 10q26 | miR-21 | PTEN | suppr | 30675129 | |||||||
| 23 | linc00052 | miR-330-3p | subcutaneous xenograft | + | suppr | 30712321 | |||||||
| 24 | TUSC7 | 3q13.31 | miR-371a-5p | EMT | subcutaneous xenograft | + | suppr | 30714151 | |||||
| 25 | linc-pint | TGF-β1 | suppr | 30944652 | |||||||||
| 26 | linc01197 | nucl | β-catenin | TCF4; Wnt pathway | FOXO1 | subcutaneous xenograft | + | suppr | 31027497 | ||||
| 27 | PXN-AS1 | miR-3046 | PIP4K2B | subcutaneous xenograft | + | suppr | 31488171 | ||||||
| 28 | GAS5 | GR | CDK4/CCND2; NF-kB | SOX2 | KPC mice | + | suppr | 31740660 | |||||
| 29 | linc01111 | Cyto b | miR-3924 | DUSP1 | SAPK/JNK | subcutaneous xenograft | + | + | suppr | 31767833 | |||
| 30 | CASC2 | miR-24 | MUC6 | ITGB4/FAK; EMT | subcutaneous xenograft | + | suppr | 31894271 | |||||
| 31 | linc00673 | miR-504 | HNF1A | subcutaneous xenograft | + | suppr | 31949497 | ||||||
| 32 | linc00261 | miR-552-5p | FOXO3 | Wnt/EMT | liver metastasis model | + | suppr | 32020223 | |||||
| 33 | linc285194 | miR-34a | E-cadherin | suppr | 32303144 | ||||||||
| 34 | linc00261 | nucl | E-cadherin | FOXA2 | suppr | 32414223 | |||||||
| 35 | linc00261 | miR-23a-3p | suppr | 32590069 | |||||||||
| 36 | DGCR5 | miR-27a-3p | BNIP3 | p38 MAPK | subcutaneous xenograft | + | suppr | 32626951 | |||||
| 37 | linc00671 | 17q21.31 | cyto | EMT | subcutaneous xenograft | + | suppr | 32801328 | |||||
| 38 | MTSS1-AS | nucl | MZF1 | suppr | 32929338 | ||||||||
| No | Lnc | Source | No. of Patients | Express | Cut-Off | Variables | Reference PMID | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | Differentiation | T | Lymphatic (N) | Distance (M) | TNM | Vessel Invasion | Other | |||||||
| 1 | GAS5 | tissue | 23 | down | 24026436 | |||||||||
| 2 | BC008363 | tissue | 30 | down | 25200694 | |||||||||
| 3 | ENST00000480739 | tissue | 35 | down | + | + | 25314054 | |||||||
| 4 | LOC285194 | tissue | 85 | down | + | + | + | 25550852 | ||||||
| 5 | linc00673 | tissue | 74 | down | 27213290 | |||||||||
| 6 | HMlincRwNA717 | tissue | 150 | down | + | + | + | + | 27338046 | |||||
| 7 | ENSG00000218510 | tissue | 80 | down | + | + | + | 27628540 | ||||||
| 8 | linc-pint | plasma | 59 | down | 27708234 | |||||||||
| tissue | 61 | down | ||||||||||||
| 9 | MEG3 | tissue | 30 | down | + | + | + | 28320094 | ||||||
| 10 | CASC2 | tissue | 110 | down | + | 28865121 | ||||||||
| 11 | DGCR5 | tissue | 30 | down | MEL a | 29207609 | ||||||||
| 12 | GAS5 | tissue | 22 | down | 29225772 | |||||||||
| 13 | MEG3 | tissue | 25 | down | 29328401 | |||||||||
| 14 | XLOC_000647 | tissue | 48 | down | MEL | + | + | 29386037 | ||||||
| 15 | AB209630 | tissue | 53 | down | 29526843 | |||||||||
| 16 | BC032020 | tissue | 20 | down | 29532883 | |||||||||
| 17 | PCTST | tissue | 48 | down | MEL | + | 29978472 | |||||||
| 18 | KCNK15-AS1 | tissue | 69 | down | 30032148 | |||||||||
| 19 | linc00261 | tissue | 229 | down | + | + | 30210701 | |||||||
| 20 | linc00671 | tissue | 229 | down | + | + | 30210701 | |||||||
| 21 | linc00671/00261/SNHG9 | plasma | 229 | down | 30210701 | |||||||||
| 22 | SNHG9 | tissue | 229 | down | + | + | + | 30210701 | ||||||
| 23 | NONHSAT105177 | tissue | N/A b | down | 30237397 | |||||||||
| 24 | GAS5 | tissue | 60 | down | 30388621 | |||||||||
| 25 | GLS-AS | tissue | 30 | down | MEL | + | + | + | 30563888 | |||||
| 26 | TUSC7 | tissue | 94 | down | + | + | + | + | perineural invasion | 30714151 | ||||
| 27 | linc-pint | plasma | 46 | down | + | 30944652 | ||||||||
| 28 | linc01197 | tissue | 18 | down | 31027497 | |||||||||
| 29 | PXN-AS1 | tissue | 50 | down | + | + | + | 31488171 | ||||||
| 30 | linc01111 | tissue | 60 | down | + | + | 31767833 | |||||||
| 31 | linc01111 | plasma | 57 | down | 31767833 | |||||||||
| 32 | CASC2 | tissue | 20 | down | 31894271 | |||||||||
| 33 | linc00673 | tissue | 30 | down | MEL | + | 31949497 | |||||||
| 34 | DAPK1 | tissue | 60 | down | + | + | + | 31966799 | ||||||
| 35 | linc00261 | tissue | 54 | down | MEL | + | + | + | + | + | perineural invasion | 32020223 | ||
| 36 | linc00261 | tissue | 42 | down | 17.66 | + | + | 32414223 | ||||||
| 37 | linc00671 | tissue | 60 | down | + | + | 32801328 | |||||||
| 38 | MTSS1-AS | tissue | 132 | down | + | + | + | 32929338 | ||||||
| No | Lnc | Source | No. of Patients | Expression | Detection Method | AUC a | Survival | Prognostic Biomarker | Reference PMID |
|---|---|---|---|---|---|---|---|---|---|
| 1 | GAS5 | tissue | 23 | down | qRT-PCR | 24026436 | |||
| 2 | BC008363 | tissue | 30 | down | qRT-PCR | OS b | 25200694 | ||
| 3 | ENST00000480739 | tissue | 35 | down | qRT-PCR | OS | + | 25314054 | |
| 4 | LOC285194 | tissue | 85 | down | qRT-PCR | OS | + | 25550852 | |
| 5 | linc00673 | tissue | 74 | down | qRT-PCR | 27213290 | |||
| 6 | HMlincRNA717 | tissue | 150 | down | qRT-PCR | OS | + | 27338046 | |
| 7 | ENSG00000218510 | tissue | 80 | down | qRT-PCR | 0.931 | OS | + | 27628540 |
| 8 | linc-pint | plasma | 59 | down | qRT-PCR | 0.87 | 27708234 | ||
| tissue | 61 | down | FISH and qRT-PCR | OS | + | ||||
| 9 | MEG3 | tissue | 30 | down | qRT-PCR | 28320094 | |||
| 10 | CASC2 | tissue | 110 | down | qRT-PCR | OS/DFS c | + | 28865121 | |
| 11 | DGCR5 | tissue | 30 | down | qRT-PCR | 0.735 | OS | 29207609 | |
| 12 | GAS5 | tissue | 22 | down | qRT-PCR | 29225772 | |||
| 13 | MEG3 | tissue | 25 | down | qRT-PCR | OS | 29328401 | ||
| 14 | XLOC_000647 | tissue | 48 | down | qRT-PCR | OS | + | 29386037 | |
| 15 | AB209630 | tissue | 53 | down | qRT-PCR | OS | 29526843 | ||
| 16 | BC032020 | tissue | 20 | down | qRT-PCR | 29532883 | |||
| 17 | PCTST | tissue | 48 | down | qRT-PCR | OS | + | 29978472 | |
| 18 | KCNK15-AS1 | tissue | 69 | down | qRT-PCR | 30032148 | |||
| 19 | 00261 | tissue | 229 | down | qRT-PCR | 0.5712 | OS | 30210701 | |
| 20 | 00671 | tissue | 229 | down | qRT-PCR | 0.6057 | OS | 30210701 | |
| 21 | 00671/00261/SNHG9 | plasma | 229 | down | qRT-PCR | 30210701 | |||
| 22 | SNHG9 | tissue | 229 | down | qRT-PCR | 0.5983 | OS | 30210701 | |
| 23 | NONHSAT105177 | tissue | N/A d | down | RNA-FISH | 30237397 | |||
| 24 | GAS5 | tissue | 60 | down | IHC and qRT-PCR | 30388621 | |||
| 25 | GLS-AS | tissue | 30 | down | northern blot and qRT-PCR | OS | 30563888 | ||
| 26 | TUSC7 | tissue | 94 | down | qRT-PCR | OS | + | 30714151 | |
| 27 | pint | plasma | 46 | down | qRT-PCR | 0.8934 | 30944652 | ||
| 28 | 01197 | tissue | 18 | down | qRT-PCR | OS/DFS | 31027497 | ||
| 29 | PXN-AS1 | tissue | 50 | down | qRT-PCR | OS | 31488171 | ||
| 30 | 01111 | tissue | 60 | down | ISH and qRT-PCR | OS | 31767833 | ||
| 31 | 01111 | plasma | 57 | down | qRT-PCR | 31767833 | |||
| 32 | CASC2 | tissue | 20 | down | qRT-PCR | 31894271 | |||
| 33 | 00673 | tissue | 30 | down | qRT-PCR | OS | 31949497 | ||
| 34 | DAPK1 | tissue | 60 | down | qRT-PCR | 31966799 | |||
| 35 | 00261 | tissue | 54 | down | qRT-PCR | OS | 32020223 | ||
| 36 | 00261 | tissue | 42 | down | qRT-PCR | OS | 32414223 | ||
| 37 | linc00671 | tissue | 60 | down | qRT-PCR | OS | 32801328 | ||
| 38 | MTSS1-AS | tissue | 132 | down | qRT-PCR | OS | 32929338 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Hou, S.; Ye, Z.; Wang, W.; Hu, X.; Hang, Q. Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance. Cancers 2022, 14, 2115. https://doi.org/10.3390/cancers14092115
Li J, Hou S, Ye Z, Wang W, Hu X, Hang Q. Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance. Cancers. 2022; 14(9):2115. https://doi.org/10.3390/cancers14092115
Chicago/Turabian StyleLi, Jiajia, Sicong Hou, Ziping Ye, Wujun Wang, Xiaolin Hu, and Qinglei Hang. 2022. "Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance" Cancers 14, no. 9: 2115. https://doi.org/10.3390/cancers14092115
APA StyleLi, J., Hou, S., Ye, Z., Wang, W., Hu, X., & Hang, Q. (2022). Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance. Cancers, 14(9), 2115. https://doi.org/10.3390/cancers14092115






